Abstract
Spectrin and protein 4.1R crosslink F-actin, forming the membrane skeleton. Actin and 4.1R bind to one end of β-spectrin. The adjacent end of α-spectrin, called the EF domain, is calmodulin-like, with calcium-dependent and calcium-independent EF hands. The severely anemic sph1J/sph1J mouse has very fragile red cells and lacks the last 13 amino acids in the EF domain, implying that the domain is critical for skeletal integrity. To test this, we constructed a minispectrin heterodimer from the actin-binding domain, the EF domain, and 4 adjacent spectrin repeats in each chain. The minispectrin bound to F-actin in the presence of native human protein 4.1R. Formation of the spectrin-actin-4.1R complex was markedly attenuated when the minispectrin contained the shortened sph1J α-spectrin. The α-spectrin deletion did not interfere with spectrin heterodimer assembly or 4.1R binding but abolished the binary interaction between spectrin and F-actin. The data show that the α-spectrin EF domain greatly amplifies the function of the β-spectrin actin-binding domain (ABD) in forming the spectrin-actin-4.1R complex. A model, based on the structure of α-actinin, suggests that the EF domain modulates the function of the ABD and that the C-terminal EF hands (EF34) may bind to the linker that connects the ABD to the first spectrin repeat.
Introduction
The red blood cell (RBC) membrane skeleton is formed principally by α2β2-spectrin heterotetramers that crosslink short protofilaments of F-actin at the distal (tail) ends of spectrin with the aid of protein 4.1R, which binds to both proteins. Actin and protein 4.1R bind to calponin homology (CH) domains in the actin-binding domain (ABD) at the N-terminus of the spectrin β-chain.1 The adjacent, C-terminal end of α-spectrin, called the EF domain, contains an N-terminal pair of calmodulin-like, Ca2+-responsive EF hands, termed EF12, that bind Ca2+ with affinities in the low millimolar range,2 and a C-terminal pair of Ca2+-insensitive EF hands, EF34, similar to those in α-actinin,3 which extend to the C-terminus of the α-spectrin protein. Because Ca2+ is not bound at the micromolar concentrations that exist inside red cells and because Ca2+ has no apparent effect on the binding of erythrocyte spectrin to actin, the EF domain is generally assumed to be inert and vestigial in red cells. However, the sph1J/sph1J mouse, which has severe hereditary spherocytosis and unstable red cell membranes, makes a mutant α-spectrin that lacks the last 13 amino acids of the EF domain and the protein.4 The mutant protein is overexpressed by several-fold but is poorly incorporated into the RBC membrane skeleton, showing that the domain has some important but undiscovered function.
To test this possibility, we constructed a minispectrin heterodimer from the actin-binding domain, the EF domain, and 4 adjacent spectrin repeats in each chain. Like native spectrin, the minispectrin interacts weakly with F-actin alone in a pelleting assay but interacts strongly in the presence of protein 4.1R. Formation of the minispectrin-actin-4.1R ternary complex is greatly attenuated when the 13 C-terminal amino acids of α-spectrin are deleted, as in the sph1J mouse, confirming that the EF domain plays a major and previously unappreciated role in promoting spectrin-actin binding.
Methods
Buffers
Buffers included phosphate-buffered saline (150mM NaCl, 5mM Na phosphate, 0.5mM ethyleneglycoltetraacetic acid, pH 8.0) and binding buffer (120mM KCl, 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.5mM ethyleneglycoltetraacetic acid, 0.5mM dithiothreitol, pH 7.5).
Recombinant spectrin peptides
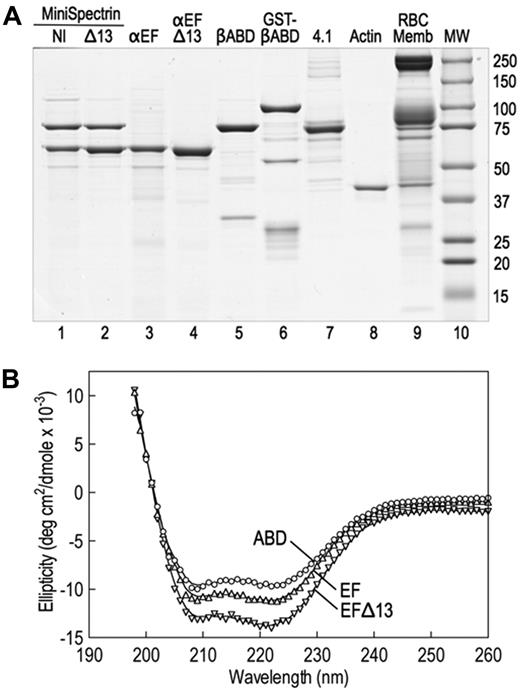
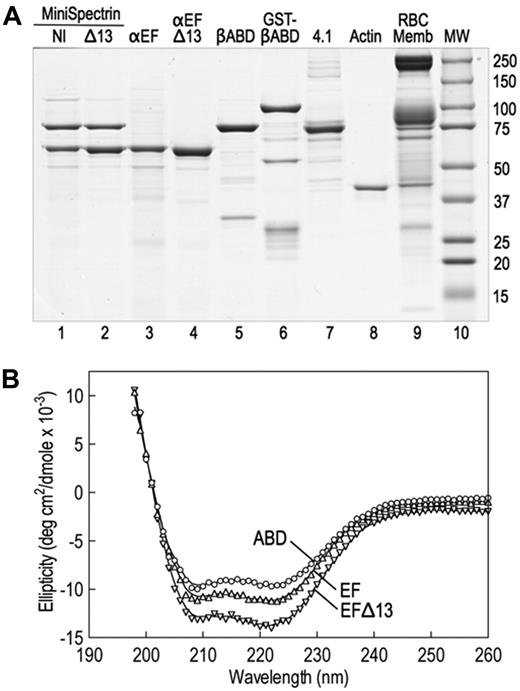
The spectrin glutathione S-transferase (GST) fusion proteins were made by subcloning polymerase chain reaction amplicons into GST expression vectors (GE Healthcare). cDNA clones of human spectrin AI (SPTA1; GenBank M61877), using the corrected carboxyterminal sequence (GenBank AF060556), and human spectrin BI (SPTB, GenBank J05500) were used as templates. α18-21EF (70 918 Da, amino acids 1805-2418) and α18-21EFΔ13 (69 390 Da, amino acids 1805-2417) were subcloned into pGex-6P. βABD1-4 (86 396, amino acids 1-743) was cloned into pGex2T. All constructs were sequenced to verify their sequence. The proteins were expressed in Escherichia coli BL21. A small overnight culture grown at 37°C was diluted 1:20 and grown further at 30°C until the OD600 was 0.5 to 0.7. Expression was induced with 1mM isopropyl-β-D-thiogalactopyranoside, and the cultures were grown at 30°C for an additional 2 hours. The bacterial pellets were stored at −80°C. Proteins were isolated at 4°C using the protocol of Harper et al,5 with modifications. The pellet from a 500-mL culture was resuspended in 10 mL of 130mM NaCl, 10mM sodium phosphate, pH 7.4, 5mM ethylenediaminetetraacetic acid, 5 μg/mL leupeptin, 5 μg/mL pepstatin, and 5μM diisopropylfluorophosphate and sonicated 3 times for 30 seconds on ice; 1 mL of packed glutathione (GSH) Sepharose beads (GE Healthcare) was added to the centrifuged (30 minutes, 180 000g) supernatant, and the suspension was tumbled at 4°C for 1 hour. The protein-bound GSH beads were washed 4 times in phosphate-buffered saline, and, when desired, the recombinant proteins were cleaved from GST with PreScission protease (pGex-6P) according to the manufacturer's directions (GE Healthcare) or with thrombin (pGex-2T), 40 U/mL beads at 25°C for 4 hours. Protease digestions were terminated with 5μM diisopropylfluorophosphate. Uncleaved GST proteins were eluted from GSH beads with 10mM GSH for 4 hours at 4°C and dialyzed versus 2 changes of 2 L of binding buffer for 48 hours. Before binding experiments, all proteins were dialyzed against binding buffer. Examples of the recombinant fusion proteins are shown in Figure 1A.
