Abstract
Serum response factor (Srf) is a MADS–box transcription factor that is critical for muscle differentiation. Its function in hematopoiesis has not yet been revealed. Mkl1, a cofactor of Srf, is part of the t(1;22) translocation in acute megakaryoblastic leukemia, and plays a critical role in megakaryopoiesis. To test the role of Srf in megakaryocyte development, we crossed Pf4-Cre mice, which express Cre recombinase in cells committed to the megakaryocytic lineage, to SrfF/F mice in which functional Srf is no longer expressed after Cre-mediated excision. Pf4-Cre/SrfF/F knockout (KO) mice are born with normal Mendelian frequency, but have significant macrothrombocytopenia with approximately 50% reduction in platelet count. In contrast, the BM has increased number and percentage of CD41+ megakaryocytes (WT: 0.41% ± 0.06%; KO: 1.92% ± 0.12%) with significantly reduced ploidy. KO mice show significantly increased megakaryocyte progenitors in the BM by FACS analysis and CFU-Mk. Megakaryocytes lacking Srf have abnormal stress fiber and demarcation membrane formation, and platelets lacking Srf have abnormal actin distribution. In vitro and in vivo assays reveal platelet function defects in KO mice. Critical actin cytoskeletal genes are down-regulated in KO megakaryocytes. Thus, Srf is required for normal megakaryocyte maturation and platelet production partly because of regulation of cytoskeletal genes.
Introduction
Serum response factor (Srf), a nearly ubiquitous transcription factor that is expressed in all hematopoietic cell types, is a founding member of the MCM1-agamous-deficiens-Srf (MADS) domain-containing family of transcription factors, and binds to so called CArG sites (CC/AT-rich/GG, with CCTTATATGG emerging as a major consensus sequence).1 More than 200 CArG boxes control expression of more than 150 Srf target genes, including genes of the cytoskeleton as well as several immediate-early genes, for example the proto-oncogene c-fos,2 and also bcl2,3 whereby Srf participates in apoptosis, cell growth, differentiation, and cell-cycle regulation. Srf is a downstream target of many pathways; for example, the mitogen-activated protein kinase pathway that acts through the ternary complex factors (TCF) as well as Rho signaling,4 which promotes actin polymerization. Srf plays an important role in the regulation of smooth, skeletal, and cardiac muscle genes5-8 during development and in adult life including aging.9,10 Srf can be activated to promote transcription in response to extracellular signals that induce its association with specific cofactors. The 2 families of Srf cofactors are the TCF proteins (Elk1, SAP1, SAP2),11 and the myocardin family of proteins that includes myocardin, megakaryoblastic leukemia 1 (Mkl1), and Mkl2.1,12,13 In some cell types, TCF proteins and Mkl1 compete for binding to Srf to activate or inhibit distinct transcription targets.14,15 In Drosophila, Srf interaction with Mkl (MAL-D) promotes cytoskeletal strength during cellular migration.16,17 In murine embryonic stem cells, Srf is crucial for actin cytoskeletal organization and focal adhesion assembly.18 Murine embryos lacking Srf fail to form mesoderm and thus die early in development.19
Prior work in our laboratory has focused on acute megakaryoblastic leukemia with the t(1;22) translocation, involving fusion between an RNA binding motif protein 15 (RBM15) and Mkl1. Mkl1 is known to act as a cofactor for Srf-mediated gene activation in muscle differentiation.12,20 We have demonstrated a physiologic role for Mkl1 in megakaryopoiesis.21 Mkl1 expression increases with normal megakaryocyte (Mk) differentiation, and promotes Mk polyploidization. Mkl1 knockout (KO) mice have normal hematopoietic stem cells and megakaryocyte-erythroid progenitors (Pre-Meg-E). However, there is a dramatic increase in the number of CD41+ megakaryocytes with most of these having low (2N) ploidy. There is a significant decrease in the number of polyploid megakaryocytes and thrombocytopenia. Mkl1 requires Srf to enhance polyploidization during megakaryocytic differentiation. Based on these findings, we have examined the effects of the Mkl1 cofactor Srf in megakaryocyte development.
We show, that Srf deletion specifically in the megakaryocytic lineage leads to macrothrombocytopenia, whereas bone marrow (BM) and spleen show significant accumulation of abnormal megakaryocytes. Examination of candidate Srf target genes reveals that several actin cytoskeletal regulatory proteins are down-regulated in Srf KO megakaryocytes as a likely cause for the observed phenotype.
Methods
BM collection and MegaCult-C assays
All procedures involving mice were performed in compliance with relevant laws and institutional guidelines and were approved by the Yale University Institutional Animal Care and Use Committee. Pf4-Cre mice (C57BL/6-Tg(Pf4-cre)Q3Rsko/J), which express a codon-improved Cre recombinase under control of the mouse platelet factor 4 (Pf4) promoter,22 and Srffl/fl mice (B6.129-Srftm1Ddg/J)8 were obtained from The Jackson Laboratory and crossed to obtain Pf4+/Srffl/fl (KO), Pf4+/Srffl/wt heterozygous (HET), and Pf4+/Srfwt/wt wild-type (WT) mice or Pf4−/Srffl/fl, Pf4−/Srffl/wt, or Pf4−/Srfwt/wt (all WT) mice. Rosa26-Stop-LoxP-eYFP mice were obtained from The Jackson Laboratory and bred to Pf4-Cre mice to generate described genotypes with and without the enhanced yellow fluorescent protein (YFP) transgene. Age- and sex-matched WT, HET, and KO littermates were used in these studies. After anesthesia and humane killing, femurs, tibias, spines, and spleens of littermates were excised. For histology sections, femurs and spleens were fixed overnight in 4% paraformaldehyde. Femurs were then decalcified in Decalcifier I solution (Surgipath) for 2 hours. Specimens were dehydrated in 70% ethanol and processed by the Research Histology Facility at Yale Medical School for 5-μm longitudinal paraffin sections, hematoxylin and eosin staining, and immunohistochemistry for von Willebrand Factor (anti-VWF; Dako). To quantify megakaryocytes in BM and spleen, paraffin sections stained with anti-VWF were examined using an Olympus BX51 microscope equipped with objective lenses (40/0.75, 20/0.4, 10/0.25, and 4/0.1) and an Olympus Q-Color5 camera (Olympus America) and QCapture Pro Version 5.1 software (QImaging). Total megakaryocyte number was counted in each 4X observation field. BM cell suspensions were harvested by flushing with Iscove modified Dulbecco medium/2% fetal bovine serum followed by filtering through a 100-μm nylon strainer to remove bone debris. For colony-forming unit–megakaryocyte (CFU-Mk) MegaCult-C assays, BM mononuclear cells were cultured for 6 to 8 days according to the manufacturer's protocols (StemCell Technologies); 50 ng/mL human thrombopoietin (TPO), 50 ng/mL human interleukin-11 (IL-11), 10 ng/mL murine IL-3, and 20 ng/mL human IL-6 (PeproTech) were used in these assays. Colonies containing more than 3 megakaryocytes (acetylcholinesterase-positive cells) were counted as CFU-Mks. Duplicate assays were performed for each mouse. At least 3 mice were analyzed for each experiment.
