Abstract
Acute myeloid leukemia (AML) is characterized by molecular heterogeneity that is not fully reflected in the current classification system. Recent insights point toward a significant role of aberrant DNA methylation in leukemogenesis. Therefore, we investigated the prognostic impact of DNA methylation in AML. To screen for promoter methylation in AML we applied a combination of base-specific cleavage biochemistry and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), a powerful methodology allowing for quantitatively investigating DNA methylation status in a large series of both promoter regions and leukemia samples. We analyzed 92 genomic regions in 182 patient samples, correlated findings with clinical and molecular data, and validated the results in an independent cohort of 74 AML samples. Using this approach, we were able to identify novel leukemia subgroups based on distinct DNA methylation patterns. Furthermore, we defined a methylation-based outcome predictor for patient survival (P < .01) that in multivariable analysis provided independent prognostic information (hazard ratio, 1.52; 95% CI, 1.06-2.16). Here, we report the first large-scale methylation-based outcome predictor in AML, and thereby our findings support the use of genomic methylation markers for improved molecular classification and prognostication in adult AML.
Introduction
Despite recent progress, acute myeloid leukemia (AML) still remains a highly fatal disease. Even among AML patients achieving a complete remission many will relapse and die of their disease. More intensive consolidation treatments, such as allogeneic stem cell transplant, might prevent relapse, but are themselves associated with high treatment-related mortality.1 Therefore, it is crucial to identify patients who need more aggressive therapy to prevent relapse and at the same time avoid overtreatment of patients with good prognosis.
Currently, the most important outcome predictors in AML include age, white blood cell count, a history of a preceding malignancy, and cytogenetics.1 The correlation between karyotype and outcome has been studied extensively and identified patient cohorts at low (eg, t(8;21) inv(16)/t(16;16), t(15;17)), intermediate (eg, cytogenetically normal [CN-AML]) or high (eg, complex karyotypes) risk.2-4 However, treatment stratification is still imperfect, in particular for CN-AML, the largest cytogenetic group (40%–50% of cases). In CN-AML, molecular markers such as aberrations involving the NPM1, CEBPA, FLT3, or MLL genes have been shown to be clinically relevant.5,6 Despite the advances in molecular genetics, the current classification system does not fully reflect the heterogeneity of AML.
Gene expression profiling has been used in an attempt to improve the molecular AML classification. Initial studies have provided remarkable results by identifying novel AML subgroups and prognostic gene expression signatures.7-11 Furthermore, a recent gene expression study observed differential expression of genes coding for the DNA methylation enzymes (regulators) DNMT3A and DNMT3B.7 Because DNA methylation is recognized as a key regulatory element of gene expression,12-14 these findings point to a potential pathogenic role of aberrant DNA methylation patterns in distinct subgroups of AML patients.15 In accordance, aberrant methylation patterns have already been described in AML,16,17 and recent studies further support a crucial role of epigenetic changes (epigenetics) in leukemia biology,18-21 as well as a potential impact in patient outcome.22 However, so far a lack of suitable technologies to quantitatively evaluate promoter methylation of numerous genes in large sample sets has prevented extensive exploration of the role of DNA methylation in hematologic malignancies and its impact in outcome prediction.
We recently introduced a novel technique based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), which enables large-scale quantitative DNA methylation analysis,23 and this technology has already been successfully applied to evaluate methylation patterns of single genes in leukemia.24,25 Here, we report the application of this technology to analyze DNA methylation in 92 genomic target regions covering more than 2000 individual CpG sites in a large set of AML patient samples (n = 256). The results of our study led to the identification of methylated genes of potential pathogenic relevance, the discovery of novel clinically relevant subclasses, as well as the definition of a methylation-based outcome predictor. These findings demonstrate the existence of distinct promoter methylation patterns that in the future might provide a novel basis for improved outcome prediction in AML.
