Abstract
Antiphospholipid syndrome (APS) is an autoimmune thrombophilia characterized by arterial/venous thrombosis and/or pregnancy morbidity in the presence of antiphospholipid antibodies that mainly recognize β2 glycoprotein I (β2GPI). To investigate potential platelet ligands of β2GPI, platelet membrane proteins from healthy persons and patients with APS were passed through a β2GPI-affinity column. By using mass spectrometry, platelet factor 4 (PF4) appeared as the dominant β2GPI binding protein. PF4 could bind in vitro, with high-affinity, recombinant β2GPI, and the binding was abrogated by soluble β2GPI. Coprecipitation experiments further confirmed this interaction. In silico molecular docking showed that PF4 tetramers can bind 2 β2GPI molecules simultaneously. Size exclusion chromatography confirmed that anti-β2GPI antibodies selectively interact with complexes composed of (β2GPI)2–(PF4)4. In addition, as shown by the β2GPI antigenicity evaluation, the reactivity of APS sera was higher against PF4–β2GPI complex than against β2GPI alone. On complex formation, anti-β2GPI–β2GPI–PF4 significantly induced platelet p38MAPK phosphorylation and TXB2 production, mainly through F(ab′)2 fragments of antibodies. In summary, this study makes evident that β2GPI forms stable complexes with PF4, leading to the stabilization of β2GPI dimeric structure that facilitates the antibody recognition. This interaction can probably be involved in the procoagulant tendency of APS.
Introduction
Antiphospholipid syndrome (APS) is an immune-mediated, acquired thrombophilia characterized by arterial or venous thrombosis and pregnancy morbidity, in association with persistently elevated levels of antiphospholipid (aPL) antibodies.1-3 The main antigenic targets of aPL antibodies are proteins that bind to anionic phospholipids, such as β2 glycoprotein I (β2GPI)4,5 and prothrombin.6 β2GPI is a 50-kDa plasma protein composed of 5 complement control protein modules, which are termed domains I to V.7,8
Deregulated activation of endothelial cells,9,10 platelets,11-14 and monocytes15 by anti-β2GPI–β2GPI complex, disruption of fibrinolysis, and activation of coagulation cascade and complement have been proposed16-19 to explain thrombotic predisposition in APS. Because β2GPI lacks intracellular domains,7 initiation of cell signaling remains obscure. Dimers of β2GPI, mimicking the anti-β2GPI–β2GPI complexes, were shown to bind activated platelets through apolipoprotein E receptor 2′ and to induce platelet aggregation.12 The GPIbα subunit of the GPIb/IX/V receptor has been shown to bind β2GPI and to induce thromboxane B2 production in vitro.14
Our study was based on the following nil hypothesis: proteins derived from platelet membrane extracts, which bind to a β2GPI affinity column, will be those that potentially interact with β2GPI in vivo. Using appropriate techniques, we describe that platelet factor 4 (PF4), derived from platelet a-granules, interacts specifically with β2GPI.
PF4 is a member of the C-X-C chemokine family with a molecular mass of approximately 7800 Da.20 PF4 is secreted by activated platelets but has also the ability to bind to the platelet surface.20,21 PF4 has high affinity (KD = 5-20nM) for heparin and other anionic glycosaminoglycans (eg, endothelial cell surface or platelet surface GAGs).22
PF4 has a proven precoagulant role: (1) inhibition of heparin-dependent acceleration of thrombin inactivation by antithrombin,23 (2) potentiation of platelet aggregation,20 (3) aggregation defect of platelets in a PF4 knockout mice in response to low doses of thrombin,24 and (4) impaired platelet thrombus formation that was corrected by infusing PF4.24 However, PF4 appears to have also anticoagulant effects via acceleration of proteolytic generation of activated protein C, by thrombin–thrombomodulin complex.25,26
PF4–heparin complex formation is highly immunogenic and induces anti-PF4–heparin antibodies in a minority of heparin-treated patients. In the presence of these autoantibodies, the complexes activate platelets mainly through Fcγ receptors,27-29 inducing heparin-induced thrombocytopenia and thrombosis syndrome (HITT).
PF4 seems to be a common denominator in both syndromes, APS and HITT, which share similar clinical manifestations such as thrombocytopenia and thrombosis. In this report, we demonstrate that β2GPI binds PF4 and, using in silico analysis and size exclusion chromatography, we indicate the stoichiometry of this interaction. The β2GPI–PF4 complex is strongly recognized by APS patients' sera. Furthermore, there is evidence for platelet activation by anti-β2GPI–β2GPI–PF4 or by β2GPI–PF4 complexes.
Methods
Affinity purification of β2GPI and anti-β2GPI antibodies from sera of patients with APS
A combination of a cardiolipin-affinity and a mono-SHR cation exchange column (Pharmacia) was applied as previously described.30,31 The laboratory and the clinical characteristics of all the patients' sera used (n = 7) are shown in Table 1 (patients 1-7). All the eluted protein peaks were evaluated in both anticardiolipin and anti-β2GPI enzyme-linked immunoabsorbent assay (ELISA). They were also subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by silver nitrate staining. The identification of β2GPI was evaluated by Western blot analysis with mouse anti-β2GPI mAb (clone ID2; AbD Serotec).
Platelet membrane preparation/membrane proteins extraction
With the approval of the ethics commission of the National University of Athens, fresh platelet-rich plasma was isolated from 3 healthy donors (HDs) and 7 patients with APS (Table 1; patients 1-7) by platelet apheresis. Informed consent was obtained in accordance with the Declaration of Helsinki. HDs had not taken any medication during the previous 10 days. Platelet membranes were purified by using the protocol of Moebius et al,32 based on ultracentrifuges in a discontinuous sorbitol gradient.
Affinity purification of platelet membrane proteins bound to β2GPI–CNBr–sepharose column
Affinity-purified β2GPI from APS sera was coupled to CNBr-activated Sepharose 4B (GE Healthcare). The column was equilibrated with PBS at a flow rate 0.35 mL/min. Each platelet membrane protein extract in a final concentration of 0.5 mg/mL (n = 10) was loaded onto the column and was run through β2GPI–sepharose at a flow rate 0.10 mL/min. After washing with 5 column volumes with PBS, the captured proteins were eluted, using 0.1M glycine-HCl, pH 2.7. The eluents were neutralized by using 0.5M NaOH in PBS and dialyzed overnight versus PBS. Eluted proteins were analyzed by SDS-PAGE on a 12% gel. Gels were stained either with silver nitrate or with Coomassie Brilliant Blue R250, and the protein bands were analyzed by mass spectrometry (MS).
Nano-HPLC/MS for protein identification
High-performance liquid chromatography (HPLC)/MS/MS analysis was performed in a mass spectrometer equipped with a nano electrospray ionization source. The electrospray source was coupled online with an LC/HPLC system. The tryptic peptides were separated on a reversed-phase C18 column and electrosprayed into the LCQ mass spectrometer. Proteins were identified with the use of TurboSEQUEST (Thermo Scientific), which uses the MS and MS/MS spectra of peptide ions to search against the IPI (International Protein Index) human protein database.
