Abstract
The identification of transcriptional regulatory networks, which control tissue-specific development and function, is of central importance to the understanding of lymphocyte biology. To decipher transcriptional networks in T-cell development and differentiation we developed a browsable expression atlas and applied a novel quantitative method to define gene sets most specific to each of the represented cell subsets and tissues. Using this system, body atlas size datasets can be used to examine gene enrichment profiles from a cell/tissue perspective rather than gene perspective, thereby identifying highly enriched genes within a cell type, which are often key to cellular differentiation and function. A systems analysis of transcriptional regulators within T cells during different phases of development and differentiation resulted in the identification of known key regulators and uncharacterized coexpressed regulators. ZBTB25, a BTB-POZ family transcription factor, was identified as a highly T cell–enriched transcription factor. We provide evidence that ZBTB25 functions as a negative regulator of nuclear factor of activated T cells (NF-AT) activation, such that RNA interference mediated knockdown resulted in enhanced activation of target genes. Together, these findings suggest a novel mechanism for NF-AT mediated gene expression and the compendium of expression data provides a quantitative platform to drive exploration of gene expression across a wide range of cell/tissue types.
Introduction
T cells undergo thymic development and differentiate into distinct subsets defined by cytokine production and effector functions. This development depends upon Notch and Wnt signaling pathways and transcription factors including GATA3, MYB, RUNX1, IKZF1, and TCF7 (for review see Rothenberg et al1 and Singer et al2 ). In contrast to B cells, where PAX5 and EBF1 were identified as B-lineage specific in expression and function, many of the known T-cell regulators are not restricted to the T lineage.1 In addition, several factors that have critical roles in T-cell development, such as, MYB, GFI1, STAT5B, TOX, and POU2F1 are stably expressed throughout development.3 These observations lead several investigators to hypothesize that T lineage–specific factors remain to be discovered, and several studies have attempted to identify these novel Transcription factors (TFs).4-6 However, these studies focused on changes between different T-cell subsets or between T cells and a few limited numbers of non–T-cell controls. Given that transcriptional steady state abundance is best quantified with respect to other cells, we hypothesized that T cell–specific factors will emerge only in an extensive dataset that includes a large number of immune and nonimmune cells and tissues.
We compiled a large dataset of 557 publicly available microarrays that covers 126 normal primary cells/tissues and reveals expression patterns of approximately 12 000 genes. A novel benchmarking system was devised that enhances the signal to noise ratio and is a measure of cell/tissue specificity. This scoring system is comparable between genes and allows ranking in each cell/tissue profiled based on specificity level. We used this compendium to study the transcriptional control of T-cell development and differentiation. A systems level analysis of 1373 TFs recovered many of the known T-lineage regulators and identified several potentially novel factors. We identify several potentially novel regulators and validate ZBTB25, BTB-POZ family member, functions as a transcriptional repressor of nuclear factor of activated cells (NF-AT) in T cells. We demonstrate that silencing of ZBTB25 results in enhanced expression of NF-AT target genes in response to T-cell receptor (TCR) engagement. In addition, we demonstrate the ability to expand this dataset further by including profiled cell lines and identify genes enriched in hematologic malignancies compared with normal tissues and other cancers.
Methods
Microarrays and the enrichment score
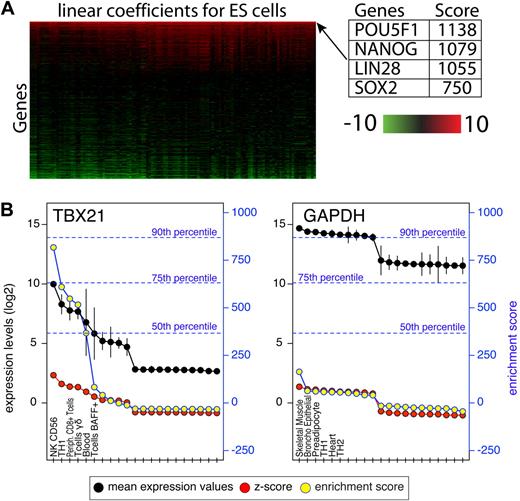
The Gene Expression Omnibus7 and ArrayExpress8 collections were scanned for experiments in which normal primary human cells or tissues were profiled. Experiments that were performed on Affymetrix platforms for which the raw files were available were selected and grouped by platform accession numbers. Raw Affymetrix files were processed using R Version 2.6.2 (The R Foundation for Statistical Computing) and Bioconductor modules Version 2.1.9 Microarray normalization was performed using the GCRMA module and present/absent calls were calculated using Affymetrix MAS5 package in Bioconductor. For the purpose of computing the enrichment scores, only probes with at least 1 present call across the entire dataset for which the expression value was above log2(100) were retained. We refer to each set of replicates representing a cell type or tissue as a group. Each group was compared pairwise to all other groups using the Limma module of Bioconductor.10 Limma uses linear models and Bayes methods to assess differential expression. For each group we used Limma and compared that group to each of the other 125 groups in the panel, generating 125 linear model coefficients for each probe and 125 associated P values. P values were adjusted using the Bonferroni correction. The linear model coefficient is a measure of difference between 2 groups. The enrichment score for each probe was defined as the sum of all linear model coefficients for which the adjusted P values were less than .05. This process is illustrated in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and a heat map of linear model coefficients for transcription factors in embryonic stem cells is shown in Figure 1A. Probes highly expressed in only 1 group within the panel will result in very high enrichment scores due to the sum of large statistically significant coefficient.