CD measurements
Stock solutions of the recombinant proteins were diluted in 10mM sodium phosphate buffer, pH 7.4, 0.5mM ethyleneglycoltetraacetic acid to the following concentrations: α18-21EF to 0.05 mg/mL, α18-21EFΔ13 to 0.0323 mg/mL, and βABD1-4 to 0.0387 mg/mL. The circular dichroism (CD) profiles of these samples were obtained from 260 nm to 195 nm in 3.0-mL cuvettes with a 1.0-cm path length in an AVIV Circular Dichroism Spectrophotometer, Model 62DS (Aviv Biomedical Inc). Protein concentrations of the stock protein solutions were obtained from absorption at 280 nm using molar extinction coefficients estimated from the amino acid analysis of each peptide. The measured ellipticities (Δϵ) were corrected to molar ellipticities (θ, in units of degrees cm2/dmol) using calculated mean residue weights of 115.5, 115.5, and 116.3 for α18-21EF, α18-21EFΔ13, and βABD1-4, respectively. The spectra of the recombinant proteins are shown in Figure 1B and indicate that the proteins are folded and predominantly α-helical.
Minispectrin heterodimers
Minispectrins were prepared as described by Begg et al.6 Equimolar amounts of the minispectrin peptides were incubated for 1 hour on ice and chromatographed on a prepacked Sephacryl S-200 column (GE Healthcare, catalog no. 17-1166-01) at 10 mL/hr; 2-mL fractions were collected. The elution profile was monitored by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Fractions with equimolar amounts of α- and β-spectrin peptides were pooled and used for binding.
Protein 4.1R
Human erythrocyte protein 4.1R was extracted with 2M Tris from erythrocyte membranes depleted of band 6, spectrin, and actin. The extraction procedure of Dotimas et al7 was used with one unit of deidentified human blood. The 2M Tris extract was dialyzed after extraction against 3 changes of 2 L of 5mM Na phosphate, 0.5mM ethyleneglycoltetraacetic acid, pH 7.4, centrifuged (30 minutes, 180 000g), and loaded on a 4-mL Q-Sepharose column (GE Healthcare) equilibrated with the dialysis buffer. The column was washed with 60 mL of 50mM NaCl in column buffer, and protein 4.1R was eluted with 50 mL of 220mM NaCl in column buffer. After evaluation by SDS-PAGE, the 4.1R was dialyzed against binding buffer.
Rabbit skeletal muscle actin
Rabbit muscle acetone powder was purchased from Pel-Frez. The actin was prepared by the method of Spudich and Watt,8 with the difference that F-actin was extracted with 800mM KCl to dissociate the troponin rather than 0.6mM KCl. The actin concentration was determined using the extinction coefficient of 0.617 (1 mg/mL, 1 cm light path) at 290 nm.
SDS-PAGE and protein determinations
SDS gel electrophoresis was performed in 9% Laemmli gels.9 The concentration of protein 4.1R was determined with the Bio-Rad dye-binding assay using bovine serum albumin as the reference standard. The concentrations for the recombinant proteins were determined using extinction coefficients calculated from the amino acid sequence with the DNASTAR program Lasergene.
Radioactive labeling of proteins
Proteins were labeled with [125I]-Bolton Hunter reagent,10 according to the manufacturer's directions (PerkinElmer Life and Analytical Sciences, catalog no. NEX-120).
GST pull-down binding experiments
The protein components and buffers were mixed in a final volume of 300 μL in a 1.5-mL tube and tumbled at 25°C for 60 minutes. The binding buffer, described in “Buffers”, also contained 0.5 μg/mL leupeptin, pepstatin, and Pefabloc, 1 mg/mL bovine serum albumin, 2mM MgCl2, and 0.5mM adenosine triphosphate. A total of 40 μL of GSH beads, 1:1 in binding buffer, were added to each sample and tumbled an additional 60 minutes at 25°C. The total reaction was loaded in an Ultrafree-MC centrifugal filter device (Millipore, catalog no. UFC30GVNB), placed in a 10 × 73-mm polystyrene tube, and centrifuged for 25 minutes at 2000g in a tabletop Beckman centrifuge. The pellets were washed once with 100 μL of binding buffer, and the bound proteins were eluted from the beads with 100 μL of 1 × SDS gel sample buffer. After centrifugation through the filter, the samples were either analyzed on SDS gels or, if labeled with 125I, counted in a γ-counter.
F-actin pelleting experiments
These experiments were identical to those in “GST pull-down binding experiments,” except that the final reaction volume was 200 μL. After the one-hour binding reaction, 2 70-μL aliquots at each point were centrifuged in Beckman cellulose propionate tubes in a Beckman 42.2Ti rotor at 50 000g for 20 minutes to sediment the F-actin and associated proteins. Free [125I]-protein was measured from a 20-μL aliquot taken from the supernatant meniscus of each tube and counted in a γ-counter. Two 20-μL aliquots of the reaction before centrifugation were counted and used as the value for [125I]-protein added.
Structure prediction
We used the PSIPRED server (http://bioinf4.cs.ucl.ac.uk:3000/psipred), the Advanced Protein Secondary Structure Prediction Server (http://imtech.res.in/raghava/apssp), and the PROF Secondary Structure Prediction System (http://www.aber.ac.uk/∼phiwww/prof) to predict secondary structures from amino acid sequences. The program K2D2 was used to estimate the structure of peptides from far ultraviolet CD measurements (http://www.ogic.ca/projects/k2d2).
Results
The minispectrin mimics full-length spectrin in interactions with actin and protein 4.1R
We constructed 2 minispectrins by combining a GST fusion protein containing the actin-binding domain of β-spectrin and the first 4 spectrin repeats (GST-βABD1-4) with the last 4 repeats in the α-spectrin subunit followed by the EF domain. One minispectrin had the normal EF domain (α18-21EF). The other lacked the C-terminal 13 amino acids that are deleted in the sph1J mutation (α18-21EFΔ13). Both peptides were stably expressed (Figure 1A lanes 3-4). Because spectrin repeats β1 to β4 and α18 to α21 contain the sites that nucleate the side-to-side interaction of α- and β-spectrins to form the heterodimer,6,11 the 2 partial spectrin chains react spontaneously to form a minispectrin heterodimer (Figure 1A lanes 1-2). The peptides used to form the dimer contain some additional minor bands (Figure 1A lanes 3-5), but the minispectrin is pure after chromatography on Sephacryl G-200 (Figure 1A lanes 1-2). The minispectrin peak is symmetric and elutes at a mass of 209 000 Da on a column calibrated with globular proteins. This is reasonably close to the expected mass of 184 000 Da, especially if the minispectrin is elongated, as would be expected given the attached spectrin repeats. The data suggest that the minispectrin is homogeneous and is a monomer.