Biotinylation of proteins
Recombinant human PF4 (R&D Systems), recombinant human β2GPI (Fitzgerald International), and BSA were biotinylated with the use of Sulfo-NHS-Biotin (Pierce) according to the manufacturer's instructions. The excess nonreacted biotin was removed with overnight dialysis versus PBS. Biotinylated PF4 (biot-PF4) and biot-β2GPI was tested by Western blot with the use of both monoclonal anti-PF4 (clone 170106; R&D Systems) and monoclonal anti-β2GPI, respectively, or with AP-streptavidin.
Direct binding of biot-PF4 to β2GPI coated onto microtiter plates
Direct binding assays were performed as previously described,14 with some modification, by using MaxiSorp immunoplates (Nalge Nunc International). Briefly, wells were coated with 50 μL of recombinant β2GPI, β-lactoglobulin, or BSA as negative controls (10 μg/mL) in 50mM carbonate-bicarbonate buffer (pH 9.6) and were incubated overnight at 4°C. After 5 times washing with PBS/0.1% Tween 20 (PBST), wells were blocked with 2% BSA/PBST for 2 hours at room temperature (RT). They were washed 5 times and were then incubated overnight at 4°C with 50 μL of various concentrations of biot-PF4 or biot-BSA (0-5 μg/mL) in 1% BSA/PBS. After 5 additional washes, the plates were incubated with AP-streptavidin (dilution 1:1000) for 1 hour. After the addition of p-nitrophenyl-phosphate substrate (pNPP), the optical density (OD) was measured at 410 nm.
Competitive (homologous) inhibition of biot-PF4 binding to β2GPI coated onto microtiter plates, after preincubation with recombinant β2GPI
Maxisorp immunoplates were coated overnight with 50 μL of recombinant β2GPI (10 μg/mL) in carbonate-bicarbonate buffer. The plates were washed, blocked, and then washed again as before. Preincubated biot-PF4 (1 μg/mL) with increasing concentrations of β2GPI or β-lactoglobulin (0-13.5 μg/mL) for 30 minutes at 37°C were then added to the wells and incubated overnight at 4°C. The wells were washed and incubated with AP-streptavidin. After the addition of pNPP substrate solution, the OD was measured.
NeutrAvidin-agarose precipitation
Biot-β2GPI or non–biot-β2GPI, as negative control, was incubated with PF4 in equal molarities (0.1 and 0.2mM in PBS) for 30 minutes at 37°C. After incubation, 10 μL NeutrAvidin-agarose beads (ThermoScientific), diluted 1:1 in PBS/0.1% SDS, were added to the mixtures and further incubated for 1 hour at RT with rotation. The avidin-agarose beads were washed 5 times with PBS/0.1%SDS, and the resin-bound complexes were eluted from agarose with reducing 2-fold Laemmli buffer. Eluted proteins were subjected to SDS-PAGE on a 13.5% gel and analyzed by immunoblot. The membranes were blocked with 5% nonfat milk-TBST (0.1%) for 2 hours and then incubated with monoclonal anti-PF4 (0.5 μg/mL) for 1 hour at RT. Membranes were washed and were then incubated with HRP-conjugated goat anti–mouse IgG antibody (Jackson ImmunoResearch) diluted 1:2500 in blocking buffer. Peroxidase reaction was visualized with the ECL Plus system (Pharmacia Biotech).
Study of antigenicity of β2GPI–PF4 complex
The antigenicity of β2GPI–PF4 complex in comparison with the antigenicity of β2GPI alone was tested with a modified anti-β2GPI ELISA.33 Briefly, polystyrene microtiter plates were coated overnight with 50 μL of recombinant β2GPI (10 μg/mL). The plates were washed, blocked, and washed again as described before. They were then incubated overnight either with 50 μL of PF4 (2.0 μg/mL in blocking buffer/2% BSA/PBS) or with blocking buffer alone. After extensive washing, the wells were incubated with sera from patients with APS (dilution 1:1000) in 2% BSA/PBS for 2 hours at RT. Afterward, the plates were washed and incubated with AP-conjugated goat anti–human IgG antibody (Jackson ImmunoResearch) diluted 1:2000 in blocking buffer. After the addition of pNPP substrate, the OD was measured.
Docking procedure and interaction analysis
The crystal structures of human β2GPI and PF4 tetramer were imported to ICM-Pro software (Molsoft Inc). To investigate the possible orientation of PF4 tetramer on the β2GPI surface, docking simulations were performed. Four different properties were taken into account (van der Waals, electrostatic, hydrogen bonding, and hydrophobic potential).34 The PF4 tetramer was represented explicitly, and full flexibility was allowed during the docking around the rotatable bonds. The Merck molecular force field parameters were assigned to atoms of both proteins.35 The interaction energy of PF4 with β2GPI was calculated,36 and the final energy value of every conformation was recorded. The energy of every new trial conformation was then compared with the ones obtained in previous iterations and ranked accordingly. Further refinement of the best conformations was carried out, where the regions of the β2GPI protein, interacting with the PF4 and the PF4 molecule itself, were computed explicitly with full side chain flexibility.
Contact areas were calculated as the difference between solvent-exposed areas of PF4 tetramer and β2GPI alone and solvent-exposed area of the PF4–β2GPI complex. Electrostatic analysis was performed with the use of the REBEL algorithm.37 The binding energy was evaluated by a previously described approximation.38
Determination of protein complexes by high-performance size-exclusion chromatography
A Superose 6 10/300 GL column (GE Healthcare) was connected to an ÄKTA HPLC system and was equilibrated according to the manufacturer's instructions. Mass standards were from a commercial molecular weight calibration kit (GE Healthcare). PF4 alone (0.13 mg/mL in PBS) and β2GPI alone (0.4 mg/mL in PBS) were injected individually to the column, using PBS as eluent at a flow rate 0.5 mL/min. Subsequently, preincubated complexes, for 30 minutes at 37°C, of PF4 plus β2GPI (0.13 and 0.4 mg/mL, respectively) and PF4 plus β2GPI plus anti-β2GPI (0.13 and 0.4 and 0.6 mg/mL, respectively) were injected to the column as previously. Size-exclusion chromatography (SEC)–purified fractions were collected, concentrated in a SpeedVac, and analyzed by immunoblot with anti-PF4 mAb.
Measurement of platelet thromboxane B2 metabolite production as an indication of platelet activation
This protocol was based on described experiments from Shi et al14 with several modifications. Polystyrene microtiter plates were coated with 200 μL protein-A (100 μg/mL) in carbonate-bicarbonate buffer and incubated for 1 hour at RT. After washing 4 times with PBST, the resident binding sites were blocked with 2% BSA/PBST for 2 hours. The wells were then incubated for 1 hour with 100 μL of anti-β2GPI or isotype control IgG1k (1 μg/well) in 42mM calcium free-HEPES-Tyrode (HT) buffer (pH 7.3). After 4 times washing, wells were incubated for 1 hour with 100 μL of β2GPI or BSA (0.7 μg/well). Finally, the plate was washed 4 times, and 100 μL of PF4 (0.5 μg/well) or HT buffer alone was added to relevant wells and incubated overnight at 4°C.
The next day, fresh platelets were isolated from the venous blood of HDs as described before12 and were resuspended in calcium-free HT buffer (2 × 108 platelets/mL). Platelets (500 μL) with 500 μL of calcium-free HT and 250 μL of buffer containing 20mM CaCl2 were dispensed into polypropylene tubes, and 50 μL of this platelet preparation was added to the relevant ELISA well (after washing the plate 4 times with PBST).