Probe mapping
Affymetrix individual probes in each probe set were matched to the human genome (HG18) using Blast-like alignment tool with a tile size of 5. Probes were allowed to have a maximum of 2 mismatches with no gaps. Probes were mapped to exons of annotated transcript of known genes for which a National Center of Biotechnology Information GeneID exists. Only probe sets in which at least half of the individual probes matched an exon were accepted. Mapping results for each probe are available on the EnrichmentProfiler Web page.
Methods describing transcription factor binding site prediction, plasmids, antibodies, compilation of gene lists, reporter assays, RNAi, knockdown studies and associated references are available in the supplemental Methods.
Results
The Gene Expression Omnibus,7 ArrayExpress,8 and the scientific literature were text-mined for experiments in which normal primary human cells and tissues were profiled on single channel Affymetrix platforms. Experiments for which raw Affymetrix CEL files were available were grouped by microchip identification number. Affymetrix U133A platform was used here but this approach can be applied to other platforms. When cells obtained from a single subject were not available, experiments where multiple subjects were pooled were used. This typically occurs when a very specific population of cells is sorted and multiple individuals are required to obtain enough cells for profiling. In such cases, some genes vary in expression levels between subjects; however, our goal was to identify genes with high tissue specificity, such as transcription factors that define cell state or genes that have a key functional role in a specific cell type. Overall, 557 arrays of normal (non-disease state) primary cell types/tissues that represent 126 cell types/tissues were compiled (supplemental Table 1). This collection of arrays was processed, normalized and filtered as a single experiment (see “Microarrays and the enrichment score”).
To quantify the level of gene enrichment per cell/tissue, individual arrays within each group were treated as replicates. In such a large dataset, every gene is differentially expressed to some extent, thus we devised a novel benchmarking system which scores each probe in 1 tissue relative to all other tissues. The bioconductor linear models for microarray data (LIMMA) module was used to compare each group pairwise to each of the other 125 groups.10 This analysis resulted in 125 linear model coefficients per probe for each tissue and is illustrated in supplemental Figure 1. The linear model coefficient is a measure of difference between 2 groups. Larger differences result in higher coefficient values and are typically associated with more significant P values. We defined an enrichment score for each probe as the sum of all coefficients that were statistically significant (Bonferroni corrected P < .05). To illustrate this scoring system, the coefficient matrix of TFs in embryonic stem (ES) cells is shown as a heat map in Figure 1. Each row contains 125 coefficients that typically range from −10 (green) to +10 (red). The rows are sorted by their sum from high to low. POU5F1 (OCT4), NANOG, LIN28, and SOX2 are the top 4 scoring TFs in ES cells and were previously shown to be sufficient to reprogram human somatic cells into ES cells.11 The coefficients for these TFs are high across the entire row implying that their expression levels in ES cells were significantly higher compared with any other tissues present in the panel. Therefore, the sum of these coefficients resulted in a very high score that reflects their level of specificity. Here, we refer to this score as an enrichment score. One important attribute of this scoring system is that the actual enrichment score is comparable between genes and within each gene. To demonstrate the difference between enrichment scores and expression levels, mean expression values, z-scores and enrichment scores for GAPDH and TBX21 are shown in Figure 1B. GAPDH is expressed at very high levels in all cells profiled here, while TBX21, a known regulator of Th1 cells, is highly expressed in a subset of lymphocytes. Because the enrichment score is a result of a statistical analysis, it provides an improved signal/noise ratio compared with mean expression levels or a z-score transformation, which are commonly used. The z-score indicates the number of standard deviations above/below the mean, however, it does not take into account the variability within each group (depicted by vertical black lines). This is best illustrated by the sharp drop in the enrichment score observed for TBX21 in whole blood due to the large standard deviation within this group, which is not reflected in the z-score. In addition, the enrichment score, unlike the z-score, is comparable between genes. Using a distribution of the highest enrichment score for each probe the degree of specificity can be estimated. For instance, only 10% of the genes score above 869 (90th percentile), indicating that the TBX21 enrichment observed in NK CD56+ T cells is in fact high, while the highest enrichment score of 163 observed for GAPDH is low. In Figure 2 the enrichment profiles of FOXP3, POU5F1, PHF7, and ATF5 are shown. FOXP3 and POU5F1 are known to determine cell fate in regulatory T cells (Tregs)12 and ES cells,13 respectively, and PHF7 and ATF5 have been shown to have important functional roles in testes14 and liver,15 respectively. These TFs have very high enrichment scores in these cells. Throughout the figures we use a color-coding scheme that reflects the quantile of the score as shown in Figure 2.