Native and recombinant proteins used in this study. (A) Coomassie blue-stained Laemmli SDS-PAGE. The assembled and purified minispectrins are in lanes 1 and 2. (B) CD spectra of the recombinant α18-21EF (EF), α18-21EFΔ13 (EFΔ13), and β1-4ABD peptides. The spectra display the troughs at 208 nm and 222 nm characteristic of α-helical peptides, indicating that the recombinant proteins are folded. Structure estimation predicts approximately 75% α-helix for the βABD1-4 peptide, approximately 80% for α18-21EF, and approximately 85% for α18-21EFΔ13. However, the latter estimate is rough because the error in the estimate is high. Very little β-structure (< 2%) is predicted. The data approximate what would be expected from the known structures of the highly α-helical spectrin repeats and EF hands that constitute most of the α18-21EF peptide and the spectrin repeats and CH domains that make up much of the βABD1-4 peptide. The fact that the mutant peptide α18-21EFΔ13 appears to be even more helical than α18-21EF suggests that the C-terminal 13 amino acid deletion substantially alters the structure of the EF domain.
Native and recombinant proteins used in this study. (A) Coomassie blue-stained Laemmli SDS-PAGE. The assembled and purified minispectrins are in lanes 1 and 2. (B) CD spectra of the recombinant α18-21EF (EF), α18-21EFΔ13 (EFΔ13), and β1-4ABD peptides. The spectra display the troughs at 208 nm and 222 nm characteristic of α-helical peptides, indicating that the recombinant proteins are folded. Structure estimation predicts approximately 75% α-helix for the βABD1-4 peptide, approximately 80% for α18-21EF, and approximately 85% for α18-21EFΔ13. However, the latter estimate is rough because the error in the estimate is high. Very little β-structure (< 2%) is predicted. The data approximate what would be expected from the known structures of the highly α-helical spectrin repeats and EF hands that constitute most of the α18-21EF peptide and the spectrin repeats and CH domains that make up much of the βABD1-4 peptide. The fact that the mutant peptide α18-21EFΔ13 appears to be even more helical than α18-21EF suggests that the C-terminal 13 amino acid deletion substantially alters the structure of the EF domain.
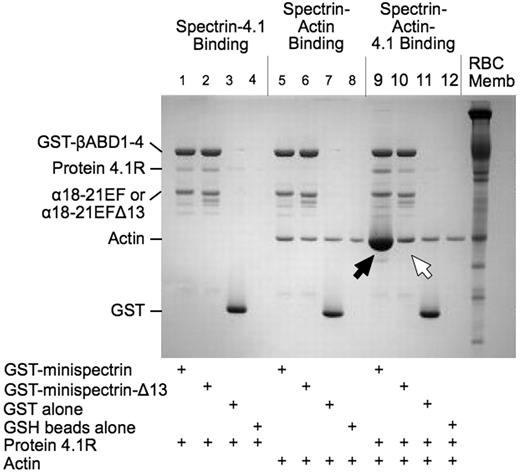
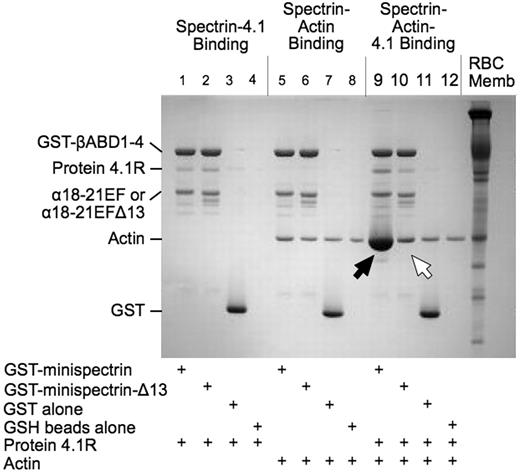
As shown in Figure 2, when the normal minispectrin is retrieved on GSH beads after incubation with F-actin and protein 4.1R, large amounts of actin are bound (Figure 2 lane 9, black arrow). Very little actin is bound in the absence of protein 4.1R (lane 5). There is also an increase in the amount of protein 4.1R bound (lane 9), which may reflect the increased binding affinity of 4.1R in the ternary complex or some binding of protein 4.1R to actin alone.12 When the binding is measured the opposite way, by pelleting F-actin and assaying bound [125I]-labeled minispectrin, a similar result is observed (Figure 3A): robust binding in the presence of 4.1R and minimal binding without. Binding approaches a plateau at a level corresponding to use of all the 4.1R in the system. The addition of 20μM to 1mM free Ca2+ to the reaction had no effect on binding (data not shown). The binary and ternary complex binding curves are similar to the curves obtained by one of us (S.E.L.) previously with intact, native erythrocyte spectrin,13 showing that the minispectrin satisfactorily approximates the intact molecule.
Interactions of normal and sph1J mutant minispectrins with actin and protein 4.1R assessed by a GST pulldown assay. Minispectrin-4.1R (lanes 1-4): 4.1R binds equally to both the normal and mutant minispectrins and does not bind to GST alone or to the GSH beads. Minispectrin-actin (lanes 5-8): The results show a slight interaction of the normal minispectrin with actin (more actin is bound in lane 5 than in lane 6). The mutant minispectrin does not interact (ie, the amount of actin bound is the same as the nonspecific binding observed in the controls). Minispectrin-actin-4.1R (lanes 9-12): A large amount of actin binds to the normal minispectrin (lane 9, black arrow), but only a small amount binds to the minispectrin lacking the C-terminal 13 amino acids of the α-spectrin EF domain (lane 10, white arrow).
Interactions of normal and sph1J mutant minispectrins with actin and protein 4.1R assessed by a GST pulldown assay. Minispectrin-4.1R (lanes 1-4): 4.1R binds equally to both the normal and mutant minispectrins and does not bind to GST alone or to the GSH beads. Minispectrin-actin (lanes 5-8): The results show a slight interaction of the normal minispectrin with actin (more actin is bound in lane 5 than in lane 6). The mutant minispectrin does not interact (ie, the amount of actin bound is the same as the nonspecific binding observed in the controls). Minispectrin-actin-4.1R (lanes 9-12): A large amount of actin binds to the normal minispectrin (lane 9, black arrow), but only a small amount binds to the minispectrin lacking the C-terminal 13 amino acids of the α-spectrin EF domain (lane 10, white arrow).
Minispectrin-actin-4.1R binding assessed by an F-actin pelleting assay. (A) Effect of protein 4.1R on minispectrin-actin interactions. F-actin (2.38μM as G-actin) was combined with increasing amounts of [125I]-minispectrin, with or without protein 4.1R (0.38μM), and the bound minispectrin was measured after pelleting the F-actin. (B) Comparison of the amount of ternary complex formed by normal and mutant minispectrins. F-actin (2.38μM as G-actin) and protein 4.1 (0.38μM) were combined with increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13, and the bound minispectrin was measured after pelleting the F-actin. Residual binding of the minispectrin to F-actin in the absence of 4.1R was subtracted from each curve to give 4.1R-specific binding. Note the marked ineffectiveness of the minispectrin bearing the α-spectrin C-terminal deletion found in the sphIJ mouse with unstable red cell membranes and severe hemolytic anemia (minispectrin-ΔC13).