The plates were then shaken for 1 minute and incubated for 25 minutes at 37°C. Therefore, thromboxane A2 (TXB2) production was terminated with 50 μL of indomethacin (0.3mM), and the plates were centrifuged at 3000 rpm for 5 minutes. TXB2 levels were measured in supernatants with the Amersham Thromboxane B2 Biotrak Assay.
Measurement of platelet TXB2 production, in the absence of anti-β2GPI antibodies
Wells were coated with 100 μL β2GPI (1 μg/well) for 1 hour at RT. After washing and blocking, the wells were treated as above. Fresh isolated platelets were then added, and TXB2 levels were measured as described in “Measurement of platelet thromboxane B2 metabolite production as an indication of platelet activation.”
Measurement of TXB2 production from platelets primed with suboptimal doses of thrombin
The production of TXB2 induced by anti-β2GPI, β2GPI, and PF4 was also assessed in platelets treated with low doses of thrombin. Thus, 50-μL aliquots of washed platelets were incubated in microtubes for 10 minutes at 37°C with 0.005 U/mL thrombin, before their application to the relevant ELISA wells, as described in the previous 2 sections. Platelets used as positive control, treated with 1 U/mL thrombin, and platelets used as negative control, treated with 0.005 U/mL thrombin, were added into 2 empty ELISA wells.
Immunoblot analysis for phosphorylation of p38MAPK in platelets
On the basis of previously described protocols,39 100-μL aliquots of washed platelets in HT buffer were incubated for 30 minutes at 37°C with 3 μg/mL anti-β2GPI (or isotype control) and 2 μg/mL β2GPI (or BSA) in the presence or absence of 1.5 μg/mL PF4. Platelets were then incubated with 0.005 U/mL thrombin for 10 minutes at 37°C. As positive control, platelets treated with 1 U/mL thrombin in HT buffer were used.
Platelets were lysed with 30 μL of RIPA buffer plus protease inhibitors and centrifuged at 3000g for 15 minutes. Lysates were heated for 5 minutes, and 5.5 μg of total protein from each sample was subjected to SDS-PAGE in 12% gel. After electrophoresis and transfer to PVDF, the membranes were blocked and then incubated with phospho-p38MAPK (Thr180/Tyr182) rabbit anti–human monoclonal antibody or p38MAPK rabbit polyclonal antibody (Cell Signaling). Afterward, membranes were incubated with HRP-conjugated goat anti–rabbit IgG, and peroxidase reaction was visualized by the ECL Plus system. The bands were quantitated with the use of ImageJ 1.42q software (National Institutes of Health).
Determination of the principal antibody fragment [Fc portion or F(ab′)2 fragment] for anti-β2GPI–induced effects on platelets
The platelet receptor for the Fc domain of IgGs (FcγRIIa) was blocked by anti-FcγRII mAb IV.3 (StemCell Technologies). Subsequently, the assays for the phosphorylation of p38MAPK and the production of TXB2 were repeated as described. Briefly, platelets in HT buffer were incubated with 10 μg/mL mAb IV.3 for 10 minutes at 37°C and were then treated with subactivating doses of thrombin. After that, 50 or 100 μL of platelets was added to the relevant ELISA wells or microtubes, respectively, which contained, depending on the experiment, immobilized protein A and either anti-β2GPI antibody or isotype control, plus β2GPI or BSA, with or without PF4. Phosphorylation of p38MAPK and measurement of TXB2 were evaluated as described earlier.
Statistical analysis
KD value was estimated by using the saturation binding analysis module of GraphPad Prism (GraphPad Software Inc). Data from the antigenicity study were analyzed by the Student paired t test (2-tailed). Data from TXB2 measurements were analyzed by the Mann-Whitney U test. P values less than .05 were considered significant.
Results
Identification of purified β2GPI and anti-β2GPI antibodies
All the eluted protein peaks, from the affinity purification, represented aPL antibodies (anticardiolipin and anti-β2GPI) except from the last peak, which contained only autologous β2GPI. The purity of β2GPI was confirmed by silver staining, which showed only one band. This band was confirmed to be β2GPI by Western blotting with an anti-β2GPI mAb. In addition, purified anti-β2GPI antibodies recognized recombinant β2GPI, using an anti-β2GPI ELISA, as previously described (data not shown).
β2GPI-coupled to CNBr-activated sepharose selectively binds PF4 (CXCL4) or PF4var (CXCL4L1) from 10 purified platelet membrane protein preparations
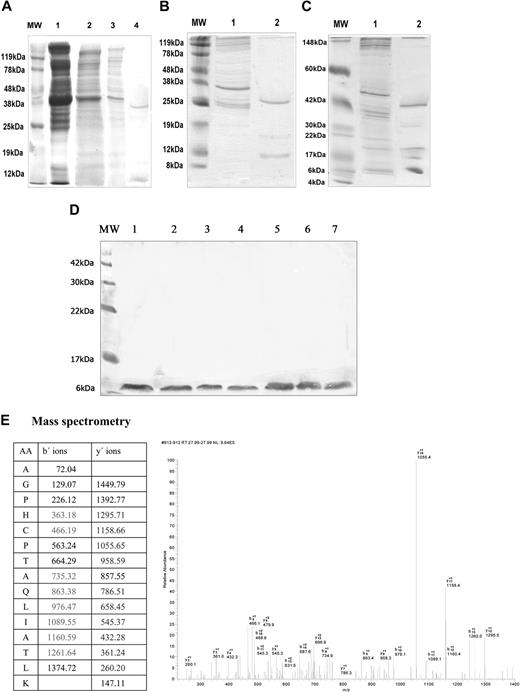
Platelet membrane protein extracts from 3 HDs (HD1, HD2, and HD3) and 7 patients with APS (APS1-APS7) were run through a β2GPI–sepharose affinity column. The eluted proteins were analyzed by SDS-PAGE and silver staining. As shown by MS analysis, the eluents derived from the HD1 platelet preparations (Figure 1A) consisted of a 39-kDa and an approximately 8-kDa protein band, representing actin and PF4/PF4 variant (PF4/PF4var), respectively. MS could not differentiate between PF4var and PF4, because the 2 proteins differ in only 3 amino acids in the C terminal α-helix of the mature protein. The eluents from HD2 (Figure 1B) consisted of a 30-kDa and an approximately 9-kDa band, representing tropomyosin-1a and PF4/PF4var, respectively. The eluents from HD3 (Figure 1C) consisted of protein bands of 40 kDa, 30 kDa, 19 kDa, 15 kDa, and 8 kDa, representing actin, tropomyosin-1a, myosin regulatory light chain, myosin light chain, and PF4/PF4var, respectively. The eluents from the platelet preparations of patients with APS (Figure 1D) consisted of a single 8-kDa protein, corresponding to PF4/PF4var. The results from MS regarding PF4/PF4var are presented in Figure 1E.