Methods
Patients
A total of 182 DNA samples derived from 98 peripheral blood and 84 bone marrow specimens from adult AML patients were provided by the German-Austrian AML Study Group (AMLSG) with patient informed consent and institutional review board approval from all participating centers, in accordance with the Declaration of Helsinki. Patients were entered into 1 of 2 treatment protocols (AMLSG-HD98A and AMLSG-HD98B) between February 1998 and November 2001); all patients received intensive induction and consolidation therapy.26,27 The median follow-up time was 534 days overall (1939 days for survivors); conventional cytogenetic banding, fluorescent in situ hybridization analysis, and NPM1, CEBPA, FLT3, and MLL mutational analyses were performed as previously described5,28 at the central reference laboratory for cytogenetic and molecular diagnostics of the AMLSG. Detailed clinical, cytogenetic, and molecular cytogenetic information are provided in the supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The validation set samples (n = 74) were also derived from the AMLSG-HD98A and AMLSG-HD98B trials (enrollment between February 1998 and May 2004; see also supplemental Table 2).
MALDI-TOF-MS–based DNA methylation analysis
MALDI-TOF-MS–based DNA methylation analysis was performed as previously described.23 In brief, the target gene regions were amplified using bisulfite-modified DNA and the primer pairs described in supplemental Table 3. The primers were designed to be located in a CpG island overlapping the 5′ region of the corresponding gene. If no CpG was located 5′ of the gene, the closest CpG Island was used for design. One of the polymerase chain reaction primers was tagged so that the polymerase chain reaction product carries the T7 recognition sequence. This allowed the generation of a single-stranded RNA copy of the template, which was then cleaved base specifically. The cleavage products were analyzed using MALDI-TOF. All reactions were carried out according to the manufacturer's specification (Sequenom Inc).23
Genes subject to methylation analysis in this study have been selected based on our previous gene expression profiling study in AML. In brief, we selected the top 48 genes associated with outcome represented by 50 genomic regions7 ; we included an additional 32 genes known to be aberrantly methylated in cancer or to be associated with leukemogenesis (eg, CDKN2A or CEBPA) represented by 42 genomic regions (for a detailed list of genes, see supplemental Table 3). The promoter regions of these 80 genes (represented by 92 genomic regions; ie, amplicons) included more than 2170 CpG sites for each sample. The CpG sites were analyzed in 1320 informational units that contained either individual CpG sites or short stretches of subsequent CpG sites (CpG units).
Gene expression profiling
Statistical methods
All statistical analyses were carried out using the R statistical environment (available at CRAN, http://cran.r=project.org/).30 We used the gregmisc package for 2-dimensional clustering, the hclust package for hierarchic cluster analysis, the survival package for Cox regression analysis and Kaplan-Meier estimates, and the SuperPC package for supervised principle components analysis.31 In brief, methylation patterns associated with outcome (overall survival) were determined using a semisupervised principal component analysis classification algorithm. This algorithm is available as an R implementation in the SuperPC package. We used this package for patient stratification in the training data according to the recommendations by the inventors of the algorithm.31 To limit search space and consequently avoid overfitting of the data, we searched for the first component associated with outcome. As the algorithm requires a threshold to segregate patients who have been assigned a score on a continuous scale, we determined this threshold by cross validation as recommended by the Stanford group (see also http://www-stat.stanford.edu/∼tibs/superpc/tutorial.html).32 The applicability of the classification model was then verified within the test set samples followed by a validation of findings in a third independent AML cohort. As SuperPC essentially uses principal component analysis, it is not trivial to describe the correlation between model-specific features and the resulting principal components (Eigenvectors). However, the SuperPC package allows ranking of the contribution of features to the classification system and this output was used for gene-directed analysis.
Relative methylation was compared between good and poor outcome groups using the Wilcoxon signed-rank test, a nonparametric counterpart of the paired t test. The 2-way hierarchic cluster analysis clustered samples and CpG units based on pair-wise Euclidean distances and the complete linkage clustering algorithm.33 This was carried out using the heatmap.2 function of the gregmisc package using the R statistical environment.