Primary murine megakaryocyte enrichment
After lysis of mature red blood cells by BD PharmLyse (BD Biosciences), cells were cultured at 2 × 106 cells/mL in StemSpan (StemCell Technologies), 30% BIT9500 (StemCell Technologies), 50 ng/mL murine TPO (PeproTech), 2mM l-glutamine, and penicillin/streptomycin. After 4 days, the cells were fractionated on a 3% discontinuous bovine serum albumin (BSA; Sigma-Aldrich) gradient as described.21 The fully mature polyploid megakaryocytes are mainly in the pellet below the 3% BSA fraction. For some assays, murine BM was lineage-depleted (lin−) with the BD IMag Mouse Hematopoietic Progenitor (Stem) Cell Enrichment Set, according to the manufacturer's protocol (BD Biosciences).
Microarray analysis and real-time RT-PCR
Total RNA was isolated using the RNeasy Micro Kit (QIAGEN) according to the manufacturer's protocol. Genomic DNA was digested with RNase-free DNase I (QIAGEN). The Mouse Illumina (mouseRef-8, v1.1; Illumina) expression beadchip targeting 24 613 transcripts was used for the RNA expression profiling experiments. RNA was labeled and hybridized to the array using standard Illumina protocols by the Keck Microarray Resource Center at the Yale School of Medicine (http://keck.med.yale.edu/microarrays/). The array was scanned using an iScan, and the data exported using BeadStudio 3.2 software. The microarray data were processed using the BioConductor R2.10.0 lumi-package to perform background correction, variance stabilizing transformation, and quantile normalization.23 Probes that were called present in 2 or less samples were excluded from analysis. Differentially expressed genes were identified using the BioConductor Limma package.24 Overrepresented functional classes of differentially expressed genes were identified using the DAVID Web site.25 Microarray data were deposited into the Gene Expression Omnibus database (data series GSE21859). For real-time reverse-transcriptase polymerase chain reaction (RT-PCR) analysis, cDNA was prepared using Superscript III Reverse Transcriptase (Invitrogen) with random primers (Invitrogen). Gene expression levels were determined using Applied Biosystems TaqMan Expression Assays for Srf, Coronin 1a (Coro1a), IL-1 receptor-associated kinase 2, calponin 2 (Cnn2), myosin light chain 6 (Myl6), and filamin a (Flna). TaqMan gene expression assay for Pol2ra was used as an internal control for normalization. PCR was performed on a CFX96 C1000 thermal cycler (Bio-Rad). Gene expression was analyzed by the comparative threshold method.
Murine peripheral blood counts
Mice were anesthetized with isoflurane (Halocarbon) followed by retro-orbital bleeding (∼ 100 μL). The blood was evacuated into glass capillary tubes treated with 5 μL of 0.5M ethylenediaminetetraacetic acid to prevent clot formation. The complete blood count of blood samples was performed using a Hemavet 950FS (Drew Scientific) according to the manufacturer's protocol.
Flow cytometric analysis of DNA content and surface markers
DNA content of megakaryocytes was determined by staining whole BM cells (in some cases BM was first subjected to lineage depletion described in “Primary murine megakaryocyte enrichment”) with fluorescein isothiocyanate (FITC)–conjugated anti-CD41 antibody (BD Biosciences) followed by fixation with 50% ethanol at 4°C for 2 hours, followed by digestion with 20 μg/mL RNase for 20 minutes at 37°C, and staining with 1 μg/mL propidium iodide (Sigma-Aldrich). To determine mouse BM LSK (lin−c-kit+Sca+), Pre-Meg-E (megakaryocyte-erythroid progenitors, lin−c-kit+CD150+CD105−CD41−), and megakaryocyte progenitor (lin−c-kit+CD41+) cells,26 freshly obtained BM cells were stained with phycoerythrin (PE)–cyanin 5 anti-CD150 (BioLegend), FITC anti-CD41 (BD Biosciences), Alexa Fluor 647 anti–Sca-1, allophycocyanin-H7 anti-CD117 (BD Biosciences), PE-cyanin 7 anti-CD105 (BioLegend), biotin-labeled lineage cell detection cocktail (Miltenyi Biotec), and anti–biotin-PE (Miltenyi Biotec). In experiments in which enhanced YFP expression was measured, e-fluor 350-conjugated anti-CD41 antibody (BioLegend) was used instead. Flow cytometric analysis was performed with an LSRII flow cytometer (BD Biosciences) and FlowJo Version 8.8.7 software (TreeStar).
Proplatelet formation assays
Lin− murine BM was cultured overnight in StemSpan (StemCell Technologies), 30% BIT9500 (StemCell Technologies), and recombinant murine stem cell factor (100 ng/mL) and TPO (50 ng/mL). The next day, megakaryocyte progenitors (lin−c-kit+CD41+) were sorted on a FACSAria and cultured for an additional 3 days in TPO containing medium either on fibronectin-coated tissue culture plates or in 3% methylcellulose under direct visualization in the Incucyte system (Essen Instruments), which allows automated monitoring of live cells in culture. Adherent and proplatelet forming cells were enumerated at specific time points.