SDS-PAGE and Coomassie (or silver) stain of unbound and eluted proteins, from β2GPI affinity column, derived from different platelet membrane preparations. (A) Proteins from HD1 platelet preparation; MW indicates molecular weight marker; lane 1, complete platelet lysate; lane 2, platelet membrane protein extract; lane 3, unbound proteins; lane 4, eluted proteins (39 kDa and approximately 8 kDa). (B) Proteins from HD2 platelet preparation; MW indicates molecular weight marker; lane 1, unbound proteins; lane 2, eluted proteins (30 kDa and approximately 9 kDa). (C) Proteins from HD3 platelet preparation; MW indicates molecular weight marker; lane 1, unbound proteins; lane 2, eluted proteins (40 kDa, 30 kDa, 19 kDa, 15 kDa, 8 kDa). (D) Proteins from platelet preparations from patients with APS; MW indicates molecular weight marker; lanes 1 to 7, single eluted protein (8 kDa) from patients with APS 1 to 7. (E) MS/MS analysis of the 8-kDa protein eluted from β2GPI affinity column. Generated fragments of precursor ion m/z 761.35 correspond to the peptide AGPHCPTAQLIATLK. Experimentally detected y- and b-ions are marked in bold (color).
SDS-PAGE and Coomassie (or silver) stain of unbound and eluted proteins, from β2GPI affinity column, derived from different platelet membrane preparations. (A) Proteins from HD1 platelet preparation; MW indicates molecular weight marker; lane 1, complete platelet lysate; lane 2, platelet membrane protein extract; lane 3, unbound proteins; lane 4, eluted proteins (39 kDa and approximately 8 kDa). (B) Proteins from HD2 platelet preparation; MW indicates molecular weight marker; lane 1, unbound proteins; lane 2, eluted proteins (30 kDa and approximately 9 kDa). (C) Proteins from HD3 platelet preparation; MW indicates molecular weight marker; lane 1, unbound proteins; lane 2, eluted proteins (40 kDa, 30 kDa, 19 kDa, 15 kDa, 8 kDa). (D) Proteins from platelet preparations from patients with APS; MW indicates molecular weight marker; lanes 1 to 7, single eluted protein (8 kDa) from patients with APS 1 to 7. (E) MS/MS analysis of the 8-kDa protein eluted from β2GPI affinity column. Generated fragments of precursor ion m/z 761.35 correspond to the peptide AGPHCPTAQLIATLK. Experimentally detected y- and b-ions are marked in bold (color).
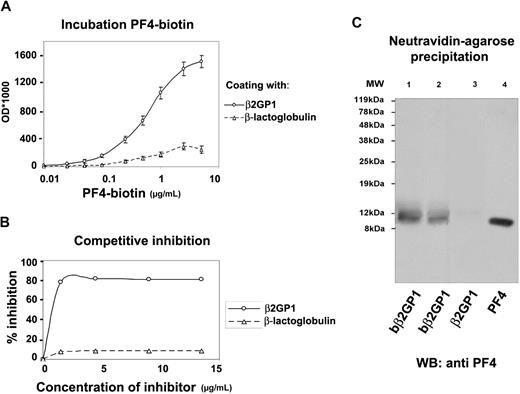
Recombinant PF4 directly binds to recombinant β2GPI in a dose-dependent manner
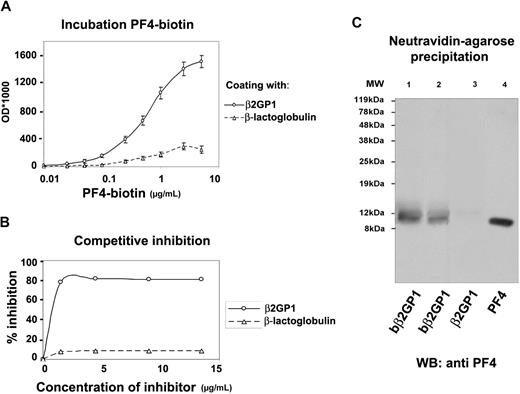
To investigate the binding capacity of PF4 to β2GPI, direct binding assays were performed. Fixed concentrations (10 μg/mL or 0.2mM) of β2GPI and β-lactoglobulin or BSA (as negative controls) were incubated with increasing concentrations (0-5 μg/mL or 0-625nM) of biot-PF4. The results (Figure 2A) showed that PF4 bound to recombinant β2GPI in a dose-dependent manner with a KD of 105.3nM (± 9.8nM). Taking into account that PF4 tetramers tend to interact simultaneously with 2 β2GPI molecules (see “Docking of PF4 tetramer to β2GPI”), the KD of the interaction can further reduce to 53nM (± 4.9nM). On the contrary, biot-PF4 did not bind to irrelative proteins such as β-lactoglobulin or BSA, suggesting the selectivity of the interaction.
In vitro binding assays in solid phase and in solution. (A) Saturation binding of biotinylated PF4 (biot-PF4) to β2GPI; Immunoplates coated with a constant concentration (10 μg/mL) of recombinant β2GPI or β-lactoglobulin were incubated with various concentrations (0-5 μg/mL) of biot-PF4. After the addition of AP-streptavidin, the OD was measured at 410 nm. Values are the mean ± SD. (B) Homologous inhibition of biot-PF4 binding to β2GPI-immobilized onto microtiter plates, after preincubation with recombinant β2GPI. Preincubated biot-PF4 (1 μg/mL) with increasing concentrations of soluble β2GPI or β-lactoglobulin (0-13.5 μg/mL) for 30 minutes at 37°C was added to immobilized β2GPI onto microtiter plates and incubated overnight at 4°C. After AP-streptavidin and pNPP substrate, OD values were measured at 410 nm and converted to percentage of inhibition. (C) Precipitation with NeutrAvidin-agarose beads and Wester blot analysis with monoclonal anti-PF4 antibody. Biotinylated recombinant β2GPI (biot-β2GPI) and nonbiotinylated β2GPI (β2GPI), used as negative control, were incubated with PF4 in equal molarities (0.1mM and 0.2mM). After incubation, avidin-agarose beads were added to the mixtures, and the resin-bound complexes were eluted with boiling. Eluted proteins were analyzed by SDS-PAGE and were immunoblotted with monoclonal anti-PF4. MW indicates molecular weight marker; lane 1, eluent from biot-β2GPI–PF4–resin sample (0.2mM); lane 2, eluent from biot-β2GPI–PF4–resin sample (0.1mM); lane 3 (negative control), eluent from β2GPI–PF4–resin sample (0.2mM); lane 4, recombinant PF4.
In vitro binding assays in solid phase and in solution. (A) Saturation binding of biotinylated PF4 (biot-PF4) to β2GPI; Immunoplates coated with a constant concentration (10 μg/mL) of recombinant β2GPI or β-lactoglobulin were incubated with various concentrations (0-5 μg/mL) of biot-PF4. After the addition of AP-streptavidin, the OD was measured at 410 nm. Values are the mean ± SD. (B) Homologous inhibition of biot-PF4 binding to β2GPI-immobilized onto microtiter plates, after preincubation with recombinant β2GPI. Preincubated biot-PF4 (1 μg/mL) with increasing concentrations of soluble β2GPI or β-lactoglobulin (0-13.5 μg/mL) for 30 minutes at 37°C was added to immobilized β2GPI onto microtiter plates and incubated overnight at 4°C. After AP-streptavidin and pNPP substrate, OD values were measured at 410 nm and converted to percentage of inhibition. (C) Precipitation with NeutrAvidin-agarose beads and Wester blot analysis with monoclonal anti-PF4 antibody. Biotinylated recombinant β2GPI (biot-β2GPI) and nonbiotinylated β2GPI (β2GPI), used as negative control, were incubated with PF4 in equal molarities (0.1mM and 0.2mM). After incubation, avidin-agarose beads were added to the mixtures, and the resin-bound complexes were eluted with boiling. Eluted proteins were analyzed by SDS-PAGE and were immunoblotted with monoclonal anti-PF4. MW indicates molecular weight marker; lane 1, eluent from biot-β2GPI–PF4–resin sample (0.2mM); lane 2, eluent from biot-β2GPI–PF4–resin sample (0.1mM); lane 3 (negative control), eluent from β2GPI–PF4–resin sample (0.2mM); lane 4, recombinant PF4.