Results
Biologic findings
We analyzed a total of 256 diagnostic AML samples for promoter methylation of 92 genomic regions selected based on previously identified prognostic and epigenetically modified genes. All samples were enriched to a fraction of blast cells of more than 80% after density gradient centrifugation. Although in general the reproducibility of the quantitative methylation analysis was quite good (supplemental Figure 1), we investigated whether the methylation patterns might be influenced by the sample source by calculating the mean methylation value for each CpG unit across all samples derived from peripheral blood and across all cases derived from bone marrow. As there was no significant difference between the peripheral blood– and bone marrow–derived sample groups (P = .55, t test; supplemental Figure 2) we did not further discriminate between these 2 groups.
Next, we performed an unsupervised 2-dimensional hierarchic cluster analysis to explore associations among AML samples and the relationship of the relative methylation of CpG units within and between genes (Figure 1). Most samples with an inv(16) were located in one cluster (highlighted in yellow), most t(11q23) cases grouped together (highlighted in green) and translocation t(15;17) and t(8;21) cases also clustered mainly in one group (highlighted in pink). Other unsupervised cluster-defined groups were cytogenetically less well defined. However, a group consisting of 2 normal and 1 complex karyotype cases presented with largely hypermethylated DNA (highlighted in blue), thereby they were the most different from the methylation patterns of all other samples.
Hierarchic cluster analysis of AML samples and their methylation profiles. Two-way hierarchic cluster analysis of 182 acute myeloid leukemia (AML) samples (rows) and DNA methylation of CpG units in 92 promoter regions (columns). DNA methylation values are depicted by a pseudocolor scale as indicated (methylation increases from red [nonmethylated] to white [methylated]). Samples with overall poor data quality were removed before clustering. Gray denotes data of poor quality. Samples are color-coded according to the underlying cytogenetic aberration (legend depicted top left). Most samples with an inv(16) were located in one cluster (highlighted in yellow); most t(11q23) cases grouped together (highlighted in green) and translocation t(15;17) and t(8;21) cases did also cluster mainly in one group (highlighted in pink). A small set of 3 samples splits off early in the dendrogram showing generally hypermethylated DNA (bottom 3 rows, highlighted in blue). CpG units form 2 large clusters that are characterized mainly by the level of methylation. The larger group (group 1, highlighted in pink) is characterized mainly by unmethylated CpG units, the smaller one (group 2, highlighted in yellow) mainly by methylated CpG units. In each cluster a group of CpG units can be separated that shows more variable methylation values across the AML samples (as indicated). The black bars on top of the heatmap denote the top 10% of CpG units with the highest variance across the AML samples (> 90th percentile).
Hierarchic cluster analysis of AML samples and their methylation profiles. Two-way hierarchic cluster analysis of 182 acute myeloid leukemia (AML) samples (rows) and DNA methylation of CpG units in 92 promoter regions (columns). DNA methylation values are depicted by a pseudocolor scale as indicated (methylation increases from red [nonmethylated] to white [methylated]). Samples with overall poor data quality were removed before clustering. Gray denotes data of poor quality. Samples are color-coded according to the underlying cytogenetic aberration (legend depicted top left). Most samples with an inv(16) were located in one cluster (highlighted in yellow); most t(11q23) cases grouped together (highlighted in green) and translocation t(15;17) and t(8;21) cases did also cluster mainly in one group (highlighted in pink). A small set of 3 samples splits off early in the dendrogram showing generally hypermethylated DNA (bottom 3 rows, highlighted in blue). CpG units form 2 large clusters that are characterized mainly by the level of methylation. The larger group (group 1, highlighted in pink) is characterized mainly by unmethylated CpG units, the smaller one (group 2, highlighted in yellow) mainly by methylated CpG units. In each cluster a group of CpG units can be separated that shows more variable methylation values across the AML samples (as indicated). The black bars on top of the heatmap denote the top 10% of CpG units with the highest variance across the AML samples (> 90th percentile).