Minispectrin-actin-4.1R binding assessed by an F-actin pelleting assay. (A) Effect of protein 4.1R on minispectrin-actin interactions. F-actin (2.38μM as G-actin) was combined with increasing amounts of [125I]-minispectrin, with or without protein 4.1R (0.38μM), and the bound minispectrin was measured after pelleting the F-actin. (B) Comparison of the amount of ternary complex formed by normal and mutant minispectrins. F-actin (2.38μM as G-actin) and protein 4.1 (0.38μM) were combined with increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13, and the bound minispectrin was measured after pelleting the F-actin. Residual binding of the minispectrin to F-actin in the absence of 4.1R was subtracted from each curve to give 4.1R-specific binding. Note the marked ineffectiveness of the minispectrin bearing the α-spectrin C-terminal deletion found in the sphIJ mouse with unstable red cell membranes and severe hemolytic anemia (minispectrin-ΔC13).
Deletion of the C-terminal 13 amino acids of α-spectrin greatly decreases F-actin binding
The minispectrin containing the sph1J mutation binds far less F-actin than the normal minispectrin in the presence of protein 4.1R (Figure 2 lane 10, white arrow) and essentially no F-actin in its absence (compare lane 6 with the controls in lanes 7 and 8). Similarly, in the pelleting assay with all 3 components, the mutant minispectrin binds to F-actin with much lower apparent affinity than the normal minispectrin does (Figure 3B). In this experiment, binding of the normal minispectrin to F-actin saturates at a value less than the concentration of protein 4.1R in the reaction, indicating that some of the protein 4.1R is inactive. In our experience, there is considerable variation in the ability of different preparations of purified, native 4.1R to promote spectrin-actin binding; however, the same preparation of 4.1R is used in both of the reactions in Figure 3B and in all other experiments where normal and mutant minispectrins are compared, so the relative ineffectiveness of the mutant minispectrin, compared with the normal, is unchanged. In addition, we performed multiple replicates of all experiments using different preparations of protein 4.1R to ensure the results were reproducible.
When the assay is reversed and the binding of [125I]-labeled protein 4.1R to F-actin is measured at increasing minispectrin concentrations (Figure 4A) or the binding of [125I]-labeled minispectrin to F-actin is measured at increasing 4.1R concentrations (Figure 4B), the mutant minispectrin is again much less effective. In both cases, the mutant protein exhibits a lower apparent affinity and a lower binding capacity at saturation.
Protein 4.1 and minispectrin-dependent ternary complex formation. (A) Minispectrin-dependent binding of [125I]-protein 4.1 to F-actin. [125I]-Protein 4.1 (0.38μM), F-actin (2.38μM as G-actin), and increasing amounts of either minispectrin or minispectrin-Δ13 were combined, and the bound 4.1R was measured after pelleting the F-actin. (B) Protein 4.1-dependent binding of [125I]-minispectrin or [125I]-minispectrin-Δ13 to F-actin. Increasing amounts of protein 4.1R were added to the [125I]-labeled minispectrins (0.54μM) and F-actin (2.38μM), and each bound minispectrin was measured after pelleting the F-actin. Binding of the mutant minispectrin is impaired under both conditions.
Protein 4.1 and minispectrin-dependent ternary complex formation. (A) Minispectrin-dependent binding of [125I]-protein 4.1 to F-actin. [125I]-Protein 4.1 (0.38μM), F-actin (2.38μM as G-actin), and increasing amounts of either minispectrin or minispectrin-Δ13 were combined, and the bound 4.1R was measured after pelleting the F-actin. (B) Protein 4.1-dependent binding of [125I]-minispectrin or [125I]-minispectrin-Δ13 to F-actin. Increasing amounts of protein 4.1R were added to the [125I]-labeled minispectrins (0.54μM) and F-actin (2.38μM), and each bound minispectrin was measured after pelleting the F-actin. Binding of the mutant minispectrin is impaired under both conditions.
The α-spectrin deletion does not affect spectrin heterodimer assembly or the binding of protein 4.1 R by spectrin but does affect the spectrin-actin interaction
One trivial possibility to explain the results shown in Figures 3 and 4 is that the C-terminal deletion in the sph1J mutation alters the assembly of the minispectrin heterodimer or inhibits its interaction with protein 4.1R. Neither is the case. Indeed, the mutant α-spectrin peptide (α18-21EFΔ13) actually has approximately 2 to 3 times higher affinity for the β-spectrin peptide (GST-βABD1-4) than does the normal α-spectrin peptide (α18-21EF) (Figure 5A). The same pattern was observed in 3 independent experiments. GST-βABD1-4 bound the mutant peptide, α18-21EFΔ13, with a dissociation constant (Kd) of approximately 0.2μM and bound the normal peptide, α18-21EF, with a Kd of approximately 0.5μM. As expected, the binding saturates at a molar ratio of approximately 1:1 (average 1:1.1).
Heterodimer assembly and protein 4.1R binding. (A) Assembly of the minispectrin heterodimer. GST-βABD1-4 (0.87μM) was mixed with increasing amounts of [125I]-α18-21EF or α18-21EFΔ13 (0.14-2.12μM), and the complex was retrieved on GSH beads as described in “GST pull-down experiments” (B) Protein 4.1R binding to minispectrin. Increasing amounts of [125I]-protein 4.1 (0.18-1.36μM) were added to GST-minispectrin or GST-minispectrin-Δ13 (each 0.54μM). In each experiment, binding of the [125I]-ligand to GSH beads alone was subtracted from the values with added spectrin. (C) Minispectrin binding to F-actin in the absence of protein 4.1R. Increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13 were added to F-actin (2.38μM as G-actin), and the bound minispectrin was measured after pelleting the F-actin. Results from 3 independent experiments are combined. Note that the diminished ability of the mutant minispectrin to form the ternary complex is not a failure to assemble the minispectrin molecule or to bind protein 4.1R, but a failure to bind to F-actin.
Heterodimer assembly and protein 4.1R binding. (A) Assembly of the minispectrin heterodimer. GST-βABD1-4 (0.87μM) was mixed with increasing amounts of [125I]-α18-21EF or α18-21EFΔ13 (0.14-2.12μM), and the complex was retrieved on GSH beads as described in “GST pull-down experiments” (B) Protein 4.1R binding to minispectrin. Increasing amounts of [125I]-protein 4.1 (0.18-1.36μM) were added to GST-minispectrin or GST-minispectrin-Δ13 (each 0.54μM). In each experiment, binding of the [125I]-ligand to GSH beads alone was subtracted from the values with added spectrin. (C) Minispectrin binding to F-actin in the absence of protein 4.1R. Increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13 were added to F-actin (2.38μM as G-actin), and the bound minispectrin was measured after pelleting the F-actin. Results from 3 independent experiments are combined. Note that the diminished ability of the mutant minispectrin to form the ternary complex is not a failure to assemble the minispectrin molecule or to bind protein 4.1R, but a failure to bind to F-actin.
Similarly, the mutant and normal minispectrins bind protein 4.1R equivalently (Figure 5B; Figure 2 lanes 1-2). The stoichiometry at saturation averages 1:1.25, perhaps because of some oligomerization of the protein 4.1R. The affinity of the 4.1R-minispectrin interaction (Kd ∼ 0.2μM) is considerably greater than the affinity of the isolated actin-binding domain for 4.1R calculated from the data of An et al (Kd ∼ 1.2μM).1 It is close to the Kd values measured for the interaction of protein 4.1R with intact erythrocyte spectrin by Tyler et al (Kd = 0.1μM)14 and Wolfe et al (Kd = 0.2μM)15 using pelleting assays, but much less than the Kd of 0.012μM that Podgórski and Elbaum obtained using fluorescence and microcalorimetry techniques.16 Whichever number is correct, it appears that the α-spectrin EF domain or the adjacent spectrin repeats enhance the binding of protein 4.1R to the actin-binding domain by a factor of approximately 5 to 10.