Competitive inhibition experiments
To study the specificity of the β2GPI–PF4 interaction, homologous inhibition experiments were performed. In particular, preincubated biot-PF4 (1 μg/mL) with increasing concentrations of β2GPI or β-lactoglobulin (0-13.5 μg/mL) was tested for its binding capacity to β2GPI immobilized onto solid phase. In fact, as shown in Figure 2B, the level of inhibition by β2GPI reaches up to approximately 80%. This result strongly suggests that PF4 interacts with soluble β2GPI, abrogating its binding to β2GPI-coated plates.
NeutrAvidin-agarose precipitates biot-β2GPI–PF4 complexes formed in solution
Equal molarities of biot-β2GPI (or nonbiotinylated β2GPI as negative control) and PF4 were incubated with NeutrAvidin-agarose resin, and the resin-bound protein complex was immunoblotted by monoclonal anti-PF4. As shown in Figure 2C, PF4 is detected in the eluents deriving from biot-β2GPI–PF4–resin samples (lanes 1-2), whereas it is absent in the eluent from β2GPI–PF4–resin sample (lane 3 negative control). This result shows that β2GPI binds PF4 in solution, and this interaction is specific because PF4 is not detected in the negative control.
Interaction between β2GPI and PF4 increases the antigenicity of β2GPI
Because PF4–heparin complexes exhibit high antigenicity, we sought to examine whether PF4–β2GPI complexes are, in a similar manner, more antigenic than β2GPI alone. A modified anti-β2GPI ELISA was performed, and the OD values of sera from patients with APS (n = 10; see Table 1, patients 1-10), after binding to β2GPI alone or to β2GPI–PF4 complex, were compared. All the APS serum samples have been tested for the presence of anti-PF4 antibodies, using an anti-PF4 ELISA protocol, and it was found that none of the sera contained anti-PF4 antibodies. As shown in Figure 3, the reactivity of APS sera was higher against PF4–β2GPI complex than against β2GPI alone. In particular, sera from patients with APS recognized PF4–β2GPI complex with a mean OD increase of approximately 32%, compared with β2GPI alone (t = 8.540; P < .001). Thus, the formation of complexes not only retains but actually enhances the antigenicity of β2GPI alone.
Study of antigenicity of β2GPI–PF4 complex. Coated plates either with β2GPI alone (10 μg/mL) or with β2GPI + PF4 (10 μg/mL and 2 μg/mL, respectively) were incubated with sera from patients with APS (n = 10). After the addition of secondary antibody, OD values were measured at 410 nm.
Study of antigenicity of β2GPI–PF4 complex. Coated plates either with β2GPI alone (10 μg/mL) or with β2GPI + PF4 (10 μg/mL and 2 μg/mL, respectively) were incubated with sera from patients with APS (n = 10). After the addition of secondary antibody, OD values were measured at 410 nm.
Docking of PF4 tetramer to β2GPI
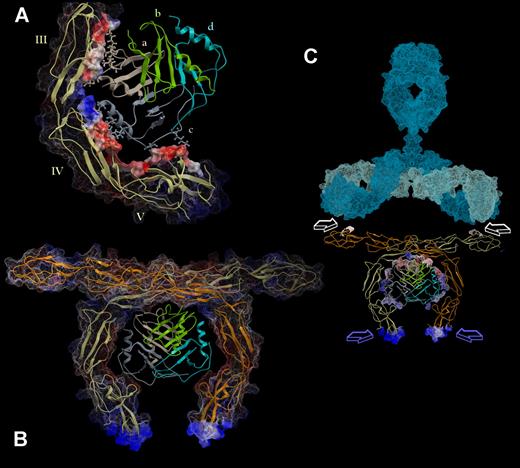
Docking solutions suggested that PF4 binds to negatively charged patches of domains III to V in the inner curve of β2GPI (Figure 4A-B). In this regard, the positively charged surface of PF4 tetramer interacts with the negatively charged regions of β2GPI domains III to V. In addition, hydrogen bonds are formed between residues R148, E225, E226, K231, and E259 of β2GPI and residues D7, K14, R22, R20, K66, Q56, and K14 of PF4, as shown in Figure 4C.
Structural analysis of β2GPI–PF4 complex. This model was extracted from the dynamics docking simulation and then extensively minimized. Among the 70 best ranked structures extracted from the simulation trajectory, this model exhibited the most favorable binding energy. (A) The molecular surface representation of β2GPI is colored by its electrostatic potential (red indicates electronegative; blue, electropositive; gray, hydrophobic), whereas PF4 is shown in ribbons colored by its secondary structure. (B) The interaction interface of PF4 is colored by its electrostatic potential. It possesses electrostatic charges complementary to that of the interaction surface of β2GPI (shown as orange ribbons). (C) Representation of the hydrogen bonds formed between domains III, IV, and V of β2GPI (shown as green ribbons) and PF4 (shown as yellow ribbons). The residues involved in these interactions are displayed as CPK-colored ball/stick and labeled in the color of the corresponding chain. (D) Steric clash analysis of the N-glycans at residues Asn143, Asn174, and Asn234 of β2GPI (as determined by x-ray crystallography in PDB structure 1QUB) and PF4. The oligosaccharides are shown as blue sticks with dotted surface, whereas PF4 is displayed as a yellow ribbon. Only a bulky-complex sugar at Asn164 could interfere in ligand binding.
Structural analysis of β2GPI–PF4 complex. This model was extracted from the dynamics docking simulation and then extensively minimized. Among the 70 best ranked structures extracted from the simulation trajectory, this model exhibited the most favorable binding energy. (A) The molecular surface representation of β2GPI is colored by its electrostatic potential (red indicates electronegative; blue, electropositive; gray, hydrophobic), whereas PF4 is shown in ribbons colored by its secondary structure. (B) The interaction interface of PF4 is colored by its electrostatic potential. It possesses electrostatic charges complementary to that of the interaction surface of β2GPI (shown as orange ribbons). (C) Representation of the hydrogen bonds formed between domains III, IV, and V of β2GPI (shown as green ribbons) and PF4 (shown as yellow ribbons). The residues involved in these interactions are displayed as CPK-colored ball/stick and labeled in the color of the corresponding chain. (D) Steric clash analysis of the N-glycans at residues Asn143, Asn174, and Asn234 of β2GPI (as determined by x-ray crystallography in PDB structure 1QUB) and PF4. The oligosaccharides are shown as blue sticks with dotted surface, whereas PF4 is displayed as a yellow ribbon. Only a bulky-complex sugar at Asn164 could interfere in ligand binding.