Attributes of the enrichment score. (A) A heatmap representation of LIMMA linear coefficients for ES cells. The heatmap depicts linear coefficients derived from a pairwise comparison of expression values in ES cells and every other cell type/tissue in the panel. For illustration purposes only transcription factors are shown, are sorted vertically by the enrichment score (sum of coefficients in each row). (B) Expression values (black) and their standard deviation (black vertical lines), z-scores (red) and enrichment scores (yellow) for TBX21 and GAPDH. For illustration purposes only the top 10 and bottom 10 expressing tissues are shown. The left y-axis indicates expression values on log2 scale and z-score values. The right blue y-axis indicates enrichment scores. A distribution of enrichment scores was obtained from the highest score for each probe on the array and the percentiles of that distribution are shown in blue dotted lines.
Attributes of the enrichment score. (A) A heatmap representation of LIMMA linear coefficients for ES cells. The heatmap depicts linear coefficients derived from a pairwise comparison of expression values in ES cells and every other cell type/tissue in the panel. For illustration purposes only transcription factors are shown, are sorted vertically by the enrichment score (sum of coefficients in each row). (B) Expression values (black) and their standard deviation (black vertical lines), z-scores (red) and enrichment scores (yellow) for TBX21 and GAPDH. For illustration purposes only the top 10 and bottom 10 expressing tissues are shown. The left y-axis indicates expression values on log2 scale and z-score values. The right blue y-axis indicates enrichment scores. A distribution of enrichment scores was obtained from the highest score for each probe on the array and the percentiles of that distribution are shown in blue dotted lines.
Enrichment profiles of the top 10 tissues for 4 transcription factors. POU5F1, FOXP3, PHF7, and ATF5 are known for their role in embryonic stem cells (ES cells), T regulatory cells, testes, and liver, respectively. Bars are color-coded for the enrichment score relative to the distribution of highest and lowest score per gene. The values in the color scheme on top represent the percentiles of these distributions.
Enrichment profiles of the top 10 tissues for 4 transcription factors. POU5F1, FOXP3, PHF7, and ATF5 are known for their role in embryonic stem cells (ES cells), T regulatory cells, testes, and liver, respectively. Bars are color-coded for the enrichment score relative to the distribution of highest and lowest score per gene. The values in the color scheme on top represent the percentiles of these distributions.
Because transcriptional regulators are primary lineage determining factors in cellular differentiation, our next goal was to identify TFs that govern T-cell development and differentiation. In the next section, we harness the power of this dataset to the discovery of TFs and genes that play a role in T-cell development and differentiation. We demonstrate the ability to recover known genes, indicate potential novel genes and validate a novel regulator in T cells.
T-cell development and differentiation
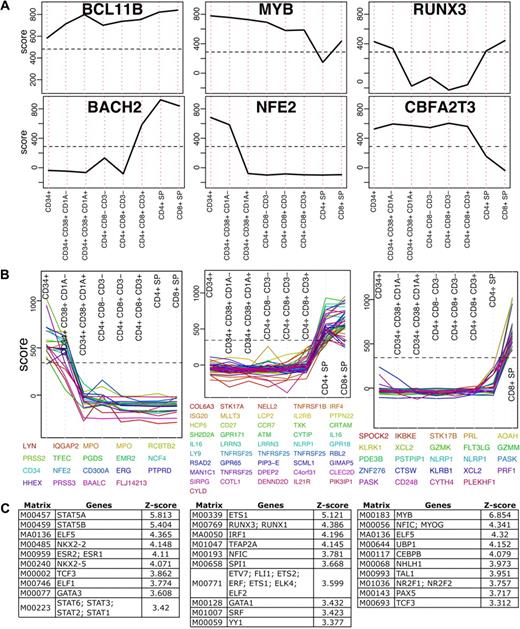
T cells develop from pluripotent precursors that migrate to the thymus where they proliferate, develop and differentiate (for review see Rothenberg et al1 and Singer et al2 ). The dataset presented here, although not complete, includes several developmental stages. Due to the properties of the enrichment score, TFs relevant to thymocyte development can be identified by applying simple filters. First, a distribution of the highest score per probe in thymocyte development was calculated and the 99th percentile of that distribution was used as a cutoff. This filter identifies the most highly enriched genes, including those that do not change across development. BCL11B is a transcription factor in this category, which maintained very high enrichment scores across all development stages. BCL11B is a C2H2-type zinc finger TF which was previously shown essential for thymocyte development16 (Figure 3A). Next, we selected the top scoring 2.5% genes in thymocyte development and filtered those for genes with the largest change (85th percentile of maximum-minimum) across development. We identified 16 TFs that matched these criteria (Figure 3A and supplemental Figure 2). Importantly, gene deletion of fourteen TFs were previously shown to result in T-cell developmental phenotypes confirming that the expression analysis is capable of recovering genes important for transcriptional control of T cells with high precision (supplemental Table 2).
Transcription factors in T-cell development. (A) Enrichment profiles of 6 selected TFs across T-cell development stages (see supplemental Figure 2 for all TFs). These TFs score above the 97.5th percentile (indicated by a horizontal dashed line) in at least one development stage and their change across the development is larger than the 85th percentile. (B) All genes scoring above the 97.5th percentile that change across development above the 85th percentile were clustered to 40 clusters using K-means. Here 3 representing clusters are shown (see supplemental Figure 3 for all clusters). (C) Transcription factor binding site analysis results for each of the 3 clusters shown in panel B. Position weight matrices are ranked by z-score and only the top 10 results in each are shown.