The data in Figure 5A-B suggest that the C-terminal deletion of α-spectrin in the sph1J mouse must primarily affect the binary interaction of the minispectrin with F-actin, and Figure 5C supports this deduction. Spectrin-actin binding is slight at the concentrations we were able to use, but clearly, the mutant minispectrin binds much less well to F-actin than the normal construct, and indeed hardly binds at all. The estimated Kd of the normal minispectrin-actin interaction is approximately 1μM in Figure 5C and 1.8μM in Figure 3A. Raae et al17 also constructed a minispectrin and obtained a Kd of 2.5μM for its interaction with F-actin. These 3 values are equivalent to an association constant (Ka) of 4 to 10 × 105 M−1, which is somewhat higher than the value of 2.5 × 105 M−1 obtained for the association of the isolated actin-binding domain with F-actin by An et al1 and much higher than the value of 5 × 103 M−1 estimated for the interaction of intact spectrin with F-actin in our earlier work.13
The isolated α- and β-spectrin peptides do not bind to F-actin in the presence of protein 4.1 under conditions where the minispectrins bind
This is shown in Figure 6. Neither α18-21EF nor βABD1-4 binds significantly to F-actin at the concentrations of each peptide used in the assay (0.38μM). Although it may seem surprising that βABD1-4 does not bind under these conditions, previous studies show that it takes 10 to 20μM βABD (amino acids 1-301) to inhibit ternary complex formation by half.1 Assuming βABD1-4 behaves like βABD, virtually no binding would have been expected at 0.38μM. In contrast, when the same concentrations of α18-21EF and βABD1-4 are combined in a minispectrin, considerable ternary complex forms (Figure 6). The data emphasize the great effect that adding the EF domain to the actin-binding domain has on formation of the ternary complex.
Comparison of the binding of [125I]-spectrin α18-21EF, [125I]-βABD1-4, [125I]-minispectrin, and [125I]-minispectrin-Δ13 to F-actin in the presence of protein 4.1R. The spectrin fragments and protein 4.1 were all 0.38μM. F-actin was 2.38μM in G-actin monomers. Error bars represent the range of 2 to 5 independent experiments for each protein. Neither the isolated α-spectrin EF domain nor the β-spectrin actin-binding domain binds to actin at these concentrations, whereas the assembled minispectrin binds robustly.
Comparison of the binding of [125I]-spectrin α18-21EF, [125I]-βABD1-4, [125I]-minispectrin, and [125I]-minispectrin-Δ13 to F-actin in the presence of protein 4.1R. The spectrin fragments and protein 4.1 were all 0.38μM. F-actin was 2.38μM in G-actin monomers. Error bars represent the range of 2 to 5 independent experiments for each protein. Neither the isolated α-spectrin EF domain nor the β-spectrin actin-binding domain binds to actin at these concentrations, whereas the assembled minispectrin binds robustly.
Discussion
The N-terminal end of β-spectrin is well known to bind F-actin and protein 4.1R through its CH domains,1 but the neighboring EF domain at the C-terminal end of α-spectrin has no known physiologic function. The domain contains 2 proximal EF hands (EF12) that resemble calmodulin structurally. They bind Ca2+ ions at nonphysiologic concentrations (Kd ∼ 0.5mM),1,18 transforming from a closed to open conformation, like calmodulin, in the process.19 There are also 2 distal EF hands (EF34)2 that belong to the Ca2+-independent class and are found in the same position in α-actinin, a related spectrin family member.3
The sph1J/sph1J mouse,4 which has severe hereditary spherocytosis and unstable red cell membranes, makes a mutant α-spectrin that lacks the last 13 amino acids of the EF domain, which extends to the C-terminus of the protein. The deletion removes the final α-helix of EF4. This helix interacts intimately with the initial α-helix of EF3 in the nuclear magnetic resonance structure of EF34 from α-actinin,3 so it is very probable that the deletion disrupts the overall structure of EF34 in α-spectrin. This is consistent with the CD spectra, which show a significant difference between α18-21EF and α18-21EFΔ13 (Figure 1B). EF34 folds independently of EF12,2 so it is possible that the structure and function of EF12 are not perturbed. This fits with our preliminary finding that Ca2+ binding, which is a function of EF12,2 is not significantly altered in the recombinant EF domain containing the sph1J mutation (data not shown). The mutant spectrin is overexpressed by several-fold but is poorly incorporated into the RBC membrane skeleton,4 showing that the domain has some important function.
We have recently found one such function. The EF domain binds protein 4.2, a 72-kDa peripheral membrane protein that associates with band 3, the erythrocyte anion channel.20 Spectrin-4.2 binding is amplified by micromolar amounts of Ca2+, and the Ca2+-sensitive binding but not the Ca2+-independent binding is blocked by Ca2+-calmodulin, which binds to the EF domain.20 Band 3, the red cell anion exchanger, also binds protein 4.2, but that association does not interfere with the interaction of protein 4.2 and α-spectrin. The data imply that a portion of band 3 is associated with the actin junctional complex and forms a second site for the attachment of the membrane skeleton to the lipid bilayer, complementing the spectrin-ankyrin-band 3 attachment at the other end of spectrin. Other recent reports show that band 3 interacts with adducin21 and is part of a macromolecular complex that attaches to protein 4.1 in vivo,22 supporting this idea.
However, loss of spectrin-4.2 binding would not be sufficient to explain the severe hemolysis that afflicts the sph1J/sph1J mouse because complete loss of protein 4.2 in mice23 or humans24 only produces mild hemolysis, spherocytosis, and anemia. This led us to test the hypothesis that the EF domain also contributes to the critical actin-binding function of spectrin, and the present studies clearly show this is true. The EF domain greatly amplifies binding of a minispectrin to F-actin in the presence of protein 4.1R (Figure 6), and this effect is muted by deletion of the last 13 amino acids in α-spectrin (Figure 3B), as in the sph1J mutant. The mutation does not significantly alter assembly of the minispectrin heterodimer (Figure 5A) or binding of protein 4.1R to the minispectrin (Figure 5B), but it does impair the binary interaction between the minispectrin and F-actin (Figure 5C).
Interestingly, 25 years ago, Cohen and Langley showed that β-chains isolated from native erythrocyte spectrin would not bind F-actin in the presence of protein 4.1R unless they were first recombined with α-spectrin chains.25 Unfortunately, this observation was overlooked or forgotten, perhaps because it was not the primary message of the paper. Isolated α-spectrin chains did not bind to F-actin in the presence of 4.1R in their hands, nor did the isolated minispectrin α-chains (α18-21EF) in ours (Figure 6). The maximal concentration of α18-21EF we tested was only 1.5μM. It is possible a higher concentration might bind, but we think it is more probable that the EF domain acts indirectly to regulate the actin-binding domain rather than as an accessory actin binding partner.
One problem in trying to answer this question is that we do not have direct information about the structural arrangement of the proteins in the spectrin-actin-4.1R complex. But it is probable that spectrin will resemble other spectrin family members that bind F-actin. α-Actinin is a well-studied example that features an N-terminal actin-binding domain containing 2 tandem CH domains, and a C-terminal EF domain containing EF hands homologous to EF12 and EF34 of spectrin, separated by 4 spectrin repeats. Because α-actinins are a homodimer of antiparallel chains, each end of α-actinin contains neighboring CH domains and EF domains. The arrangement of these is known from cryoelectron microscopy combined with homology modeling, using crystal or nuclear magnetic resonance structures of isolated domains.26,27 Additional information is available from structures of the actin-binding domains of other spectrin family members, such as fimbrin,28 utrophin,29 dystrophin,30 and plectin.31
Figure 7 is a model for spectrin based on the structure of skeletal muscle α-actinin,26 which contains Ca2+-dependent and Ca2+-independent EF hands such as those in spectrin.3 In the model, the 2 CH domains are in the “closed” conformation, where the CH domains are in close contact (Figure 7B). The CH domains can also adopt a more splayed-out “open” arrangement.27 It is not known which arrangement exists in spectrin, but in either case, the EF domain remains in contact with the CH domains and on the side away from actin (Figure 7B). Thus, the model supports the concept that the EF hands exert their influence on actin binding through the CH domains and do not bind directly to actin.