Platelet function testing
Platelet activation assays were adapted from Nieswandt et al.27 Briefly, 50 μL of whole blood was collected into a heparinized glass capillary tube from the retro-orbital plexus into 200 μL of 20-U/mL heparin in Tris-buffered saline (20mM Tris-HCL, 137mM NaCl). This suspension was further diluted with 1 mL of 2mM CaCl2 in modified Tyrodes-HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (5 mM HEPES, 140 mM NaCl, 2.7 mM KCl, 5.5mM dextrose, 0.42mM Na2HPO4, 12mM NaHCO3). In 5-mL polystyrene fluorescence-activated cell sorter (FACS) tubes (BD Biosciences), 5 μL of pan-platelet marker, glycoprotein IIb (GPIIb)/IIIa-FITC (M022-1; Emfret Analytics), and 5 μL of either isotype control rat immunoglobulin G–PE (P-190-2; Emfret Analytics) or platelet activation marker, activated GPIIb/IIIa-PE (JON/A-PE, M130-2; Emfret Analytics) was added. Then, 4 μL of agonist, 1mM adenosine 5′-diphosphate (ADP, catalog no. A6646; Sigma-Aldrich), or control (Tris-buffered saline) was added. After 26 μL of whole blood suspension was added, samples were vortexed for 2 seconds, incubated for 15 minutes at 25°C in the dark, diluted with phosphate-buffered saline (PBS) to a total volume of 400 μL, and analyzed on a BD Biosciences FACSCalibur immediately thereafter. Analysis was performed using FlowJo Version 8.8.7 software. A standard platelet gate was established based on size and granularity.
Bleeding time measurement
The 3-week-old mice were anesthetized with isoflurane and 0.5 cm of their tails cut off with a sharp razor blade. Tails were held in 37°C PBS, and bleeding from the tail timed from time of cut until the end of bleeding.
Immunofluorescent staining and confocal analysis
Megakaryocyte progenitors (lin−c-kit+CD41+) were cultured in TPO containing medium for 3 days and subsequently allowed to adhere to glass coverslips coated with fibronectin for 3 hours. Coverslips were rinsed with PBS, fixed with 3.7% formaldehyde, and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich). Cells were blocked with PBS/2%BSA and incubated with mouse antitubulin antibody (1:200, anti–β-tubulin; Sigma-Aldrich), Phalloidin–tetramethylrhodamine B isothiocyanate (1:200; Sigma-Aldrich), and FITC-conjugated rat anti–mouse CD41 antibody for 1 hour. Bound antibody was detected using Alexa Fluor 647–labeled goat anti–mouse secondary antibody. For platelet analysis, platelets were separated from whole blood by centrifugation at 200g, fixed in 4% formaldehyde in platelet buffer (145mM NaCl, 10mM HEPES, 10mM glucose, 0.5mM NaHPO4, 5mM KCl, and 2mM MgCl2, [pH 7.4]), and spun unto poly-l-lysine–coated coverslips (Sigma-Aldrich). Permeabilization and staining were performed as for megakaryocyte progenitors. Fluorescent signal was visualized on a Leica SP5 confocal microscope, and data analyzed with Leica Application Suite Advanced Fluorescence Version 1.8.3 software. Platelet size was measured based on diameter of the tubulin ring.
Electron transmission microscopy
Megakaryocytes from day 13.5 fetal liver specimens were cultured and enriched as described in “Primary murine megakaryocyte enrichment.” Megakaryocyte cell pellets fixed for 2 hours in paraformaldehyde/glutaraldehyde 2.5%/2.5% buffer in sodium cacodylate buffer (Electron Microscopy) at room temperature and stored in 0.2M sodium cacodylate buffer until further processing. Platelet pellets were prepared by spinning whole blood anticoagulated with ethylenediaminetetraacetic acid at 100g for 15 minutes. Platelets were pelleted and fixed as described for megakaryocytes.
Fixed megakaryocytes and platelet pellets were post-fixed in osmium tetroxide, washed, and dehydrated through a series of alcohols, infiltrated with propylene oxide, and embedded in epoxy resin in an inverted beam capsule. Ultrathin sections were stained with uranyl acetate and lead citrate, and examined with a Tecnai G2 Spirit BioTWIN transmission electron microscope at an accelerating voltage of 80 kV. Images were recorded with an AMT 2k charge-coupled device camera.
Statistical analysis
Statistical significance was assessed (Excel Version 11.5.6) with a 2-tailed unpaired t test.
Results
Validation of megakaryocyte-specific Srf deletion
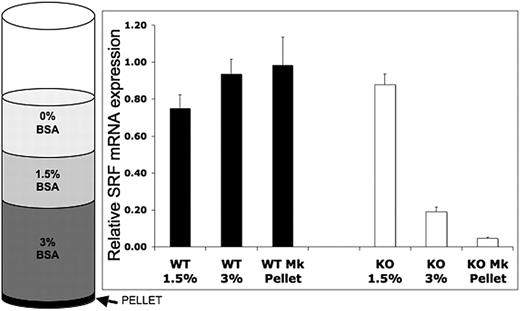
SrfF/F mice were crossed with Pf4-Cre mice, which express Cre recombinase from the Pf4 promoter resulting in excision of the LoxP-flanked exon 1 specifically in the megakaryocytic lineage.22 Pf4-Cre+/−/SrfF/F or Pf4-Cre+/+/SrfF/F (hereinafter referred to as Pf4/Srf KO) progeny are viable. Lineage-specific knockdown of Srf mRNA expression was validated by quantitative RT-PCR on BM cell fractions and by FACS analysis using a YFP reporter. For RT-PCR, BM from Pf4/Srf WT and Pf4/Srf KO mice was cultured for 4 days in TPO-containing medium, which promotes megakaryocyte differentiation. Cellular composition of the BSA-gradient fractions was verified by Wright-Giemsa staining of cytospins (data not shown). Although there is no decrease in Srf in the 1.5% BSA fraction, which includes 90% myeloid cells, Srf expression is significantly decreased in the 3% BSA fraction, in which megakaryocytes are enriched to 70%-80%, and even more so in the cell pellet, which consists of nearly all megakaryocytes (Figure 1). The timing and specificity of Srf deletion to the megakaryocytic lineage was verified by flow cytometry of YFP expression in PF4-Cre+/YFP mice in which Pf4-mediated Cre expression leads to activation of YFP expression by excision of the stop codon in front of the YFP gene under control of the Rosa promoter. YFP expression is restricted to cells in the megakaryocyte lineage, and increases with megakaryocytic differentiation. There is no YFP expression in Pre-Meg-E progenitors (lin−c-kit+CD150+CD105−CD41−), low level expression in committed megakaryocyte progenitors (lin−c-kit+CD41+), and high expression in mature megakaryocytes (lin−CD41+CD150+, supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In peripheral blood, YFP expression is restricted to platelets (determined by forward and side light scatter (FSC/SSC) properties and CD41+), whereas cells of all other lineages are YFP-negative (supplemental Figure 1B). Deletion of Srf expression was also verified by quantitative RT-PCR in cultured FACS-sorted megakaryocyte progenitors (Table 1).