The clustering of CpG units revealed 2 main groups. One larger group (group 1, highlighted in pink) was characterized by low levels of methylation (median: 10%), whereas the second group (group 2, highlighted in yellow) was showing high levels (median: 70%; Figure 1). To obtain a measure for the DNA methylation variability across samples we calculated the variance of the degree of methylation for each CpG unit. The majority of CpG units showed very low variance values (685 CpG units or 52% showed a variance < 0.01; 218 CpG units or 16% showed a variance < 0.001), indicating little variation among AML samples. However, in both groups a small subset of CpG units split off early, being characterized by higher variation of methylation levels (Figure 1). Hierarchic clustering based on only the CpG units with highest variance (the top 5% most variable CpG units) revealed similar findings, although complexity reduction, for example, prevented the detection of the hypermethylated AML subgroup (supplemental Figure 3).
As different amplification lengths and different CpG densities result in variable numbers of CpG sites that are measured for each analyzed region, this redundancy of measurements might bear the risk of introducing a bias in the unsupervised clustering analysis. Thus, we explored different methods (mean, median, principle component analysis, k-means clustering) to analyze our data (supplemental Figure 4). Importantly, we found no single approach to be superior in its capabilities to build sample clusters that are correlated to clinical features such as sex (male vs female), karyotype, or cytogenetic risk group, indicating that various methylation levels are not closely related to any of the examined features.
CpG methylation pattern–based outcome prediction in AML
A main objective of this study was the evaluation of a correlation between quantitative methylation patterns and prognosis. As the 2-dimensional unsupervised hierarchic clustering showed that patient samples clustered mainly into 2 large groups (see indicated clusters excluding the small, hypermethylated group highlighted in blue; Figure 1), Kaplan-Meier analysis of these 2 main clusters showed a small difference in overall survival (P < .05, supplemental Figure 5). This finding indicated that patient survival times might be associated with quantitative methylation patterns.
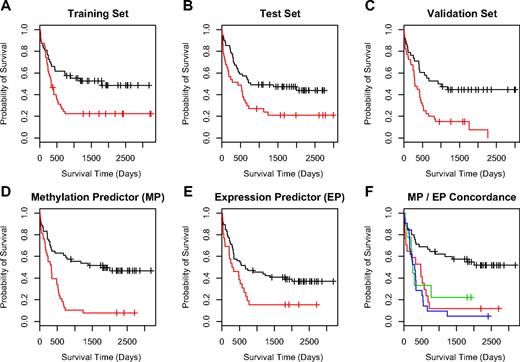
To further investigate the potential association of methylated CpG units with outcome, we used supervised principle components analysis (SuperPC),7,31 a semisupervised approach, that has been shown to yield reliable predictors for several microarray-based gene expression datasets.7,31 We first randomly separated our 182 samples into a training (n = 89) and a test (n = 93) set before applying the SuperPC algorithm to the data. With regard to clinicopathologic features such as karyotype, cytogenetic risk group, and survival time or survival status, there were no significant differences between the groups (data not shown). In the training set we built a predictive model with the resulting good and poor outcome groups showing a significant difference in overall survival (P = .006, log-rank test; Figure 2A). For validation we applied this model to the test set, where samples assigned to the good prognosis group were associated with significantly better survival compared with samples in the poor prognosis group (P = .028, log-rank test; Figure 2B). The confirmation of the predictive capabilities of the model on the test set data was encouraging and justified further validation. Thus, in an additional independent set of 74 AML samples the model assigned clinically meaningful good and poor outcome class labels as these were significantly correlated with patient survival (P < .001 log-rank test; Figure 2C).