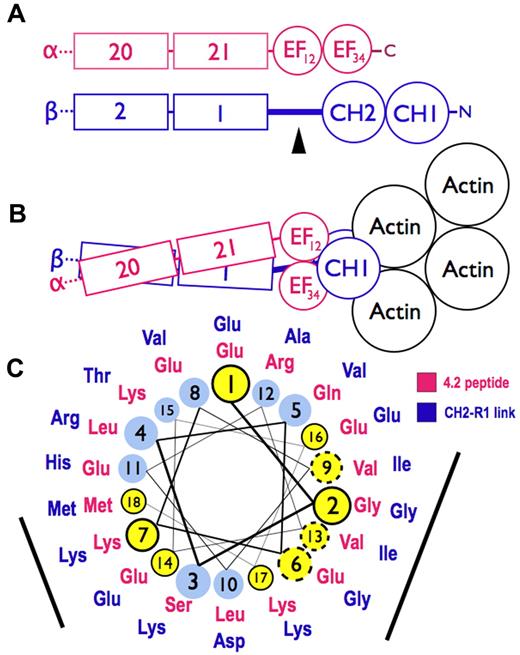
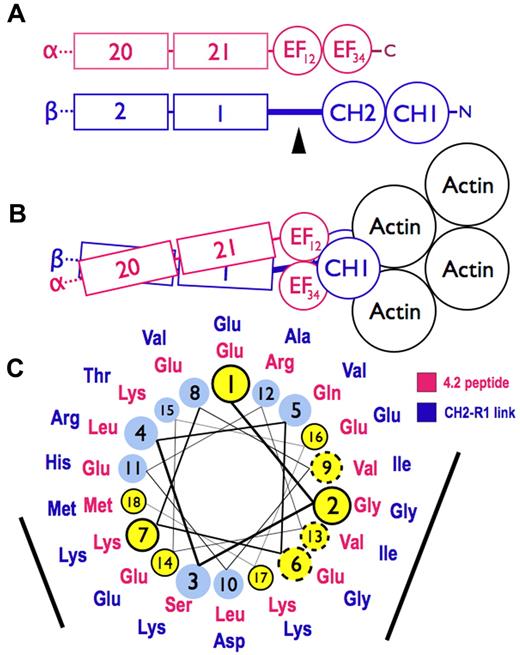
Models of spectrin-actin interactions based on the structure of α-actinin. (A) Hypothetical models, drawn roughly proportional to size, of α-spectrin (red) and β-spectrin (blue) chains near the tail end of the molecule. EF34 extends to the C-terminus of α-spectrin and is partially deleted in the sph mutation. The black arrowhead marks the segment between the CH2 domain and the first β-spectrin repeat (CH2-R1). (B) Model of the interaction of α- and β-spectrin deduced from the structure of skeletal muscle α-actinin.25 The view is from above, looking down on the membrane skeleton, with the actin filament lying parallel to the lipid bilayer. Note that in this structure the EF domains are turned 90 degrees relative to the CH domains and do not contact the actin filament. They lie near the linker separating the CH domains and the long CH2-R1 linker. The orientation of the CH domains varies from an open to closed conformation (here shown closed) in different actin-binding proteins, but the EF domain is always in contact with the CH domains and not the actin. (C) Helical wheel illustrating the similarity between the spectrin-binding peptide in protein 4.2 (red sequence),33 which binds to the EF domain of α-spectrin,19 and the CH2-R1 linker region in β-spectrin (blue sequence). Both peptides are predicted to form an α-helix. Solid black circles and dashed circles around yellow amino acid positions represent identical and conserved residues, respectively. The black bars represent 2 faces of the putative helices that are notably similar. We postulate that the CH2-R1 linker region of β-spectrin also binds to the EF hands, probably in the groove of EF34, and that this interaction regulates the protein 4.1R and/or actin binding of the adjacent CH domains.
Models of spectrin-actin interactions based on the structure of α-actinin. (A) Hypothetical models, drawn roughly proportional to size, of α-spectrin (red) and β-spectrin (blue) chains near the tail end of the molecule. EF34 extends to the C-terminus of α-spectrin and is partially deleted in the sph mutation. The black arrowhead marks the segment between the CH2 domain and the first β-spectrin repeat (CH2-R1). (B) Model of the interaction of α- and β-spectrin deduced from the structure of skeletal muscle α-actinin.25 The view is from above, looking down on the membrane skeleton, with the actin filament lying parallel to the lipid bilayer. Note that in this structure the EF domains are turned 90 degrees relative to the CH domains and do not contact the actin filament. They lie near the linker separating the CH domains and the long CH2-R1 linker. The orientation of the CH domains varies from an open to closed conformation (here shown closed) in different actin-binding proteins, but the EF domain is always in contact with the CH domains and not the actin. (C) Helical wheel illustrating the similarity between the spectrin-binding peptide in protein 4.2 (red sequence),33 which binds to the EF domain of α-spectrin,19 and the CH2-R1 linker region in β-spectrin (blue sequence). Both peptides are predicted to form an α-helix. Solid black circles and dashed circles around yellow amino acid positions represent identical and conserved residues, respectively. The black bars represent 2 faces of the putative helices that are notably similar. We postulate that the CH2-R1 linker region of β-spectrin also binds to the EF hands, probably in the groove of EF34, and that this interaction regulates the protein 4.1R and/or actin binding of the adjacent CH domains.
The way the EF hands do this is not known. One possibility, proposed by Tang et al,26 is that ligands cause the EF domains to bind the short linker connecting the CH domains, forcing them apart and altering actin binding. A second possibility is that the EF hands, specifically EF34, bind to the long linker that connects CH2 to the first spectrin repeat (R1) in beta spectrin, possibly altering the orientation of the 2 CH domains or their rearrangement on binding actin. In muscle α-actinin, EF34 binds some of the α-helical Z-repeats of titin, which lie in the groove between EF3 and EF4.3 The CH2-R1 linker in α-actinin, which is predicted to be an α-helix, is similar to the Z-repeats, and acts like an internal pseudoligand. It also binds to EF34, blocking the interaction with the titin Z-repeat.32 This autoinhibition is relieved in α-actinin by phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2), whose head group binds to CH2 and whose fatty acid tails may bind to the CH2-R1 linker.33 PI-4,5-P2 activates F-actin binding by α-actinin, and it also greatly amplifies the binding of protein 4.1R to the CH2 domain of β-spectrin.1
We suspect that this second mechanism also operates in spectrin. As noted earlier, the C-terminal EF hands in α-spectrin (EF34) are homologous to EF34 in muscle α-actinin,3,18 suggesting that they may also bind an α-helical ligand. And, as noted, we find that the EF domain binds protein 4.2.20 The 22-amino acid segment in protein 4.2 that binds spectrin (amino acids 470-492)34 lies on the surface of protein 4.235 and is probably helical according to the structure predicting algorithms indicated in “Structure prediction.” As shown in Figure 7C, it is similar to the CH2-R1 linker peptide, also predicted to be α-helical, along 2 faces of the helix. Both peptides are highly conserved in mammals. The titin Z-repeat α-helix lies in the groove of the α-actinin EF34 hands like a hot dog in a bun,3 contacting the bound helix on 2 sides, so we would expect 2 conserved surfaces on other α-helical ligands if they bind the same way. This suggests that EF34 may bind to both the CH2-R1 linker and to the spectrin binding site in protein 4.2, and, as in α-actinin, the competing interactions may constitute a regulatory switch, perhaps controlled by PI-4,5-P2, that alters the binding of actin or 4.1R to the CH2 domain. Because protein 4.1R also binds calmodulin at Ca2+-sensitive and Ca2+-insensitive sites36 and the EF domain resembles calmodulin structurally and itself binds calmodulin in the presence of Ca2+, a network of regulatory effects can be imagined.