Transcription factors in T-cell development. (A) Enrichment profiles of 6 selected TFs across T-cell development stages (see supplemental Figure 2 for all TFs). These TFs score above the 97.5th percentile (indicated by a horizontal dashed line) in at least one development stage and their change across the development is larger than the 85th percentile. (B) All genes scoring above the 97.5th percentile that change across development above the 85th percentile were clustered to 40 clusters using K-means. Here 3 representing clusters are shown (see supplemental Figure 3 for all clusters). (C) Transcription factor binding site analysis results for each of the 3 clusters shown in panel B. Position weight matrices are ranked by z-score and only the top 10 results in each are shown.
We next examined whether some TFs that do not change in expression levels could still be detected by identifying expression changes in their target genes. A similar selection process as we described for TFs was applied to all genes for which a reliable probe was available (see “Probe mapping”). Genes enriched above the 98th percentile in a specific developmental stage were selected and those that had a range larger than the median were clustered using k-means into forty groups (supplemental Figure 3). Several groups with patterns of interest that included at least 10 probes were further tested for enrichment of transcription factor binding sites in their proximal promoters (see supplemental Methods). Each of these groups was compared with 1000 random backgrounds that were used to obtain a distribution and transcription factor binding sites were ranked by z-score (standard deviation units from the mean). Results for 3 clusters are shown in Figure 3B and C. Genes up-regulated in early development were enriched for STAT5A and STAT5B binding sites. Genes that were low initially in the CD34+ stages but high in the single positives were enriched for ETS1, RUNX3 and RUNX1; and genes enriched in CD8+ cells were enriched for MYB. Although some TFs are known to be essential, in many cases the role they play in thymocyte development remains unknown. Identifying enrichment of their targets in a specific group of genes could suggest additional clues to developmental stage specific transcription. RUNX3, for instance, is known to play a role in differentiation toward CD8+ lineage commitment, which is consistent with the enrichment findings. Similarly to the TF analysis, enrichment profiles of all genes was performed and resulted in identification of known and novel genes that may play a role in differentiation. For instance, in the CD8+ SP, PLEKHF1 and the chemokine XCL2 were among the highest scoring genes based on the enrichment scoring scheme. PLEKFH1 has been previously reported as a glucocorticoid responsive gene in neuronal cells and therefore study of its biologic role in CD8+ T cells may offer insights into the immunodulatory targets of steroids in CD8+ T-cell responses. XCL2 is a chemokine that is predominantly expressed in activated T cells and was shown to induce chemotaxis in cells that express the XCR1 chemokine receptor.17 XCL2 was not previously implicated in thymocyte development or lineage commitment and may warrant further study. These results demonstrate the feasibility for genome scale datasets when combined with cell specific enrichment to infer functional connections in T-cell development.
The same strategy outlined above to T-cell development was applied to T-cell differentiation. Naive CD4+ T cells differentiate into distinct Th subsets, namely Th1, Th2, Tregs and Th17, that are defined by distinct cytokine production and effector functions. Our dataset includes expression profiles for Th1, Th2 and Tregs. To identify key regulators in Th1, Th2 and Tregs, we initially selected TFs that were enriched above the 98th percentile in at least 1 of the 4 cell types. Forty-two TFs were identified and clustered to 20 groups using K-means (supplemental Figure 4). The enrichment profiles of TFs in Th1 and Th2 cells were remarkably similar with several TFs enriched in both compared with the naive CD4+ and Tregs, including IRF4, VDR and ETS1 (supplemental Figure 5A). TBX21 and BHLHB2 were enriched in Th1 compared with Th2 cells while GATA3, CREM, BATF3, and WHSC1 were highly enriched in Th2. FOXP3 and PBXIP1 were enriched in Tregs and had similar profiles. FOXP3, TBX21, and GATA3 are hallmark TFs of Tregs, Th1, and Th2, respectively,12,18,19 and all 3 were identified by this analysis. In addition, principal component analysis confirmed that Th1 and Th2 were clustered close to one another and further from all other cells, suggesting their TF profiles are similar and unlike other T cells in the panel (supplemental Figure 5B). These expression maps illustrate a broader role for molecular similarities among transcription factors in T-cell differentiation.