Outcome predictions for AML. For development of a methylation-based outcome predictor using SuperPC, the 182 AML samples were separated into a training set (n = 89) and an independent test set (n = 93). (A) Kaplan-Meier survival analysis of the training set with the good (black line) and poor (red line) outcome group showing a significant difference in overall survival (P < .01; log-rank test). (B) The same analysis for the test sample set, where the good prognosis group was still associated with significantly longer survival times compared with the poor prognosis group (P = .028; log-rank test). (C) Kaplan-Meier analysis of the predictor using an additional independent set of 74 AML samples. For this validation set, predicted good and poor outcome classes were again significantly correlated with patient survival (P < .001; log-rank test). (D-F) A comparison of the outcome predictors based on methylation data (MP; D) and on gene expression data (EP; E). (F) Insight on the concordance of the methylation-based and expression-based predictor. The samples representing the black line belong to an outcome group (group GG) where both methods predict good survival. For the red line, both predictors label samples with poor survival outcome (group PP). For samples on the blue and green lines at least one method (expression or methylation) predicted poor outcome. In detail, the green line represents samples where MP predicted good survival, whereas EP predicted poor survival (group GP), and the blue line represents samples where MP predicted poor survival and EP predicted good survival (group PG). For both of these sample groups with discordant survival prediction, the probability of survival was in fact markedly inferior to the predictions providing concordant results in both models.
Outcome predictions for AML. For development of a methylation-based outcome predictor using SuperPC, the 182 AML samples were separated into a training set (n = 89) and an independent test set (n = 93). (A) Kaplan-Meier survival analysis of the training set with the good (black line) and poor (red line) outcome group showing a significant difference in overall survival (P < .01; log-rank test). (B) The same analysis for the test sample set, where the good prognosis group was still associated with significantly longer survival times compared with the poor prognosis group (P = .028; log-rank test). (C) Kaplan-Meier analysis of the predictor using an additional independent set of 74 AML samples. For this validation set, predicted good and poor outcome classes were again significantly correlated with patient survival (P < .001; log-rank test). (D-F) A comparison of the outcome predictors based on methylation data (MP; D) and on gene expression data (EP; E). (F) Insight on the concordance of the methylation-based and expression-based predictor. The samples representing the black line belong to an outcome group (group GG) where both methods predict good survival. For the red line, both predictors label samples with poor survival outcome (group PP). For samples on the blue and green lines at least one method (expression or methylation) predicted poor outcome. In detail, the green line represents samples where MP predicted good survival, whereas EP predicted poor survival (group GP), and the blue line represents samples where MP predicted poor survival and EP predicted good survival (group PG). For both of these sample groups with discordant survival prediction, the probability of survival was in fact markedly inferior to the predictions providing concordant results in both models.
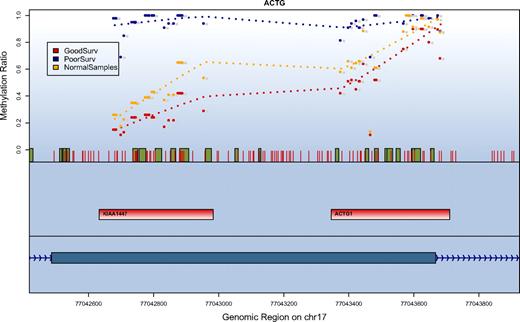
SuperPC also assigns an importance score to each of the features in the model (supplemental Table 4). The CpG units most predictive for poor outcome were derived from a CpG island–dense region located on the long arm of chromosome 17, an area that comprises the 5′ of the mRNA sequence for KIAA1447 (BAHCC1). Additional important genes on the list included homeobox genes involved in transcriptional regulation (HOXA1, HOXA4, HOXA9, HOXA10, HOXA11, HOXB2, HOXB5, and HOXD13). Comparing the methylation levels of these genes to 4 normal control samples derived from peripheral blood showed that 16 regions were statistically significantly higher methylated in the AML samples (on a level of P < .001: ESR1, HOXA1, MYOD, KRT13, NR2F2, PITX2, RBP1; on a level of P < .01: HOXA4, HOXA7, BCL11A, CKMT1, FSCN2, GYPC, KIAA1447; on a level of P < .05: HOXA11, RASSF1, CDH1, FLJ21820, SNX9, SPUVE; t test). Interestingly, 4 of the 5 most predictive genes were included in this list of hypermethylated regions. For the region on chromosome 17 with the strongest predictive value for patient survival we found that methylation levels in patients assigned to the good prognosis groups were comparable with the methylation levels observed in normal samples (Figure 3).