Assessment of megakaryocyte lineage-specific Srf deletion. Bone marrow (BM) from Pf4/Srf+/+ (n = 3) and Pf4/SrfF/F (n = 4) mice was cultured for 4 days in TPO and subjected to BSA gradient separation. Quantitative RT-PCR was performed on the isolated fractions. Mean and SD are shown. P < .01.
Assessment of megakaryocyte lineage-specific Srf deletion. Bone marrow (BM) from Pf4/Srf+/+ (n = 3) and Pf4/SrfF/F (n = 4) mice was cultured for 4 days in TPO and subjected to BSA gradient separation. Quantitative RT-PCR was performed on the isolated fractions. Mean and SD are shown. P < .01.
Loss of Srf in the megakaryocyte lineage causes thrombocytopenia with increased megakaryocytes
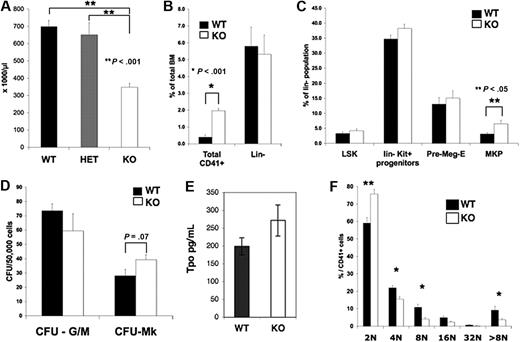
Although born at a normal Mendelian ratio, Pf4/Srf KO mice have significant thrombocytopenia with a mean (± SEM) platelet number of 460 × 103/μL (± 23 × 103/μL) versus 703 × 103/μL (± 33 × 103/μL) in KO versus littermate control WT mice, respectively (P < .01, Figure 2A). The platelet count was similar in WT versus HET (Pf4/SrfF/+) mice, and both were statistically significantly higher than in KO mice. Although there was no significant difference in the mean platelet volume as measured by the Hemavet (data not shown), an increase in platelet size was noted by confocal analysis and confirmed by electron microscopy (see “Pf4/Srf megakaryocytes and platelets have abnormal cytoskeletal organization and ultrastructure”). Both the white blood cell and red blood cell counts were unaffected in Pf4/Srf KO mice, as expected (data not shown).
Platelet counts, BM progenitor, and Mk ploidy analysis. Blood from wild-type (WT; n = 15), Pf4/SrfF/+ HET (n = 12), and Pf4/SrfF/F knockout (KO; n = 17) mice was analyzed using a Hemavet analyzer. (A) Platelet counts in Pf4/SrfF/F mice were significantly lower than in control mice (P < .001). (B-C) BM from Pf4/Srf WT (n = 8) and KO (n = 11) mice was analyzed by flow cytometry for CD41+ megakaryocytes (B, *P < .001) and progenitor populations (C, **P < .05). (D) BM from WT (n = 3) and KO (n = 4) mice was assayed for CFU-Mk. (E) Plasma TPO levels (in picograms per milliliter) were assayed from Pf4/Srf WT (n = 15) and KO (n = 10) mice. (F) Megakaryocytes from WT (n = 4) and KO (n = 5) mice were analyzed by flow cytometry for nuclear DNA content. *P < .05, **P < .01.
Platelet counts, BM progenitor, and Mk ploidy analysis. Blood from wild-type (WT; n = 15), Pf4/SrfF/+ HET (n = 12), and Pf4/SrfF/F knockout (KO; n = 17) mice was analyzed using a Hemavet analyzer. (A) Platelet counts in Pf4/SrfF/F mice were significantly lower than in control mice (P < .001). (B-C) BM from Pf4/Srf WT (n = 8) and KO (n = 11) mice was analyzed by flow cytometry for CD41+ megakaryocytes (B, *P < .001) and progenitor populations (C, **P < .05). (D) BM from WT (n = 3) and KO (n = 4) mice was assayed for CFU-Mk. (E) Plasma TPO levels (in picograms per milliliter) were assayed from Pf4/Srf WT (n = 15) and KO (n = 10) mice. (F) Megakaryocytes from WT (n = 4) and KO (n = 5) mice were analyzed by flow cytometry for nuclear DNA content. *P < .05, **P < .01.
Analysis of BM by flow cytometry revealed no differences in the number of Lin−, Lin−Kit+, or Lin−Sca+Kit+ stem/progenitor cells (Figure 2B-C) in the Pf4/Srf KO mice as expected because of stage- and lineage-specific KO of Srf. In contrast, BM had a significantly increased number of CD41+ (Figure 2B) and CD41+Kit+ (Figure 2C) megakaryocyte progenitors. There was a slight increase in bipotent Pre-Meg-E (megakaryocyte-erythroid progenitors), but this increase did not reach statistical significance. Consistent with the increase in megakaryocyte progenitors, there was a trend toward an increase in CFU-Mk numbers (Figure 2D). Overall, the increase in total CD41+ cells (4-fold over controls) was more dramatic than the increase in megakaryocyte progenitors (an ∼ 2-fold increase by flow cytometry and 1.5-fold increase by CFU-Mk assay), suggesting that there is increased proliferation and/or decreased maturation of megakaryocyte progenitors. As increased TPO levels may account for the increase in megakaryopoiesis, we tested plasma for TPO levels by enzyme-linked immunosorbent assay. Although the TPO levels were slightly increased in Pf4/Srf KO mice, the increase was not statistically significant (Figure 2E). Analysis of megakaryocyte ploidy in BM revealed that Pf4/Srf KO mice had a statistically significant shift toward lower megakaryocyte ploidy compared with WT mice (Figure 2F and supplemental Figure 2A). This shift was also the case when whole BM was cultured for 4 days before assessment of ploidy (supplemental Figure 2B). Of note, Pf4/Srf KO megakaryocytes are not impaired in reaching high ploidy.