We want to emphasize that we have no direct evidence supporting this hypothesis. Other hypotheses are easily imagined, including the possibility that the EF domain works by modifying the flexibility or structure of the actin-binding domain when the 2 are in contact in a minispectrin. Fortunately, all these speculations are testable.
Recently, 2 more missense mutations, sph3J and sph4J, have been identified in the EF domain or nearby spectrin repeats that produce a phenotype in mice similar to the sph1J mutation studied here.37 Similar mutations have not yet been described in humans with hereditary hemolytic anemias, but relatively few mutations have been identified in spectrin in hereditary spherocytosis, especially in α-spectrin, so it is possible that such mutations exist but have not yet been found. We hope the present paper will encourage investigators to look for C-terminal α-spectrin mutations in patients with hereditary spherocytosis, especially severe hereditary spherocytosis.
Presented in abstract form at the 47th Annual Meeting of the American Society of Hematology, Atlanta, GA, December 12, 2005.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Walter Gratzer and Luanne Peters for their review of the manuscript and Kelly Arnett from the Stephen Blacklow Laboratory, Brigham and Women's Hospital, Harvard Medical School for help with the circular dichroism measurements.
This work was supported by the National Institutes of Health (grant RO1 HL081608).
National Institutes of Health
Authorship
Contribution: C.K. performed the research, assisted with the design of experiments, collected and analyzed the data, and wrote the first draft of the manuscript; and S.E.L. designed the research, analyzed and interpreted the data, and wrote the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Samuel E. Lux, Department of Medicine, HU260.1, Children's Hospital Boston, 300 Longwood Ave, Boston, MA 02115; e-mail: lux@enders.tch.harvard.edu.

![Figure 3. Minispectrin-actin-4.1R binding assessed by an F-actin pelleting assay. (A) Effect of protein 4.1R on minispectrin-actin interactions. F-actin (2.38μM as G-actin) was combined with increasing amounts of [125I]-minispectrin, with or without protein 4.1R (0.38μM), and the bound minispectrin was measured after pelleting the F-actin. (B) Comparison of the amount of ternary complex formed by normal and mutant minispectrins. F-actin (2.38μM as G-actin) and protein 4.1 (0.38μM) were combined with increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13, and the bound minispectrin was measured after pelleting the F-actin. Residual binding of the minispectrin to F-actin in the absence of 4.1R was subtracted from each curve to give 4.1R-specific binding. Note the marked ineffectiveness of the minispectrin bearing the α-spectrin C-terminal deletion found in the sphIJ mouse with unstable red cell membranes and severe hemolytic anemia (minispectrin-ΔC13).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100003.jpeg?Expires=1769144925&Signature=ZSeJSpCv1phpXA4TyERV4KYJ8z~yrImqQ7Ha5eSytullYjkUDAHWIqBC~onAaHCaV8E37~hu0MrCNlIVpNO0j0Z4UIjIvO313gST9jlS~2lXHv2uBmUKTPfWj5foEil1xY7cZtov33M2j1UvofpmLqUIvdyuekHcK4aMJG4ar0Vq-W5TSCbkvARQas2PcVD8K1xb-pb15rFTU4ZR9G24JbFm81cR8CoR0SQfbTRf7-gyX5EW8qqYVYZPZ4Aj54SyIaBqf7wpPcskNG0~DYaBUjNHIR79KNy4f5XqATgimd8RyHzQeudR3RDgOwRaWg-8nRGEk5XaXwWqWwMTaG-wIw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Protein 4.1 and minispectrin-dependent ternary complex formation. (A) Minispectrin-dependent binding of [125I]-protein 4.1 to F-actin. [125I]-Protein 4.1 (0.38μM), F-actin (2.38μM as G-actin), and increasing amounts of either minispectrin or minispectrin-Δ13 were combined, and the bound 4.1R was measured after pelleting the F-actin. (B) Protein 4.1-dependent binding of [125I]-minispectrin or [125I]-minispectrin-Δ13 to F-actin. Increasing amounts of protein 4.1R were added to the [125I]-labeled minispectrins (0.54μM) and F-actin (2.38μM), and each bound minispectrin was measured after pelleting the F-actin. Binding of the mutant minispectrin is impaired under both conditions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100004.jpeg?Expires=1769144925&Signature=5DHCV8zMlUD8hzk4rK70szbEoZk4vwOSnyAq86Q7LtTM4YOPqiWv1e0tGPo-HEq-yBG2W8hPEbUHRRQ0gUWEwbmO9b9GPnABfLcxFiEeKKq9oFLGUOljRMV1k6pbVF63PbZSRR4uXatEsQvXyBFGlop~qXTuKoWANZ8zt2iM7EVddkCd2OjlKcBe9506I6RBYO-vcg6s2ppq-fHl1Tqqe6c~-bJ-94LNuk7ppUNVt8T1HqJs-3u4pn4t-w20d9foidZGhMMOEz1AuYXzx57dP8qFZ5VOrnXB6ohFOL72Etd-zFAIBJ6H0F1j7m0E-uBv8hruXhlqJku-7Ggu22di6g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Heterodimer assembly and protein 4.1R binding. (A) Assembly of the minispectrin heterodimer. GST-βABD1-4 (0.87μM) was mixed with increasing amounts of [125I]-α18-21EF or α18-21EFΔ13 (0.14-2.12μM), and the complex was retrieved on GSH beads as described in “GST pull-down experiments” (B) Protein 4.1R binding to minispectrin. Increasing amounts of [125I]-protein 4.1 (0.18-1.36μM) were added to GST-minispectrin or GST-minispectrin-Δ13 (each 0.54μM). In each experiment, binding of the [125I]-ligand to GSH beads alone was subtracted from the values with added spectrin. (C) Minispectrin binding to F-actin in the absence of protein 4.1R. Increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13 were added to F-actin (2.38μM as G-actin), and the bound minispectrin was measured after pelleting the F-actin. Results from 3 independent experiments are combined. Note that the diminished ability of the mutant minispectrin to form the ternary complex is not a failure to assemble the minispectrin molecule or to bind protein 4.1R, but a failure to bind to F-actin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100005.jpeg?Expires=1769144925&Signature=SDUdmvjpP9vrlxnHT0SPzjidfjQgonXd4ShXmJEqEmlapzNbP6KgoCkfshJeNSFZQdLL2orXi-p7n3qjvthH-r81PiPy6A-yPOB6YK53NztliNYZbr0F1oQSfQNOKzSTfQghw6lpyTNhz74emA6OrcIQQWTmMUbUAHeSQUQ2dbKMPPCVWCdLO0RNkrGvBZlheaO-6g8q8FliQeM1YpjpO7WBzQ3P4kR2yp5pxk6KLssQpH9e6bAWfXN-HPrJG0M52MKWWpYvpeYx~ah~OXLfbqvEe9vQjKTXw46GDpj66x~jlUe6D9Pf6KZxyv7rIv6Y6jhEJFJGLyvl4t7HeOD5HA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Comparison of the binding of [125I]-spectrin α18-21EF, [125I]-βABD1-4, [125I]-minispectrin, and [125I]-minispectrin-Δ13 to F-actin in the presence of protein 4.1R. The spectrin fragments and protein 4.1 were all 0.38μM. F-actin was 2.38μM in G-actin monomers. Error bars represent the range of 2 to 5 independent experiments for each protein. Neither the isolated α-spectrin EF domain nor the β-spectrin actin-binding domain binds to actin at these concentrations, whereas the assembled minispectrin binds robustly.