Genes specific to T cells
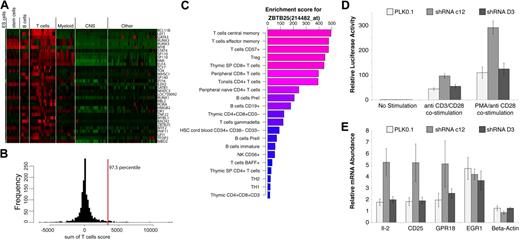
Given the high resolution of T cells in our dataset, we were interested in identifying TFs that are restricted to T cells, but not to a specific T-cell subset. To identify such TFs, the sum of enrichment scores in all T cells was calculated and the top 2.5% of that distribution was selected (Figure 4A-B). BCL11B, LEF1, and GATA3 were the top ranking TFs in T cells, although BCL11B is the only TF that appears to be T-cell specific with respect to the cells and tissues in our dataset. We confirmed this observation by reverse-transcription–polymerase chain reaction (RT-PCR) in CD4+, CD8+, CD4+CD25+ T cells, CD19+ B cells, and CD14+ monocytes (supplemental Figure 6). While these samples were obtained from pooled subjects, BCL11B expression levels in T cells were 100-fold higher than B cells and monocytes. Mice deficient in BCL11B were shown to have a disorganized thymic cortex and medulla and defects in thymocyte development.20 LEF1 and GATA3 are well-characterized TFs that have been shown key to T cells,21 however, they are not limited to the T lineage. ZBTB25 is one of the TFs that scored highly in T cells and was low elsewhere (Figure 4C). ZBTB25 belongs to the BTB/POZ-ZF transcription factor family with 60 such genes encoded in the human genome, characterized by an N-terminal POZ/BTB domain and carboxyl terminus DNA binding zinc finger motifs. Crucial roles have been revealed for several vertebrate POZ-ZF proteins including specification of CD4+ versus CD8+ lineage decisions by ZBTB7B (Th-POK) as well as germinal center formation by BCL6.22,23 A heatmap of gene enrichment profiles of POZ-ZF genes present on the U133A platform is shown in supplemental Figure 7. Such enrichment or expression maps could easily be generated for any given gene list using the EnrichmentProfiler Web interface.
T-cell–enriched transcription factors. (A) A heatmap representation of enrichment scores for genes identified as highly enriched in T cells. Genes are ranked from highest to lowest T-cell score and top scoring BCL11B is the only TF highly enriched in T cells. (B) A histogram representation of the sum of enrichment scores in T cells for all genes. The top 2.5% genes of that distribution were accepted as significantly enriched and are shown in panel A. (C) Enrichment profile of ZBTB25 showing the top 20 enriched cells/tissues. (D) Effect of ZBTB25 depletion on TCR-stimulated NF-AT signaling. ZBTB25 knockdown Jurkat E-6 stable cells were electroporated with 8 μg of NF-AT–luc reporter and 1 ng of renilla-luc reporter. After 18 hours of electroporation, cells from each sample were dispensed into 3 equal aliquots with 1 mL of complete IMDM media with or without anti-CD3 plus anti-CD28 antibodies (1 μg/mL of each) or PMA (50 ng/mL) plus anti-CD28 antibody (1 μg/mL). After another 7 hours of incubation, cells were harvested and examined for luciferase activity. The experiment was done in triplicates. (E) Effect of ZBTB25 depletion on the induction of TCR stimulated NF-AT target genes. Indicated ZBTB25 knockdown Jurkat E-6 stable cells were incubated with or without anti-CD3 plus anti-CD28 antibodies (1 μg/mL of each) for 24 hours. Then 3 sets of 1 × 106 cells from each experimental condition were harvested independently for real-time RT-PCR analysis of indicated gene expression. The experiment was done in biologic triplicates.
T-cell–enriched transcription factors. (A) A heatmap representation of enrichment scores for genes identified as highly enriched in T cells. Genes are ranked from highest to lowest T-cell score and top scoring BCL11B is the only TF highly enriched in T cells. (B) A histogram representation of the sum of enrichment scores in T cells for all genes. The top 2.5% genes of that distribution were accepted as significantly enriched and are shown in panel A. (C) Enrichment profile of ZBTB25 showing the top 20 enriched cells/tissues. (D) Effect of ZBTB25 depletion on TCR-stimulated NF-AT signaling. ZBTB25 knockdown Jurkat E-6 stable cells were electroporated with 8 μg of NF-AT–luc reporter and 1 ng of renilla-luc reporter. After 18 hours of electroporation, cells from each sample were dispensed into 3 equal aliquots with 1 mL of complete IMDM media with or without anti-CD3 plus anti-CD28 antibodies (1 μg/mL of each) or PMA (50 ng/mL) plus anti-CD28 antibody (1 μg/mL). After another 7 hours of incubation, cells were harvested and examined for luciferase activity. The experiment was done in triplicates. (E) Effect of ZBTB25 depletion on the induction of TCR stimulated NF-AT target genes. Indicated ZBTB25 knockdown Jurkat E-6 stable cells were incubated with or without anti-CD3 plus anti-CD28 antibodies (1 μg/mL of each) for 24 hours. Then 3 sets of 1 × 106 cells from each experimental condition were harvested independently for real-time RT-PCR analysis of indicated gene expression. The experiment was done in biologic triplicates.