Megakaryocytes are increased in BM and spleen and are dysmorphic
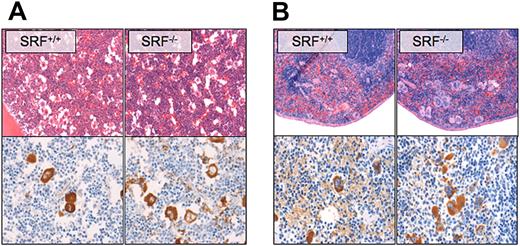
As shown by hematoxylin and eosin and VWF staining of paraffin sections, megakaryocyte numbers in BM (Figure 3A) and spleen (Figure 3B) were increased in Pf4/Srf KO mice. The morphology of the Pf4/Srf KO megakaryocytes in BM was abnormal in that megakaryocytes were found in clusters, had hypolobated nuclei, and appeared smaller with irregular edges (Figures 3A). Spleens of Pf4/Srf KO mice had a significant increase in megakaryocytes within the red pulp, again with dysmorphic appearance of the megakaryocytes (Figure 3B).
Megakaryocyte morphology and localization. Representative BM (A) and spleen (B) sections of Pf4/Srf WT and KO mice were stained with hematoxylin and eosin (top) and anti-VWF (bottom). Representative images of at least 3 mice are shown.
Megakaryocyte morphology and localization. Representative BM (A) and spleen (B) sections of Pf4/Srf WT and KO mice were stained with hematoxylin and eosin (top) and anti-VWF (bottom). Representative images of at least 3 mice are shown.
Pf4/Srf megakaryocytes have abnormal in vitro phenotype and function
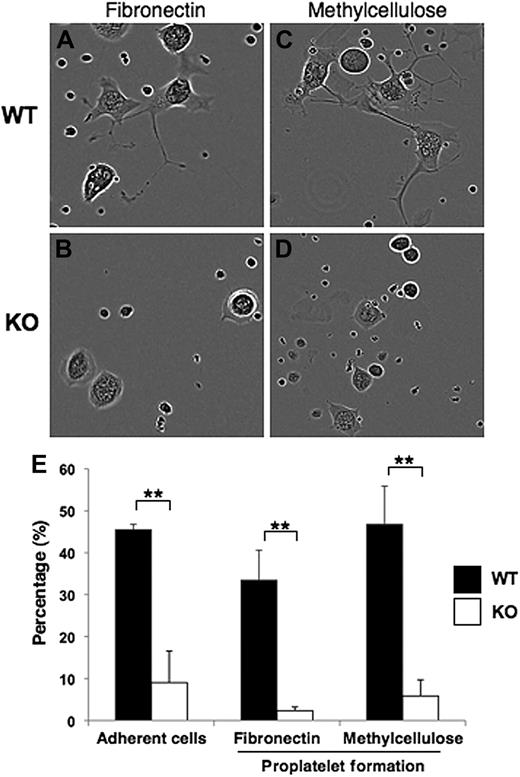
In vitro megakaryocyte assays were performed to assess adhesion and proplatelet formation on both fibronectin-coated plates (Figure 4A-B) and in semisolid (methylcellulose) medium (Figure 4C-D). Pf4-Cre–mediated deletion of Srf caused defective adherence of Pf4/Srf megakaryocytes to fibronectin (Figure 4A-B). At 3 hours after plating on fibronectin, 5% to 10% of WT megakaryocytes were adherent, whereas none of the Srf KO megakaryocytes had adhered (data not shown). By 40 hours after plating, 45% (± 1.3%) WT megakaryocytes were adherent, whereas only 9% (± 7.2%) of KO megakaryocytes had adhered (Figure 4E). Of the adherent megakaryocytes on the fibronectin-coated plates, 33% (± 7.1%) of WT megakaryocytes had formed proplatelets, whereas only 2% (± 0.9%) Srf KO megakaryocytes had formed proplatelets (Figure 4E). Similar proplatelet data were obtained for megakaryocytes matured for 40 hours in methylcellulose (Figure 4E).
Megakaryocyte adherence and proplatelet formation. (A-D) Representative images are shown for sorted CD41+c-kit+ megakaryocyte progenitors from Pf4/Srf WT and Pf4/Srf KO littermates that were cultured in TPO containing medium either on fibronectin (left) or in semisolid medium (methylcellulose, right). (E) Continuous videos were taken and analyzed for cell adherence when cultured on fibronectin (left) and for proplatelet formation by cells on fibronectin (middle) and in semisolid medium (right). The percentage of total cells analyzed over 88 hours is shown. **P < .05.
Megakaryocyte adherence and proplatelet formation. (A-D) Representative images are shown for sorted CD41+c-kit+ megakaryocyte progenitors from Pf4/Srf WT and Pf4/Srf KO littermates that were cultured in TPO containing medium either on fibronectin (left) or in semisolid medium (methylcellulose, right). (E) Continuous videos were taken and analyzed for cell adherence when cultured on fibronectin (left) and for proplatelet formation by cells on fibronectin (middle) and in semisolid medium (right). The percentage of total cells analyzed over 88 hours is shown. **P < .05.