Correlation of survival and methylation levels for chromosome 17q25 gene region. Depicted are the methylation results for a region on chromosome 17q25 with the strongest prediction for patient survival. The top panel shows methylation values in the context of their genomic location. Red vertical bars depict the location of CpG units. Green boxes indicate a conserved transcription factor binding site. For every data series the methylation values in this group are averaged and the mean is displayed for each analyzed CpG unit. Shown are the values for patients who have been classified by the methylation prediction model into a favorable outcome class (red), a poor outcome class (blue), and normal blood samples (orange). The second panel indicates the location of the amplification targets. The third (lowest) panel depicts the gene structure. The methylation ratio values indicate that patients in the good outcome class have methylation levels similar to the normal control samples.
Correlation of survival and methylation levels for chromosome 17q25 gene region. Depicted are the methylation results for a region on chromosome 17q25 with the strongest prediction for patient survival. The top panel shows methylation values in the context of their genomic location. Red vertical bars depict the location of CpG units. Green boxes indicate a conserved transcription factor binding site. For every data series the methylation values in this group are averaged and the mean is displayed for each analyzed CpG unit. Shown are the values for patients who have been classified by the methylation prediction model into a favorable outcome class (red), a poor outcome class (blue), and normal blood samples (orange). The second panel indicates the location of the amplification targets. The third (lowest) panel depicts the gene structure. The methylation ratio values indicate that patients in the good outcome class have methylation levels similar to the normal control samples.
Combined DNA methylation– and gene expression–based outcome prediction in AML
A subset of the samples (n = 92) was previously analyzed by gene expression profiling.7 Thus, we were interested to evaluate the concordance of survival-associated outcome labels between the expression and the methylation-based model in this subgroup (Figure 2D-E). Both models were in agreement for 62 cases (67%) and assigned different class labels in 30 cases (33%). In 45 cases both models predicted good survival (group GG), in 17 cases both models predicted poor survival (group PP). In 21 cases the expression-based model predicted good survival and the methylation-based model predicted poor survival (group GP) and in 9 cases the methylation-based model predicted poor survival, whereas the expression-based model assigned a favorable outcome (group PG). To evaluate the survival times of each of the 4 groups we performed a Kaplan-Meier analysis (Figure 2F). Importantly, the analysis revealed that the subgroup GG, in which both models predicted good survival, was associated with significantly longer overall survival. Furthermore, when both, or most remarkably, even when only one of the models predicted poor outcome, the probability of survival was dramatically reduced. Consequently, we assigned samples in the GG group to a good outcome class and all samples where at least one model predicted poor survival to a poor outcome class with the combined outcome predictor providing highly significant results (P < .001, likelihood ratio test).
Multivariate analysis
To evaluate the relative impact of the methylation-based outcome predictor on prognosis we performed Cox proportional hazard analysis for overall survival in the initial dataset (n = 182) as well as in the additional independent test set (n = 74). Factors included in the model were age, cytogenetic risk classification, FLT3 internal tandem duplication (ITD) mutational status, and our methylation-based outcome predictor. Finally, the analysis revealed a model including FLT3-ITD mutational status (hazard ratio [HR], 2.41; 95% CI, 1.72-3.39), cytogenetic risk classification (HR, 1.85; 95% CI, 1.43-2.39), age (HR for a difference of 10 years, 1.22; 95% CI, 1.08-1.38), and our methylation-based outcome predictor (HR, 1.52; 95% CI, 1.06-2.16) (Table 1).
Next, we also evaluated the combined predictor of gene expression– and methylation-based outcome classes. We assigned cases to the good prognosis group in which both models, the methylation- and the gene expression–based model, predicted a good outcome class. Individuals who were assigned a poor prognosis in at least one of the models were entered into the poor outcome class. This analysis revealed a final model that included our combined methylation- and the gene expression–based outcome predictor (HR, 0.381; 95% CI, 0.191-0.761), age (HR for a difference of 10 years, 1.23; 95% CI, 1.02-1.48), and FLT3-ITD status (HR, 1.92; 95% CI, 1.09-3.39).