To investigate the potential function(s) of ZBTB25, a functional shRNA approach was used. Stable Jurkat E-6 knockdown cell lines were generated using 5 lentiviral shRNAs against ZBTB25, to identify the most effective targeting sequence. By examining the knockdown efficiency of endogenous ZBTB25 of each stable cell line by RT-PCR, 3 stable cell lines were chosen for further experiments: the PLK0.1 cell line, which was generated using empty PLK0.1 vector containing no shRNA insert; the shRNA C12 cell line, which is the best knockdown cell line among the 5 ZBTB25 knockdown cell lines tested; and the shRNA D3 cell line, which did not appear to exhibit significant knockdown of ZBTB25, and thus served as a negative control to exclude off-target effects (supplemental Figure 8). To determine potential pathways that ZBTB25 might regulate in T cells, we determined the effect of ZBTB25 knockdown using a series of pathway reporters including NF-AT–luciferase and AP-1–luciferase representing the luciferase gene under the control of multimerized binding elements. ZBTB25 knockdown significantly enhanced T-cell receptor–mediated NF-AT reporter activity compared with the empty vector and shRNA (D3) controls (Figure 4D). In contrast, there was no significant difference observed in AP-1 reporter activity among these 3 cell lines after PMA/anti-CD28 costimulation (supplemental Figure 8B). To further examine the role of ZBTB25 in NF-AT signaling, we next measured the expression levels of several NF-AT target genes by RT-PCR after TCR stimulation in the context of ZBTB25 knockdown. IL2 mRNA expression levels were used to confirm NF-AT activity. Comparing activation among the 3 different cell lines after anti-CD3/CD28 costimulation, we found that ZBTB25 knockdown resulted in further induction of NF-AT targets IL2, CD25, and GPR18 (Figure 4E). In contrast, EGR1, which has been shown not to be a target of NF-AT,24 was not modulated by ZBTB25 knockdown after anti-CD3/CD28 costimulation. Taken together, these data strongly indicate that ZBTB25 knockdown augments TCR-mediated NF-AT signaling, consistent with ZBTB25 possessing a novel NF-AT transcriptional repressor activity.
Genes enriched in hematologic cancers
We next tested the application performance of EnrichmentProfiler as a resource for gene discovery in hematologic malignancies. Genome-wide transcriptional profiling of human tumors on microarrays has been used extensively over the past few years to address biologic questions, primarily questions of classification or identification of gene sets differentially expressed between 2 different conditions.25-27 However, for many cancers the preferred control, typically the cell of origin, is either unknown or difficult to obtain. Using the enrichment score presented here cancer cell–expressed genes can be profiled with respect to all other cell types and tissues in the dataset without the need of a specific control. To test this hypothesis, the primary cell dataset was extended to include the NCI-60 collection of tumor profiles, which is available on the U133A platform. Of the 98 available NCI-60 microarrays, 5 tumors for which there were no replicates were removed and the rest were classified into 15 types of cancer (supplemental Table 3). The entire dataset containing 649 arrays of cancer and primary cells/tissues was normalized and filtered as described in “Microarrays and the enrichment score.”
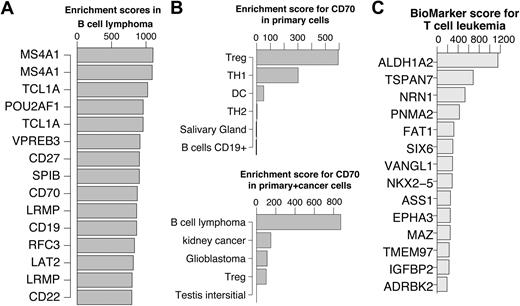
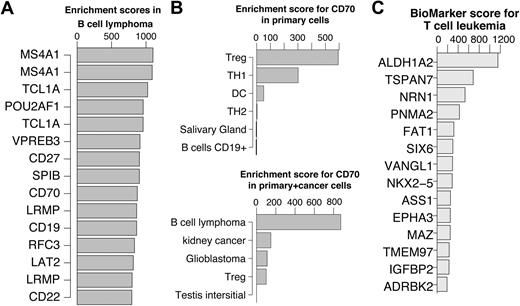
In Figure 5A, the top 15 scoring genes in B-cell lymphoma are shown. Previous studies have verified that top ranked genes are involved in B-cell lymphoma. A few examples include MS4A1 (CD20) as a marker for follicular B-cell non-Hodgkin lymphoma28 ; TCL1A, which promotes multiple classes of B-cell lymphomas in a transgenic mouse model,29 and POU2AF1, for which it was shown that the TCL1A transgenic mice no longer developed B-cell lymphoma when crossed with B-cell POU2AF1 deficient mice.30 CD27 is a receptor for CD70 which was shown to induce FOXP3 and develop intratumoral Tregs in B-cell lymphoma.31 Interestingly, the primary cell dataset profile of CD70 was slightly enriched in Tregs. This enrichment was abolished in the cancer dataset due to a very strong enrichment in B-cell lymphoma (Figure 5B). These examples suggest that key genes can be identified using the enrichment score alone without restricting the comparison to a single specific cell of origin. This approach will be most useful for cancers where the cell of origin is not clear or very difficult to obtain.
Genes enriched in hematologic cancers. (A) Top 15 enriched genes in B cell lymphoma. (B) CD70 enrichment profiles showing the top 5 cells/tissues in the primary cell dataset (top) and cancer cell dataset (bottom). Treg enrichment was abolished in the presence of cancer derived tissues due to significantly higher expression in cells derived from B-cell lymphoma. (C) Top 15 enriched genes in T-cell leukemia cells after reducing the highest enrichment score in primary T cells for each gene. For illustration purposes, in cases where multiple probes for the same genes exist, only the higher scoring probe is shown.