Pf4/Srf megakaryocytes and platelets have abnormal cytoskeletal organization and ultrastructure
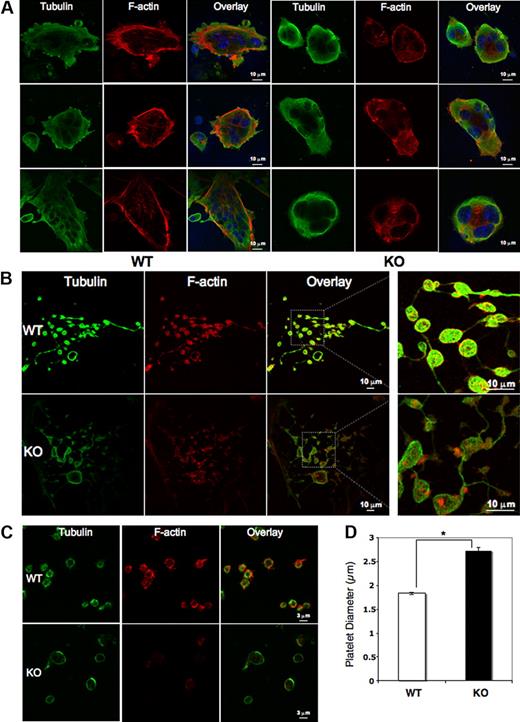
Mature megakaryocytes and platelets were analyzed for cytoskeletal tubulin and filamentous actin (F-actin) by immunofluorescence. Although the Pf4/Srf KO megakaryocytes have fewer cytoplasmic extensions, tubulin bundles are intact, forming a complex meshwork throughout the cytoplasm as in WT megakaryocytes. In contrast, Pf4/Srf KO megakaryocytes show abnormal stress fiber formation with disorganized F-actin (Figure 5A). In WT megakaryocytes, F-actin bundles appear throughout the cytoplasm and in cytoplasmic extensions, whereas in KO megakaryocytes, the F-actin staining pattern is very different with “spotty” accumulation of F-actin and lack of organized fibers. Although staining intensity of F-actin in Pf4/Srf KO megakaryocytes appears variable by immunofluorescence, only a slight difference in total F-actin is evident when assayed by flow cytometry (data not shown). Immunofluorescence of proplatelets (Figure 5B) and platelets (Figure 5C) reveals similar abnormalities. Consistent with the findings in Pf4/Srf KO megakaryocytes, KO platelets lack well-defined F-actin filaments. Of note, significant macrothrombocytopenia is evident by measurement of immunofluorescently labeled platelets: the average diameter of WT platelets was 1.8 μm (± 0.03 μm), whereas that for KO platelets was 2.7 μm (± 0.07 μm; Figure 5D). To further define the observed structural abnormalities in megakaryocytes and platelets, we performed electron transmission microscopy. Electron microscopic analysis of platelets confirms the finding of macrothrombocytopenia: in particular it reveals a population of giant platelets with a diameter from 4 μm to 7 μm, which includes approximately 8% of platelets, some of which exceed the size of red blood cells in diameter (WT platelets in Figure 6A, KO platelets in Figure 6B-D). Although the distribution and number of platelet granules appear normal, Pf4/Srf KO megakaryocytes show several ultrastructural abnormalities. The cytoplasm of Pf4/Srf KO megakaryocytes occasionally, in approximately 10% of imaged megakaryocytes, contains areas of tubules randomly arranged resembling a maze (Figure 6F-G) as opposed to the well-structured, uniform demarcation membrane system of WT megakaryocytes (Figure 6E). Although the Srf KO nuclei of the megakaryocytes appear normal, surrounded by a large cytoplasm, the cytoplasmic contents are unevenly distributed throughout the cytoplasm, including areas with missing granules and organelles (Figure 6H).
Megakaryocyte, proplatelet, and platelet cytoskeletal structure. Megakaryocytes from Pf4/Srf WT and KO mice were cultured in TPO containing medium for 4 days and subsequently plated on fibronectin-coated coverslips and stained for tubulin (green) and F-actin (phalloidin, red). 4,6′-diamidino-2-phenylindole was used to stain DNA. (A) Representative images of WT (left) and KO (right) megakaryocytes. (B) Representative images of WT (top) and KO (bottom) megakaryocytes forming proplatelets in culture. (C) Platelets from 3 Pf4/Srf WT and 3 Pf4/Srf KO mice were fixed, spun onto coverslips, and stained for tubulin and F-actin. (D) WT (n = 29) and KO (n = 34) platelet size were measured in micrometers. *P < .01.
Megakaryocyte, proplatelet, and platelet cytoskeletal structure. Megakaryocytes from Pf4/Srf WT and KO mice were cultured in TPO containing medium for 4 days and subsequently plated on fibronectin-coated coverslips and stained for tubulin (green) and F-actin (phalloidin, red). 4,6′-diamidino-2-phenylindole was used to stain DNA. (A) Representative images of WT (left) and KO (right) megakaryocytes. (B) Representative images of WT (top) and KO (bottom) megakaryocytes forming proplatelets in culture. (C) Platelets from 3 Pf4/Srf WT and 3 Pf4/Srf KO mice were fixed, spun onto coverslips, and stained for tubulin and F-actin. (D) WT (n = 29) and KO (n = 34) platelet size were measured in micrometers. *P < .01.
Transmission electron microscopy of megakaryocytes and platelets. Representative electron microscopic views of platelets and cultured megakaryocytes of Pf4/Srf WT and KO mice. Pf4/Srf KO mice contain a population of giant platelets, and Pf4/Srf KO megakaryocytes have structural alterations. (A) Representative image of a normal population of platelets from a WT mouse. (B-D) Electron micrographs showing giant platelets isolated from the blood of Pf4/Srf KO mice. Scale bar represents 500 nm (A-D). (E) Representative image of a normal megakaryocyte with a well-structured, uniform demarcation membrane system from WT mouse. (F-H) Electron micrographs of megakaryocytes with altered structures from Pf4/Srf KO mice. A small area of demarcation membrane system concentration, resembling a maze of small tubules, is observed in a population of Srf-deficient megakaryocytes (F-G). (H) Representative Pf4/Srf KO megakaryocyte micrographs in which the cellular contents are not evenly distributed throughout the cytoplasm, including areas with missing granules and organelles. Scale bar represents 2 μm (E-H).
Transmission electron microscopy of megakaryocytes and platelets. Representative electron microscopic views of platelets and cultured megakaryocytes of Pf4/Srf WT and KO mice. Pf4/Srf KO mice contain a population of giant platelets, and Pf4/Srf KO megakaryocytes have structural alterations. (A) Representative image of a normal population of platelets from a WT mouse. (B-D) Electron micrographs showing giant platelets isolated from the blood of Pf4/Srf KO mice. Scale bar represents 500 nm (A-D). (E) Representative image of a normal megakaryocyte with a well-structured, uniform demarcation membrane system from WT mouse. (F-H) Electron micrographs of megakaryocytes with altered structures from Pf4/Srf KO mice. A small area of demarcation membrane system concentration, resembling a maze of small tubules, is observed in a population of Srf-deficient megakaryocytes (F-G). (H) Representative Pf4/Srf KO megakaryocyte micrographs in which the cellular contents are not evenly distributed throughout the cytoplasm, including areas with missing granules and organelles. Scale bar represents 2 μm (E-H).