Based on these findings we also evaluated whether the methylation-based prediction model can also be applied to the subset of CN-AML patients. The model did not yield statistically significant survival prediction in either univariate (HR, 1.55; 95% CI, 0.97-2.49) or multivariate (HR, 1.62; 95% CI, 0.93-2.83) analysis in this subgroup. However, when applied to only non–CN-AML cases the model gave significant results (P < .001; log-rank test) that provided prognostic information in addition to cytogenetic subgroups (supplemental Figure 6).
Discussion
Here we present results from a large-scale candidate gene DNA methylation study in adult AML patient samples. Although our results showed that clustering according to karyotype is less pronounced based on DNA methylation data compared with cluster formation observed using gene expression data, this might be secondary to a reduced number of genomic regions analyzed in our methylation analysis. Nevertheless, the unsupervised clustering showed that cytogenetic subgroups such as cases with inv(16), t(8;21), t(15;17), and t(11q23) seem to be characterized by distinct epigenetic modifications.
Furthermore, our study provided interesting novel biologic insights, for example, the differential methylation on the long arm of chromosome 17 (17q25.3). To our knowledge, DNA methylation in this region has so far not been analyzed and associated with acute leukemia. We found highly variable DNA methylation across the analyzed AML samples in this genomic region characterized by a high density of CpG islands. Our data suggest that gene expression in this entire genomic area might be down-regulated via selective DNA methylation. This mechanism of coordinate gene suppression has recently been described by Frigola et al for colon cancer samples.34 We also found several genes with aberrant methylation patterns that are involved in transcriptional regulation in AML. These findings support earlier reports that point to a role of HOX and Polycomb as target genes in leukemia.35
In addition, another major objective of our study was to determine whether patterns of DNA methylation could be used to improve patient prognostication. Using a semisupervised approach,31 we were able to build a predictive model based on quantitative methylation patterns. This model could be confirmed by predicting clinically significant outcome classes (good vs poor overall survival) in an independent test set of AML samples as well as an additional validation dataset analyzed at a later time point. Thus, our results circumstantiate a significant association of altered DNA methylation and patient outcome and thereby suggest that the integration of DNA methylation data into a clinically relevant prediction model might be possible. Microarray expression analyses measure the abundance of mRNA, a molecule that is highly susceptible to degradation, and therefore the standardization of microarray experiments is still challenging. In contrast, changes in DNA methylation are caused by the covalent addition of methyl groups to cytosine that represents a stable DNA alteration. This epigenetic modification is conserved throughout sample preparation and therefore less prone to sample preparation-related changes. Thus, a DNA-based prognostic marker might provide a significant advantage to RNA-based methods.
However, although we have shown that it is possible to get a reliable survival prediction based on a limited set of candidate genes, it nevertheless is unlikely that the current set of genes will be the most distinctive. Our results rather represent a proof of principal providing evidence that epigenetic markers can also be valuable surrogates for patient outcome in AML. Therefore, our data should encourage future genome-wide DNA methylation studies to identify the most predictive epigenetic areas in AML. Recent technologic advances combining acceptable spatial resolution with good quantitative accuracy such as microarray- or high-throughput sequencing–based approaches might be suitable for a refined genome-wide epigenetic marker discovery.36-38 These approaches might then also be able to discriminate markers that can be used for outcome prediction within the important subgroup of CN-AML. In our model, outcome prediction was not possible within the subset of CN-AML cohort, likely because the candidate genes included in the analysis were selected based on a prognostic gene expression signature designed across all cytogenetic subgroups and not CN-AML cases only.7 Thus, although our study has shown that epigenetic biomarkers have the potential to assist current patient prognostication, future studies should aim to find an optimal set of epigenetic biomarkers and evaluate their significance in a routine clinical setting before the clinical implementation will become feasible.