Overall, the interaction interface in β2GPI is formed mainly by residues Arg135, Val136, Arg148, Thr150, Val152, and Gly163 of domain III; residues Pro224, Glu225, Glu226, Glu228, and Ser236 of domain IV; and residues Gln257, Gly258, and Glu259 of domain V. In PF4, the interaction area includes residues Leu8, Val13, Lys14, and Gln56 of chain A and residues Lys14, Ser17, Arg22, Lys66, and Glu69 of chain C of the tetramer (Figure 5A). Notably, no residues of chains B and D of the tetramer are used for the binding to β2GPI. Therefore, the residues of chains B and D that are homologous to the residues involved in PF4–β2GPI interaction are free for binding to another β2GPI molecule, allowing its dimerization. As shown in Figure 5B, this PF4-mediated dimerization of β2GPI can occur without any steric clashes. On the tripartite complex formation the high antigenic regions of domain I are placed in a way that allows their recognition by antibodies (Figure 5C).
PF4 can potentially dimerize β2GPI, favoring the recognition of the complex by autoantibodies. (A) The binding of PF4 to β2GPI involves exclusively residues of chains a and c (shown as ball/sticks in the color of their parent chain). The molecular interaction interface of β2GPI is shown as transparent surface colored by its electrostatic potential. Markedly, no residues belonging to chains b (green) and d (cyan) are used for this interaction. (B) Chains b and d of PF4 tetramer can bind an additional (to that interacting with chains a and c) β2GPI molecule, leading to dimerization of the latter. Each β2GPI monomer is shown as ribbon in different color (orange and yellow). Its molecular surface is colored according to its electrostatic potential and displayed in wire representation (for the membrane binding regions) and as dotted surface for the rest of the molecule. (C) A plausible model for the mode of antibody binding to β2GPI dimer, which is formed on the interaction with the PF4 tetramer. The immunoglobulin structure, shown in light blue (heavy chains) and cyan (light chains), was adapted from the published structure by Padlan40 (based on PDB structures 2IG2 and 1FC2). White arrows indicate the epitopes that thrombosis-associated antibodies recognize on domain I of β2GPI.41 Notably, these epitopes are arranged in a geometry that is compatible with their readily and bivalent recognition by antibody's antigen-binding sites. Blue arrows indicate the positively charged regions in domain V of β2GPI, which are involved in interaction with negatively charged membranes. The interaction interface of PF4 is displayed as dots and is colored by its electrostatic potential.
PF4 can potentially dimerize β2GPI, favoring the recognition of the complex by autoantibodies. (A) The binding of PF4 to β2GPI involves exclusively residues of chains a and c (shown as ball/sticks in the color of their parent chain). The molecular interaction interface of β2GPI is shown as transparent surface colored by its electrostatic potential. Markedly, no residues belonging to chains b (green) and d (cyan) are used for this interaction. (B) Chains b and d of PF4 tetramer can bind an additional (to that interacting with chains a and c) β2GPI molecule, leading to dimerization of the latter. Each β2GPI monomer is shown as ribbon in different color (orange and yellow). Its molecular surface is colored according to its electrostatic potential and displayed in wire representation (for the membrane binding regions) and as dotted surface for the rest of the molecule. (C) A plausible model for the mode of antibody binding to β2GPI dimer, which is formed on the interaction with the PF4 tetramer. The immunoglobulin structure, shown in light blue (heavy chains) and cyan (light chains), was adapted from the published structure by Padlan40 (based on PDB structures 2IG2 and 1FC2). White arrows indicate the epitopes that thrombosis-associated antibodies recognize on domain I of β2GPI.41 Notably, these epitopes are arranged in a geometry that is compatible with their readily and bivalent recognition by antibody's antigen-binding sites. Blue arrows indicate the positively charged regions in domain V of β2GPI, which are involved in interaction with negatively charged membranes. The interaction interface of PF4 is displayed as dots and is colored by its electrostatic potential.
As indicated by Bouma et al,7 human β2GPI contains 4 N-glycosylation sites. Clearly, glycans at Asn143, Asn174, and Asn234 of β2GPI do not affect the binding of PF4 (Figure 4D). Although the oligosaccharide at Asn164, as determined by x-ray crystallography, is extended in an angle that allows the interaction with PF4, a much larger complex glycan at this site could inhibit sterically the binding of PF4 (Figure 4D).
SEC showed the stoichiometry of PF4–β2GPI and PF4–β2GPI–/anti-β2GPI complexes in vitro
The composition of a protein complex can be calculated by measurement of the complex's mass via SEC. PF4 injected individually onto a SEC column was eluted at a protein peak of 18.3 mL, corresponding to a MW of 33 kDa, according to the MW standards (Figure 6A). This molecular mass fits well to the theoretically calculated MW of PF4 tetramers (≈ 32 kDa). Likewise, β2GPI alone is eluted at 14.7 mL, corresponding to a MW of 50 kDa (Figure 6B). Preincubated mixture of β2GPI plus PF4, of equal molarity (3.9mM), is eluted as a higher-molecular-weight peak at 12.6 mL, corresponding to an 83-kDa complex, compatible with the sum of its 33-kDa (PF4 tetramer) and 5-kDa (β2GPI) components (Figure 6C). This result provides a direct evidence of the PF4–β2GPI complex formation. A minor peak, which appeared in the elution volume of PF4, represents the molecular excess of PF4 in the solution. The effect of anti-β2GPI antibodies in β2GPI–PF4 complex formation was also investigated. In this regard, the mixture of PF4 plus β2GPI plus anti-β2GPI produced 3 peaks that appeared at elution volumes 5.1 mL (peak 1), 9.7 mL (peak 2), and 12.6 mL (peak 3) (Figure 6D). These volumes correspond to MW 290, 134, and 83 kDa, respectively. All 3 peaks contained PF4, because it was assessed by immunoblot with a specific anti-PF4 monoclonal antibody (Figure 6E), contrary to mouse IgG, which was found to be present only in peak 1 (data not shown). From these data, it can be concluded that peak 1 represents the complex of anti-β2GPI IgG with (β2GPI)2–(PF4)4 (theoretical MW = 282 kDa), whereas peaks 2 and 3 represent complexes of (β2GPI)2–(PF4)4 (theoretical MW = 132 kDa) and β2GPI–(PF4)4 (theoretical MW = 82 kDa), respectively, because of the molecular excess of β2GPI and PF4. Taken together, SEC analysis directly shows that the formation of different complexes composed of 1 or 2 molecules of β2GPI and 1 PF4 tetramer. Moreover, anti-β2GPI antibody was found to preferentially interact with the higher MW (β2GPI)2–(PF4)4 complex.
HPLC/SEC. PF4 alone (A) and β2GPI alone (B) were injected individually to a Superose 6 10/300 GL column connected to an ÄKTA HPLC system, at a flow rate 0.5 mL/min. Preincubated complexes (30 minutes at 37°C) consistent with PF4 + β2GPI (C) and PF4 + β2GPI + anti-β2GPI (D) were injected into the column as previously. (E) SEC-purified fractions from peaks 1 to 5 were concentrated in a SpeedVac and were analyzed by immunoblot with anti-PF4 mAb.
HPLC/SEC. PF4 alone (A) and β2GPI alone (B) were injected individually to a Superose 6 10/300 GL column connected to an ÄKTA HPLC system, at a flow rate 0.5 mL/min. Preincubated complexes (30 minutes at 37°C) consistent with PF4 + β2GPI (C) and PF4 + β2GPI + anti-β2GPI (D) were injected into the column as previously. (E) SEC-purified fractions from peaks 1 to 5 were concentrated in a SpeedVac and were analyzed by immunoblot with anti-PF4 mAb.