Pf4/Srf KO mice have dysfunctional platelets and prolonged bleeding times
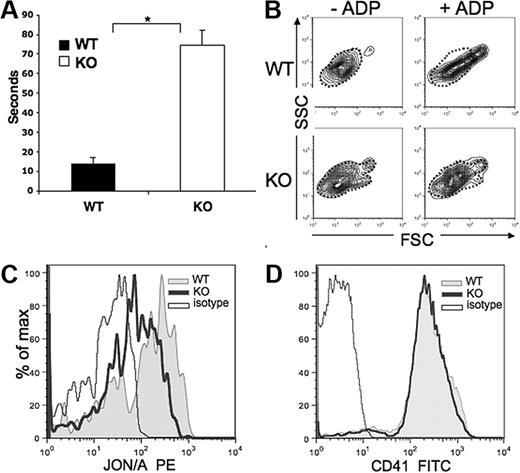
Pf4/Srf KO mice have significantly prolonged bleeding times, which in these mice with Pf4-specific Srf KO, reveals a highly specific in vivo test of platelet function (Figure 7A). Consistent with the abnormal platelet cytoskeleton, in vitro activation assays also reveal defects in the KO platelet function (Figure 7B-C). Upon exposure to the platelet agonist ADP, which binds to the platelet receptor P2Y1, activation of the GPIIb/IIIa receptor and a shape change of the platelet population are induced in WT platelets. Resting platelets have a wide distribution on SSC analysis because of their discoid shape relative to their orientation within the laser beam. When activated, platelets form pseudopods and take on a spherical shape producing a more condensed and elongated FSC/SSC signal. Although we observe this change in WT platelets, it is reduced in KO platelets (Figure 7B, dotted outline marks inactivated state). In addition, the GPIIb/IIIa receptor undergoes a conformational change upon platelet activation to allow binding of fibrinogen-inducing platelet aggregation in vivo. This change is detected by FACS analysis via selective binding of JON/A to the high affinity conformation of the receptor. Although a similar percentage of platelets bind JON/A, the mean fluorescence intensity is greatly reduced in KO versus WT platelets (Figure 7C), suggesting either decreased affinity of the antibody for the receptor on KO platelets or decreased density of GPIIb/IIIa. The latter is unlikely as CD41 (GPIIb) staining is equal in WT and KO platelets (Figure 7D).
Bleeding time and platelet function analysis. (A) Bleeding times were performed on Pf4/Srf WT (n = 8) and KO (n = 8) mice at the time of genotyping. *P < .01. (B-C) Platelet function analysis was performed by activation of platelets with ADP and analysis of FSC/SSC distribution (B) and analysis of GPIIb/IIIa distribution changes by staining with JON/A (C). (D) GPIIb density was confirmed by staining with anti-CD41. Data are representative of at least 3 independent experiments.
Bleeding time and platelet function analysis. (A) Bleeding times were performed on Pf4/Srf WT (n = 8) and KO (n = 8) mice at the time of genotyping. *P < .01. (B-C) Platelet function analysis was performed by activation of platelets with ADP and analysis of FSC/SSC distribution (B) and analysis of GPIIb/IIIa distribution changes by staining with JON/A (C). (D) GPIIb density was confirmed by staining with anti-CD41. Data are representative of at least 3 independent experiments.
Cytoskeletal regulatory genes are down-regulated in Srf KO megakaryocytes
Based on our data, we postulated that, as in muscle cells, Srf may play a key role in regulating expression of genes critical to cytoskeletal regulation and assembly. Indeed cytoskeletal genes were among the most down-regulated genes together with Srf in microarray expression analysis of mature megakaryocytes derived from FACS-sorted megakaryocyte progenitors from KO versus WT mice (Table 1 and supplemental Tables 1-2). We therefore verified expression by quantitative RT-PCR analysis both on BSA-gradient enriched megakaryocytes and on megakaryocytes derived from FACS-sorted megakaryocyte progenitors, confirming differential expression of Srf, Coro1a, Cnn2, and Flna, whereas no statistically significant difference was detected for other genes, such as IL-1 receptor-associated kinase 2, Myl6, Mkl1, and Mkl2.
Discussion
Our data show that megakaryocyte-specific KO of Srf in mice leads to significant thrombocytopenia with prolonged bleeding times explained by abnormal megakaryocyte maturation and function. We have previously described the role of Mkl1, a cofactor of SRF and fusion partner of RBM15 in the acute megakaryoblastic leukemia–specific oncoprotein RBM15-Mkl, in megakaryocyte maturation.21 Our current findings are consistent with those in Mkl1 KO mice, but KO of Srf in the megakaryocyte lineage leads to a far more profound phenotype than Mkl1 KO; compared with Mkl1 KO mice, the Pf4/Srf KO mice have a lower platelet count, and what would be best described as megakaryocyte dysplasia: megakaryocytes show abnormal morphology as evident by clustering in BM and spleen, an increase in immature megakaryocyte progenitors with concomitant decrease in ploidy, decreased adhesion and proplatelet formation, and abnormal cytoskeletal organization evident by immunofluorescence staining of the actin cytoskeleton and electron microscopic analysis of the megakaryocyte ultrastructure. As a result, mice are significantly thrombocytopenic, and platelets, which are enlarged with a considerable fraction of giant platelets, also show abnormal cytoskeletal organization and are dysfunctional resulting in prolonged bleeding times in vivo. In our model, in which SRF is deleted by Pf4-Cre mediated excision, the effect we observe may be underestimated because of the percentage of cells with complete excision increases progressively during megakaryocyte maturation, and there may be platelet formation from some megakaryocytes with preserved SRF expression. Expression analysis reveals that genes essential to cytoskeletal organization are significantly down-regulated in Srf KO megakaryocytes: Coro1a, Cnn2, and Flna. All 3 genes have been previously described to be direct targets of Srf (Sun et al28 and reviewed in Milano, Long, and Fujiwara29 ).
We have previously shown, that Mkl1 is up-regulated with megakaryocytic maturation and that Mkl1 KO mice have mild thrombocytopenia, with an increase in the absolute number of CD41+ megakaryocytes in BM, with an increase in megakaryocyte progenitors at the expense of mature megakaryocytes.21 Interestingly, bleeding times in these mice were barely abnormal (E. Cheng and D.S.K., unpublished data). Overexpression of Mkl1 in HEL cells, a bipotent human leukemia cell line capable of megakaryocytic differentiation, and in primary human CD34+ cells resulted in an increase in megakaryocyte maturation and ploidy. Recent data by Gilles et al30 show a role for Mkl1 in proplatelet formation and megakaryocyte cytoskeletal organization. Mkl1 knockdown results in abnormal megakaryocytes with decreased adhesion, terminal maturation, and proplatelet formation, potentially explained by decreased expression of Myl9 and matrix metalloproteinase 9.30 Interestingly, the study does not detect a reduction in ploidy by knockdown of Mkl1, contrary to our in vivo data in Mkl1 KO mice and our in vitro overexpression model. Incomplete knockdown of Mkl1 or technical differences may explain such discrepancy. Pf4/Srf KO mice have an increase in megakaryocytes with overall decreased ploidy. Ploidy analysis of cultured BM-derived megakaryocytes (supplemental Figure 2) though would suggest, that Srf−/− megakaryocytes can reach high ploidy, possibly less efficiently or “masked” by increased proliferation of immature megakaryocytes, as suggested by the increase in megakaryocyte progenitors.