Furthermore, as DNA methylation can also change during the course of the disease, it is likely that integrated approaches will be superior for outcome prediction. Our study has already demonstrated that a combination of DNA methylation– and gene expression–based outcome predictors can further improve prognostication in AML by adding prognostic information to well-established markers. Ultimately, a combination of methylation and gene expression markers as well as known prognostic factors such as cytogenetics2-4 and molecular aberrations5,6 will be able to refine AML classification. However, before the successful translation from bench to bedside of such an integrative prognosticator, it will need to be evaluated within prospective clinical trials.
Finally, as epigenetic alterations are potentially reversible and demethylating agents are increasingly used for the treatment of hematologic malignancies,39,40 MALDI-TOF-MS–based DNA methylation analysis might also prove beneficial in 2 additional settings. First, quantitative methylation analysis might be helpful to identify patient subgroups likely to benefit from demethylation therapy, and second methylation markers might be used to trace therapeutic success of demethylating agents during the course of the treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the AMLSG for providing leukemia specimens.
This work was supported in part by grants from the Else-Kröner-Fresenius-Stiftung (P32/2004; L.B. and K.D.) and the Stiftung Leukämie (L.B.).
Authorship
Contribution: L.B., M.E., and D.v.d.B. designed and performed research, analyzed/interpreted data, and wrote the paper; K.D., R.F.S., and M.R.N. analyzed/interpreted data; and H.D. designed research, contributed vital reagents, and analyzed/interpreted data.
Conflict-of-interest disclosure: M.E. and D.v.d.B. are shareholders and employees of Sequenom Inc. M.R.N. is a shareholder and employee of GlaxoSmithKline. The remaining authors declare no competing financial interests.
Correspondence: Mathias Ehrich, Sequenom Inc, 3593 John Hopkins Ct, San Diego, CA, 92121; e-mail: mehrich@sequenom.com.
References
Author notes
L.B. and M.E. contributed equally to this work.

![Figure 1. Hierarchic cluster analysis of AML samples and their methylation profiles. Two-way hierarchic cluster analysis of 182 acute myeloid leukemia (AML) samples (rows) and DNA methylation of CpG units in 92 promoter regions (columns). DNA methylation values are depicted by a pseudocolor scale as indicated (methylation increases from red [nonmethylated] to white [methylated]). Samples with overall poor data quality were removed before clustering. Gray denotes data of poor quality. Samples are color-coded according to the underlying cytogenetic aberration (legend depicted top left). Most samples with an inv(16) were located in one cluster (highlighted in yellow); most t(11q23) cases grouped together (highlighted in green) and translocation t(15;17) and t(8;21) cases did also cluster mainly in one group (highlighted in pink). A small set of 3 samples splits off early in the dendrogram showing generally hypermethylated DNA (bottom 3 rows, highlighted in blue). CpG units form 2 large clusters that are characterized mainly by the level of methylation. The larger group (group 1, highlighted in pink) is characterized mainly by unmethylated CpG units, the smaller one (group 2, highlighted in yellow) mainly by methylated CpG units. In each cluster a group of CpG units can be separated that shows more variable methylation values across the AML samples (as indicated). The black bars on top of the heatmap denote the top 10% of CpG units with the highest variance across the AML samples (> 90th percentile).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/3/10.1182_blood-2009-03-211003/4/m_zh89990947040001.jpeg?Expires=1768003174&Signature=T0tN-qfhKTes~qoN0voNhzhsCfWtoZdpej~UqkC9cQNUrhMyTDcVWaP1ljoQPPuC-iKucx4GwDub0dmYtLW9~zAto5e1u9ZD3dOdA9-20-q5S7COIk0GJkcTqKWH777oGeoDXaOVvUdL94QDHQgRijmu5XJqkmRQEUP3kupA~2iD3h5n3CqZCLAV-GGgfoGNhPeP0vKKZicJeemAzOhDBCkk~LY2ulsuSpQyCZnoW76wHjFZjgciFRP9Z9-OtTnm72RDlBhBpsTfFIWO0NPoYfCd4ztBAQ2HB3f3NuJ2sxOssx-rvJvanrul3AttwcTNFmwqNvtTnrb7WdIQ-7JfFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)