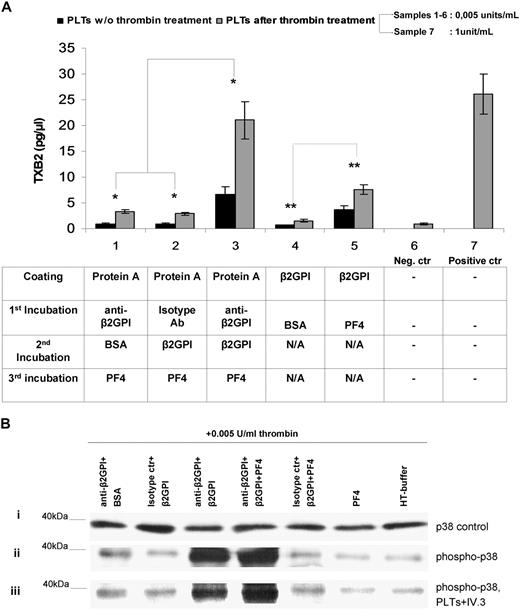
Immobilized complexes comprising anti-β2GPI antibodies, β2GPI and PF4, induce platelet activation and production of soluble TXB2
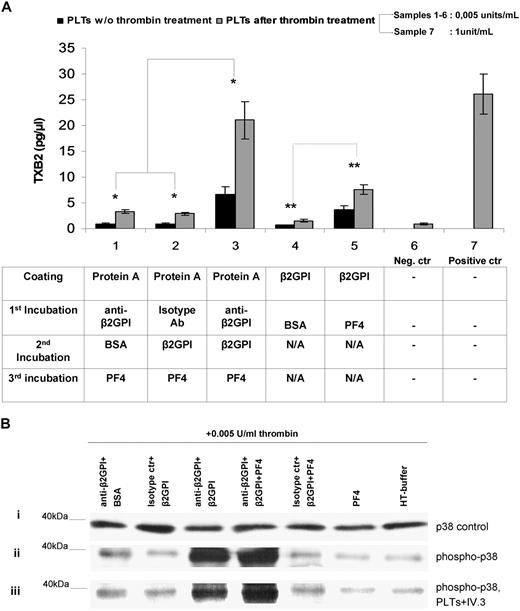
Platelets from HDs, exposed to immobilized anti-β2GPI plus β2GPI plus PF4 complexes, display significantly more TXB2 metabolite production (6.6 ± 1.45 pg/μL [platelets without thrombin priming] and 21.07 ± 3.58 pg/μL [platelets after thrombin priming]; mean ± SD; n = 7) compared with platelets exposed either to anti-β2GPI plus BSA plus PF4 (0.9 ± 0.16 pg/μL [platelets without thrombin] and 3.4 ± 0.37 pg/μL [platelets with thrombin]; n = 7) or to isotype control plus β2GPI plus PF4 (0.87 ± 0.20 pg/μL [platelets without thrombin] and 2.9 ± 0.3 pg/μL [platelets with thrombin] [n = 7; P < .005]) (Figure 7A). Moreover, as shown in bars no. 4 and no. 5 (Figure 7A), TXB2 produced by platelets exposed to immobilized β2GPI alone is notably low (0.65 ± 0.097 pg/μL [platelets without thrombin priming] and 1.6 ± 0.2 pg/μL [platelets with thrombin]), whereas the immobilized complex β2GPI plus PF4 significantly increases the production of TXB2 levels nearly 5.5-fold and 4.75-fold, respectively, in intact and thrombin-primed platelets (3.6 ± 0.4 pg/μL [platelets without thrombin priming] and 7.6 ± 0.9 g/μL [platelets with thrombin] [P < .05]). Platelets treated with high doses of thrombin (1 U/mL) without the influence of anti-β2GPI, β2GPI, or/and PF4 represent the positive control of activation and TXB2 production (26.1 ± 0.2 pg/μL). These results show that the trimolecular complex (anti-β2GPI–β2GPI–PF4) exhibit the optimum condition for platelet activation, whereas the complex β2GPI–PF4 appears to show a considerable ability to stimulate platelets in vitro. Moreover, suboptimal doses of thrombin (0.005 U/mL) increase the effect of anti-β2GPI–β2GPI–PF4 in TXB2 production nearly 3.2-fold compared with the corresponding effect in the absence of thrombin, whereas they do not alter the TXB2 production ratio between the several combinations.
Effects of anti-β2GPI–β2GPI–PF4 complex. (A) Effects of anti-β2GPI–β2GPI–PF4 complex on platelet TXB2 production in the presence or absence of thrombin priming. Washed platelets were either stimulated or not with 0.005 U/mL thrombin and were then exposed to immobilized (1) anti-β2GPI + BSA + PF4, (2) isotype control + β2GPI + PF4, (3) anti-β2GPI + β2GPI + PF4, (4) β2GPI + BSA, and (5) β2GPI + PF4. TXB2 was measured in the supernatants as described in “Measurement of platelet thromboxane B2 metabolite production.” A negative control (no. 6) comprising platelets treated with 0.005 U/mL thrombin alone, as well as a positive control (no. 7) comprising platelets treated with 1 U/mL thrombin alone, were also included. Values are the mean ± SD; n = 7. (B) Effects of anti-β2GPI–β2GPI–PF4 complex on phosphorylation of p38 MAPK. Washed platelets were treated with anti-β2GPI–β2GPI–PF4 or control combinations (see “Methods” for details) and stimulated with 0.005 U/mL thrombin. A positive control comprising platelets treated with 1 U/mL thrombin alone was also included in the experiments. Lysates of the platelets were immunoblotted by using specific antibodies for the unphosphorylated form of p38 MAPK (i) and the phosphorylated form of p38 MAPK (ii). The effect of anti-FcγRII blocking mAb IV.3 is presented (iii).
Effects of anti-β2GPI–β2GPI–PF4 complex. (A) Effects of anti-β2GPI–β2GPI–PF4 complex on platelet TXB2 production in the presence or absence of thrombin priming. Washed platelets were either stimulated or not with 0.005 U/mL thrombin and were then exposed to immobilized (1) anti-β2GPI + BSA + PF4, (2) isotype control + β2GPI + PF4, (3) anti-β2GPI + β2GPI + PF4, (4) β2GPI + BSA, and (5) β2GPI + PF4. TXB2 was measured in the supernatants as described in “Measurement of platelet thromboxane B2 metabolite production.” A negative control (no. 6) comprising platelets treated with 0.005 U/mL thrombin alone, as well as a positive control (no. 7) comprising platelets treated with 1 U/mL thrombin alone, were also included. Values are the mean ± SD; n = 7. (B) Effects of anti-β2GPI–β2GPI–PF4 complex on phosphorylation of p38 MAPK. Washed platelets were treated with anti-β2GPI–β2GPI–PF4 or control combinations (see “Methods” for details) and stimulated with 0.005 U/mL thrombin. A positive control comprising platelets treated with 1 U/mL thrombin alone was also included in the experiments. Lysates of the platelets were immunoblotted by using specific antibodies for the unphosphorylated form of p38 MAPK (i) and the phosphorylated form of p38 MAPK (ii). The effect of anti-FcγRII blocking mAb IV.3 is presented (iii).