Until now the function of Srf in megakaryopoiesis appeared defined by its role as a cofactor for Mkl1 signal transduction.21,30 However our data suggest that other regulatory pathways may play a major role in Srf physiology leading to the more profound defect in megakaryocytes lacking Srf than Mkl1. One possible explanation is that Mkl2, which is expressed in megakaryocytes (Table 1), performs redundant functions with Mk11 in megakaryocyte maturation ameliorating the phenotype in Mkl1 KO mice. Of note, as expected neither Mkl1 nor Mkl2 expression are statistically significantly different in KO versus WT Pf4/Srf megakaryocytes. Alternatively, Srf may be able to recruit additional proteins to serum response elements, such as the TCFs,31 which belong to the Ets family of transcription factors. TCFs and the myocardin family transcription factors are known to compete for Srf binding and activate different gene expression programs.15 Future work will be needed to identify the context specific binding partners of SRF in megakaryocytes.
The actin cytoskeleton and its dynamic rearrangement in response to cell signaling are essential to megakaryopoiesis and platelet formation and function.32 Mature megakaryocytes form long processes within the BM, proplatelets, from which platelets (and sometimes larger fragments) are released.33-35 Although the microtubule system is required for proplatelet elongation, actin-mediated forces are required for the bending and branching of the tubules leading to proplatelet amplification. However, little is known with regard to how actin contributes to this process and the cell signaling initiating this process.36,37 A role for filamins in thrombopoiesis and platelet function has been described. Filamins are large, actin cross-linking proteins that connect multiple transmembrane and signaling proteins to the cytoskeleton. Flna is the most abundant and widely expressed38,39 and links integrin signaling and reorganization of the actin cytoskeleton.40,41 In platelets, Flna links the GPIb-IX-V complex, the VWF receptor to the actin cytoskeleton via binding to the GPIbα subunit42-45 Complete knockout of Flna in mice leads to fetal death between days 9 and 12 because of severe cardiac structural defects,46 as well as internal bleeding.37 Disruption of the GPIb-IX-V complex leads to macrothrombocytopenia and bleeding diathesis as seen in Bernard-Soulier syndrome. GPIbα KO mice recapitulate the human disease with macrothrombocytopenia and bleeding diathesis with abnormal F-actin distribution.47 This suggests, that decreased expression of Flna may contribute to the phenotype we observe in the Pf4/Srf KO mice, in particular the macrothrombocytopenia and the decrease in proplatelet formation. However, the bleeding defect appears more prominent in GPIbα−/− mice than in Pf4/Srf KO mice, whereas Pf4/Srf KO mice show not only a prominent increase in total BM megakaryocytes, but also a left shift with concomitant decrease in ploidy.
Neither the role of Coro1a nor that of Cnn2 have been assessed in megakaryopoiesis or platelet function. Coro1a belongs to the coronin family of F-actin– and Arp2/3-binding proteins, is exclusively expressed in hematopoietic cells, and has been implicated in modulating the association of membranes with the cytoskeleton.48 Coro1a KO mouse studies show that this factor exerts an inhibitory effect on steady state F-actin formation via an Arp2/3-dependent mechanism.49 The predominant hematologic lineage affected in Coro1a KO mice are lymphocytes49 and the platelet and other blood counts are normal. However, megakaryopoiesis and platelet function have not been studied in detail.49
Calponin is an actin filament-associated regulatory protein expressed in smooth muscle and nonmuscle cells. It functions as an inhibitor of the actin-activated myosin ATPase, thereby regulating actin-myosin interaction, and the interaction of actin with other structural and regulatory proteins. It has been ascribed a role as a sensor of mechanical tension resulting in modification of the actin cytoskeleton50 whereby it may control cell motility and cytokinesis.51,52 Forced overexpression of calponin in cultured smooth muscle cells and fibroblasts transfected with viral promoter-directed expression vectors inhibited the rate of cell proliferation and the completion of cell division.52 The function of calponin and coronin, specifically megakaryopoiesis, platelet formation and function, remains to be shown. Down-regulated expression of these 3 genes suggests that in megakaryocytes as shown in muscle cells, SRF functions as a “master regulator” of the actin cytoskeleton yet again emphasizing the important role of the actin cytoskeleton in megakaryopoiesis and thrombopoiesis.
Taking our knowledge to date about Mkl1and Srf function in megakaryopoiesis, it appears that Mkl1 regulation of Srf-mediated transcriptional activation is particularly relevant to megakaryocyte maturation (constitutive Mkl1 KO has little effect until megakaryocyte commitment). However, the profound phenotype of megakaryocyte-specific Srf deletion classifies Srf as an essential transcription factor in megakaryopoiesis. Further studies will be needed to dissect signaling pathways regulating Srf-mediated transcriptional activation and divergent functions of transcriptional programs on megakaryopoiesis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Stephanie Donaldson for excellent care of and help with mice, Geoff Lyons and Zhao Zhao in the Yale Cell Sorter Facility for flow cytometric cell sorting, Madhuri Kambhampati and Susannah Kassmer for help with immunofluorescence staining, and Archibald Perkins for helpful discussion of the data. We are indebted to the Yale Center of Excellence in Molecular Hematology for assistance with cell analysis as well as bioinformatics.
This study was supported in part by National Institutes of Health (NIH) grant K08 DK073366 (S.H.). This project has been funded by NIH/National Heart, Lung, and Blood Institute Contract No. N01-HV-28186, by NIH grants HL63357, DK072442, HL68130, and CA016359, and by Connecticut grant no. 06SCB18. The Yale Center of Excellence in Molecular Hematology was funded by NIH grant DK072442.
National Institutes of Health
Authorship
Contribution: S.H. designed and performed experiments, analyzed data and wrote the manuscript; Y.G., K.H., and S.M. performed experiments and analyzed data; J.E.I. performed experiments, analyzed data, and wrote the manuscript; V.S. analyzed data; S.L. performed experiments; G.M.K. discussed the work and served as a consultant; and D.S.K. supervised the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephanie Halene, Department of Internal Medicine and Yale Cancer Center, Section of Hematology, Yale School of Medicine, 333 Cedar St, New Haven, CT 06520; e-mail: stephanie.halene@yale.edu.