Genes enriched in hematologic cancers. (A) Top 15 enriched genes in B cell lymphoma. (B) CD70 enrichment profiles showing the top 5 cells/tissues in the primary cell dataset (top) and cancer cell dataset (bottom). Treg enrichment was abolished in the presence of cancer derived tissues due to significantly higher expression in cells derived from B-cell lymphoma. (C) Top 15 enriched genes in T-cell leukemia cells after reducing the highest enrichment score in primary T cells for each gene. For illustration purposes, in cases where multiple probes for the same genes exist, only the higher scoring probe is shown.
To identify biomarkers for diagnostics and treatment purposes, enrichment scores in cancer tissues need to be compared with the primary tissue of tumor origin. To identify potential biomarkers we defined a biomarker score as the score in cancer cells/tissues minus the highest score across primary tissues of tumor origin. Based on the scheme applied to recover known and novel TFs in T-cell development, we reasoned that putative tumor specific genes could be identified by subtracting the highest score among primary tissues of tumor origin from the enrichment score in the tumor of interest.
For instance, to identify T-cell leukemia enriched genes, for each probe, the highest enrichment score in any of the primary T cells was subtracted from the T-cell leukemia enrichment score. Consequently the genes were ranked and the top 15 are shown in Figure 5C. The top scoring gene in T-cell leukemia is ALDH1A2, which was previously shown to be induced by TAL1 and LIM proteins in T-cell acute lymphoblastic leukemia.32 The second ranking gene, TSPAN7, appears to be restricted to the brain among the primary tissues, which is consistent with the current literature.33 However, it is not detectable in primary T-cell subsets except for a low expression in Thymic CD34+ T cells. This observation makes TSPAN7 an attractive T-cell leukemia biomarker candidate. TSPAN7 is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of 4 hydrophobic domains and play a role in the regulation of cell development, activation, growth, and motility. TSPAN7 was previously reported as a T-cell leukemia–specific marker and was detected as such at the mRNA levels as well as protein level.34 We confirmed this observation by quantitative PCR using T-cell leukemia cell lines, primary T cells, B cells, and monocytes (supplemental Figure 9). While TSPAN7 needs to be further studied as a potential biomarker, this example demonstrates the simplicity and robustness by which potential cancer cell/tissue enriched genes can be identified.
Discussion
This study provides a compendium of gene expression profiles that contains the largest collection of normal immune subsets assembled to date. Cell specificity is a key component in unraveling gene function and transcriptional networks. Here we devised an enrichment scoring system to quantify specificity with respect to the cells and tissues present in the compendium. The enrichment score is not a measure of expression levels. GAPDH, for instance, is highly expressed in all cells but has a very low enrichment score, which reflects a lack of specificity. Only genes highly expressed in a subset of tissues reach a high enrichment score as shown in Figure 2. The enrichment score provides several key advantages compared with the traditional z-score of mean expression levels: (1) an improved signal to noise ratio that takes into account variability within each group; (2) a score that is comparable between genes, allowing gene ranking based on their specificity in each cell type/tissue; and (3) does not require a predefined control tissue. Here we demonstrate feasibility of uncovering cell-specific transcription factors in T-cell fate decisions. This proposed schema could be adapted to study cell- and tissue-specific regulatory elements in cancer, stem cell biology, and human disease. The advantages of using the enrichment score were illustrated using multiple examples where the enrichment profiles of known genes were consistent with current knowledge. For instance, the top 4 ranking TFs in ES cells (Figure 1) were previously shown to define the ES cell state.11 The resource provided here is not limited to T cells and provides significant insights into many cells and tissues. For instance, a previously undescribed secreted hormone expression signature (eg CER1, LEFTY12, NPPB, GAL, NTS1) was observed in pluripotent embryonic stem cells, suggesting that a combinatorial control of autocrine factors maybe vital to pluripotency.
This dataset is not without limitations. Enrichment scores are limited to the genes present on the array (approximately 12 000) and existing profiles do not reflect any transcriptional changes that result from a stimulus, such as exposure to hypoxia, starvation, or cytokine stimulation, all of which are important parameters in predicting gene function.
To determine the expression pattern of TFs in the human genome we focused on T-lymphocyte biology. Previous studies of TFs in T-cell biology focused on a few T-cell subsets and differential expression within these subsets. Here we demonstrated the utility of profiling with respect to a large number of cells and tissues that represent the entire human body. While the mouse is an attractive model system to study T- and B-cell responses, it is essential to generate expression and enrichment maps in human immune cells to have the ability to compare and contrast these profiles with mouse datasets. For instance, ZBED2 is a 25-kDa human protein of unknown function containing a BED-type zinc finger domain. BED domains were previously studied in Drosophila mealanogaster BEAF and DREF proteins. These proteins were shown to bind to insulators, which are genomic elements capable of silencing distinct sectors of the chromatin. Several members of the BED domain family have been studied in humans. ZBED3 was shown to modulate the Wnt/beta-catenin signaling pathway35 and ZBED1, a human homologue of DREF, was shown to be involved in regulating cell proliferation.36 ZBED2 is conserved in Rhesus monkies, Macaques, chimpanzee, cows, horses, and dogs, but not in rodents. It is the top scoring gene in Th2 cells and the third scoring gene in Th1 cells. That high level of enrichment suggests that ZBED2 may have an important role in T-cell function that would have not been recovered by examining mouse T-cell datasets. Sixteen TFs were identified as highly enriched during T-cell development based on their high enrichment score or change across development (supplemental Figure 3). Of these 16 TFs,14 were previously deleted in mice and MYB, RUNX3, TCF7, and SOX4 had a phenotype consistent with a defect in T-cell development.37-40 BACH2 deficient mice had a B-cell defect in class switch recombination, a recombination of immunoglobulin heavy chain required to produce antibodies.41 However, another study found that BACH2 is a regulator of IL2 in cord blood CD4+ T cells.42 Both observations are supported by the enrichment profile of BACH2 in B and T cells.