Effects of PF4–β2GPI–anti-β2GPI complexes on phosphorylation of p38MAPK in platelets
On the basis of previously reported evidence about phosphorylation of p38MAPK by IgG aPL antibodies,39 we examined whether TXB2 production, induced by PF4–β2GPI–anti-β2GPI complexes, is mediated by activation of p38MAPK intracellularly. Platelets treated with 0.005 U/mL thrombin and control combinations of anti-β2GPI plus BSA or isotype control plus β2GPI or isotype control plus β2GPI plus PF4, or PF4 alone produced p38MAPK phosphorylation that did not significantly differ from that of platelets treated with 0.005 U/mL thrombin alone (Figure 7B). On the contrary, platelets treated with anti-β2GPI plus β2GPI plus PF4 complex expressed significantly increased phosphorylation of p38MAPK compared with the 5 combinations used as controls (fold increases of 10.4, 14.1, 13.6, 17.3, and 21.8, respectively). A 1.55-fold increase was observed in platelets treated with anti-β2GPI plus β2GPI plus PF4 compared with platelets treated with anti-β2GPI plus β2GPI.
p38MAPK phosphorylation and TXB2 production, triggered by PF4–β2GPI–anti-β2GPI complexes, are not inhibited by blockade of FcγRIIa receptors
As indicated in Figure 7B, blocking of platelet receptors FcγRIIa, by excess of anti-FcγRII mAb IV.3, did not prevent p38MAPK phosphorylation induced by PF4–β2GPI–anti-β2GPI complexes, in platelets pretreated with suboptimal doses of thrombin. In particular, the ratio of the bands' intensity of p38MAPK phosphorylation, before and after FcR blocking, is calculated to 0.93 (± 0.32; mean ± SD). Likewise, blocking of platelet FcγRIIa did not significantly alter the TXB2 production (data not shown).
Discussion
This study shows the details of PF4 binding to β2GPI, the major antigen recognized by aPL antibodies in patients with APS. Evidence for this binding arose from different experiments, including affinity chromatography with a β2GPI affinity column, loaded with enriched platelet membrane protein extracts, direct binding assays in solid and soluble phase, coprecipitation of β2GPI and PF4, as well as SEC.
Apart from PF4, other cytoskeletal proteins were detected in the eluted material from platelets from HDs (n = 3), whereas in platelets from patients with APS (n = 7) only PF4 was detected. One possible explanation for this difference is the higher concentration of PF4 in patients' platelet preparations, which binds and saturates β2GPI–sepharose column, abrogating the nonspecific binding of other molecules with a “sticky nature.” Interestingly, more than a decade ago, Galazka et al42 described a “mysterious” 8-kDa protein that is copresent in β2GPI preparations and disappears on treatment with SDS or urea. Because the molecular mass completely fits to the molecular weight of PF4 monomer, it is not unlikely that this protein was actually PF4.
The mode of interaction between β2GPI and PF4 was evaluated by using in silico molecular docking. It was shown that the positively charged surface of PF4 tetramer interacts with the negatively charged regions of β2GPI domains III to V, with the additional assistance of hydrogen bonds that are formed between a number of residues of β2GPI and PF4. Our analysis indicated also that an extra interaction interface is available on PF4 tetramer, allowing the binding of a second β2GPI molecule (2 PF4 monomers bind each β2GPI molecule). This finding is of particular importance because it favors β2GPI dimerization, which is a prerequisite for platelet activation.12 Our in silico study also suggests that the presence of glycans at Asn143, Asn174, and Asn234 of β2GPI does not inhibit the PF4 binding. Only a very bulky branched oligosaccharide, bound to Asn164, could preventsterically this binding. Previous studies have shown that low-molecular-mass glycans with simple structures markedly increase in systemic lupus erythematosus and other autoimmune diseases.43,44
The predicted mode of β2GPI–PF4 interaction was experimentally confirmed by SEC. It was found that PF4 tetramers form stable complexes with 1 or 2 molecules of β2GPI, whereas anti-β2GPI antibodies selectively interact with complexes comprising 2 β2GPI molecules and 1 PF4 tetramer. The latter mode of interaction fits perfectly with that predicted for anti-β2GPI antibodies by in silico modeling. In addition, as shown by the β2GPI antigenicity study, the reactivity of APS sera was higher against the PF4–β2GPI complex than against β2GPI alone.
Thus, we propose that PF4 contributes to the natural dimerization of β2GPI, leading to the stabilization of the β2GPI binding onto the phospholipid cell surfaces, enhancing also its recognition by low-affinity anti-β2GPI antibodies, which further stabilize the whole complex. The predicted quartery structure of the β2GPI–PF4 complex allows the epitopes, recognized by thrombosis-associated antibodies41 on domain I of 2 β2GPI molecules, to be arranged in a geometry that precisely fits to the 2 antigen-binding sites of an antibody.
Because platelet activation is one of the main characteristics of APS, predisposing to clinical prothrombotic tendency, we studied the functional role of these complexes in platelets. The present data conclusively show that anti-β2GPI–β2GPI–PF4 complexes induce phosphorylation of p38MAPK, after pretreatment with suboptimal doses of thrombin. The coexistence of anti-β2GPI and β2GPI was found to be a prerequisite for p38MAPK phosphorylation, although the anti-β2GPI–β2GPI–PF4 complexes appear to enhance this effect. We also show that anti-β2GPI–β2GPI–PF4 up-regulates TXB2 production comparable to platelets treated with a high dose of thrombin. This is consistent with the study by Vega-Ostertag et al39 that showed an increase in TXB2 production and p38MAPK phosphorylation in platelets treated with aPL IgG from patients with APS.
Taking into account the clinical similarities between APS and HITT as well as the fact that PF4 seems to be a common denominator in the pathogenesis of 2 syndromes, we examined whether anti-β2GPI effects on platelets are mediated through the Fc portion (such as HITT) or the F(ab′)2 fragment of antibodies. It was shown that the blocking of platelet receptors FcγRIIa by specific mAb IV.3 does not prevent p38MAPK phosphorylation and TXB2 production, concluding that these effects are not mediated by the Fc portion of anti-β2GPI antibodies.
In summary, our study is the first to describe a novel interaction between β2GPI and PF4, both in solid phase and in solution. The main significance of this interaction is the stabilization of β2GPI dimeric structure, attributed to PF4 binding, which facilitates the antibody recognition. On complex formation, PF4–β2GPI–anti-β2GPI induces platelet p38MAPK phosphorylation and TXB2 production. This activation is mainly independent on the platelet Fc receptors.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Vassiliki S. Lalioti and Ignacio V. Sandoval, Center of Molecular Biology, Autonomous University of Madrid, for supporting M.P.S. in establishing several biochemical and molecular techniques.
Authorship
Contribution: P.G.V. designed the research, supervised the experiments, and analyzed the data; M.P.S. performed the experiments, analyzed the data, and wrote the paper; J.G.R. performed the in silico structural analysis and contributed to the writing of the paper; M.S. and G.P. performed the mass spectrometry analysis and the identification of new proteins; and H.M.M. contributed in the design of some important experiments and made important comments for the design of the whole work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P. G. Vlachoyiannopoulos, Department of Pathophysiology, School of Medicine, National University of Athens, 75 M Asias St, 11527 Athens, Greece; e-mail: pvlah@med.uoa.gr.