While some TFs, as demonstrated here, can be identified through enrichment profiles, others that are widely expressed or do not vary significantly in expression may also play a role. Such TFs could still be detected by enrichment profiles of their target genes. We demonstrated this approach by clustering genes with similar enrichment profiles in T-cell development and testing their promoters for over-represented binding sites compared with suitable backgrounds. We successfully identified essential TFs including GFI1, MYB, RUNX3, ETS1, STAT5A, STAT5B, and members of the E2F family. This analysis not only provides an additional layer of confidence for TFs predicted by enrichment profiles, but also provides insight into the potential targets and the specific stages in which the TF plays a role. For instance, MYB was identified in genes highly enriched in CD8+ SP cells, while GFI1 in genes enriched in the CD34+ precursors. Novel TFs that emerged from this analysis and may play a functional role include MEF2 and NKX2-2.
ZBTB25 was identified as one of the TFs highly enriched in T cells. ZBTB25 belongs to the BTB/POZ-ZF transcription factor family with 60 such genes encoded in the human genome, characterized by an N-terminal POZ/BTB domain and carboxyl terminus DNA binding zinc finger motifs. The BTB/POZ (Broad complex Tramtrack bric-a-brac/Pox virus and zinc finger) domain is a 100 amino acid, highly conserved motif that mediates protein–protein interactions. POZ-ZF transcription factors generally interact with their cognate DNA sequences via their zinc finger motifs (the majority of which are of the Krüppel-like C2H2 type) to bring about chromatin modification and/or restructuring, typically resulting in transcriptional repression via POZ domain recruitment of corepressor complexes. Crucial roles have been revealed for several vertebrate POZ-ZF proteins including specification of CD4 versus CD8 lineage decisions by ZBTB7A (Th-POK) as well germinal center formation by BCL6. Data presented in this manuscript suggests that ZBTB25 antagonizes NF-AT induced gene expression analogous to suppression of STAT6 target genes by BCL6. These data contribute to the emerging concept that BTB/POZ domain containing TFs play an important role in cell and stage specific inhibition of T-cell function. Furthermore, a number of TFs such as ZNF22 and MITF2 are enriched in T cells, suggesting functional importance in T-cell biology.
Unraveling the complexity of CD4+ and CD8+ development and T-cell effector responses will require a systematic knowledge of the expression changes occurring during differentiation. We have developed and presented an approach that has broad applications. This study showed that expression patterns of many TFs correlate well with their function given a large enough dataset. This approach can be extended to other single channel platforms and additional datasets as data become available.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jason Eisenberg for writing the code to generate heat maps on the EnrichmentProfiler Web site; Gordon Smyth, author of LIMMA, for his constructive feedback and discussions; and Brian Seed, Mark Daly, and Xavier laboratory members for helpful discussions.
R.J.X. is supported by National Institutes of Health grants AI062773, DK83756, and DK043351. A.G. is a fellow of the Crohn's and Colitis Foundation of America.
The compendium constructed here is available for browsing and download on the EnrichmentProfiler Web page (http://xavierlab2.mgh.harvard.edu/EnrichmentProfiler). The Web page provides both enrichment scores and mean expression values for all genes present on the U133A platform. In addition, R source code for computing enrichment scores from gene expression data are available for download.
National Institutes of Health
Authorship
Contribution: Y.B. devised the enrichment method and executed all computational work including the EnrichmentProfiler public Web site; Z.C. and C.G. conducted all experimental work related to ZBTB25; C.G. participated in study design and assisted with data analysis and microarray selection; C.L. and A.G. conducted all experimental work related to TSPAN7 and BCL11B; and Y.B., Z.C., C.G., A.G., and R.J.X. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ramnik J. Xavier, The Center for Computational and Integrative Biology, Massachusetts General Hospital, 185 Cambridge St, Boston, MA 02114; e-mail: xavier@molbio.mgh.harvard.edu.
References
Author notes
Y.B. and Z.C. contributed equally to this study.
Supplemental data
The references and reported phenotypes for each transcription factor are indicated. Mouse homologs were identified using NCBI homologene (www.ncbi.nlm.nih.gov/homologene).
Group assignment for differential expression is shown where identical group names indicate these samples were grouped together and considered replicates. Raw CEL files were kindly provided by the GNF consortium (http://symatlas.gnf.org/SymAtlas).
See supplementary materials and methods (Document 1) for detailed information on criteria for compiling gene lists.