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100006.jpeg?Expires=1769144925&Signature=kC1j04vHIrfKgC4JkrTRqJIAhdPPuDOSHK4JS5ZfVIPgu1zcYWaywoia-m29vLLcxdtkgQ~cYbbsIIQeMCqUH3Nm4RiXjW-xJ9AoL7n5LC4lj~6FzjugDNHc7gM92KLxEQQ1USUrXRwSOpYREanMosjX-oRSrKOSZASP6~CRHEp5zBATWdfePji33Vlu2RoGnJBHDfeYE8bJqshJWVOnDBu8PuNsMiVyfjXjZd4Uuv6Ig57M~5IGs9fzAu9d9QGw0hFtDy4rFbgbDsEEDHT80ertQoDvwdH8~qly~doll6~lN5ZSVtH9BHIMbip2OU0SXV7G7lOej1b1iPEmkrZdPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Minispectrin-actin-4.1R binding assessed by an F-actin pelleting assay. (A) Effect of protein 4.1R on minispectrin-actin interactions. F-actin (2.38μM as G-actin) was combined with increasing amounts of [125I]-minispectrin, with or without protein 4.1R (0.38μM), and the bound minispectrin was measured after pelleting the F-actin. (B) Comparison of the amount of ternary complex formed by normal and mutant minispectrins. F-actin (2.38μM as G-actin) and protein 4.1 (0.38μM) were combined with increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13, and the bound minispectrin was measured after pelleting the F-actin. Residual binding of the minispectrin to F-actin in the absence of 4.1R was subtracted from each curve to give 4.1R-specific binding. Note the marked ineffectiveness of the minispectrin bearing the α-spectrin C-terminal deletion found in the sphIJ mouse with unstable red cell membranes and severe hemolytic anemia (minispectrin-ΔC13).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100003.jpeg?Expires=1769144926&Signature=3lJNf9zlWpYxgIBQNmAFGSgzD0hvmslMRBGg0V7odtOFBkgyy829V8Cajc0LhGBL-120eePLOB~Zp40tM21vILYhPjatjR0VU-sIvba8XcBz48q4iSC6vQ51htRHgYNEWHOk89sfnIal5kad-1t4YUxxnGIhudhtgXMAuLSG465AEnUaS1mH4g6p7013jW8X3oWfkAWo4ZTOjhLlAJl3he2jqwtV6~CsXXFPK2zdpByYLaJUMqQmgKdiO4ERW6VqK9aIRBrvTsC6qjixiEXY6sUL~ismI1HJo2qQ1JfUBRZxAMnsPQ5nm7QWMTDgB-I-S6ekuWf3RexRs5rPRFs9Mw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Protein 4.1 and minispectrin-dependent ternary complex formation. (A) Minispectrin-dependent binding of [125I]-protein 4.1 to F-actin. [125I]-Protein 4.1 (0.38μM), F-actin (2.38μM as G-actin), and increasing amounts of either minispectrin or minispectrin-Δ13 were combined, and the bound 4.1R was measured after pelleting the F-actin. (B) Protein 4.1-dependent binding of [125I]-minispectrin or [125I]-minispectrin-Δ13 to F-actin. Increasing amounts of protein 4.1R were added to the [125I]-labeled minispectrins (0.54μM) and F-actin (2.38μM), and each bound minispectrin was measured after pelleting the F-actin. Binding of the mutant minispectrin is impaired under both conditions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100004.jpeg?Expires=1769144926&Signature=X4HfowYY3lgvJvJciI8ASIujPIZDtyKt189ZYRm5bFLZKUecASAis8vC6lrGdK8zuy8wC-7BYxTLUZQKAeMHX1WiVJzOdQTyEeyyPzL17Je0zANi5hWfV~1gQc1nOW86NGJmXhNhCVyO7w3hWjm5tFNu5N-llFG~IsFiyb8F4dGn23a39CYQN5DRFtkSQ6pfKECCHbBge877Bvnwy4WWEAQb~ACfbagPu8q6kBfygyJzN~A43o8sNPpJxFEfC5t6uFSRNtZjrnQ6f9uYtMPO4a6EEbwAjhaUFUFoqwFSzw8t4RS9m8AEuu4qekAkqvXOveIGOiOYTrx1WCbqJqwRMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Heterodimer assembly and protein 4.1R binding. (A) Assembly of the minispectrin heterodimer. GST-βABD1-4 (0.87μM) was mixed with increasing amounts of [125I]-α18-21EF or α18-21EFΔ13 (0.14-2.12μM), and the complex was retrieved on GSH beads as described in “GST pull-down experiments” (B) Protein 4.1R binding to minispectrin. Increasing amounts of [125I]-protein 4.1 (0.18-1.36μM) were added to GST-minispectrin or GST-minispectrin-Δ13 (each 0.54μM). In each experiment, binding of the [125I]-ligand to GSH beads alone was subtracted from the values with added spectrin. (C) Minispectrin binding to F-actin in the absence of protein 4.1R. Increasing amounts of [125I]-minispectrin or [125I]-minispectrin-Δ13 were added to F-actin (2.38μM as G-actin), and the bound minispectrin was measured after pelleting the F-actin. Results from 3 independent experiments are combined. Note that the diminished ability of the mutant minispectrin to form the ternary complex is not a failure to assemble the minispectrin molecule or to bind protein 4.1R, but a failure to bind to F-actin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100005.jpeg?Expires=1769144926&Signature=Z1aK9ZuCE4MYslBK4OnLX8FdTlJ9cC50MVqb0w4GY-Ye6j~tWjzdnfQKSXhucpUF5JiwJiw8O-97al4G4-5QC2iaqa3SoPZC2GPW1JTxG4SebgyEi4VJ-2PUWncEy-sNKhWa-KB6iQPiXopiqkaOGcfTYa-GoBiS89M5nXQvYb4IVz5hhuhUKj7yEBQJOuItzeriLmPbX2J1Q~WQ-UOdwMCTBb1eW0vMOcEFSKA-1ICQ6SEUc6coNvcwzrWm3CQtb4NxQxZklpuCWQGAwdVYkphm1B4zYeEdwrxH8mF8IK3prpZlkVAmyaNghaCZ87GKuMZVREobMmQ4s7o0VxUfwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Comparison of the binding of [125I]-spectrin α18-21EF, [125I]-βABD1-4, [125I]-minispectrin, and [125I]-minispectrin-Δ13 to F-actin in the presence of protein 4.1R. The spectrin fragments and protein 4.1 were all 0.38μM. F-actin was 2.38μM in G-actin monomers. Error bars represent the range of 2 to 5 independent experiments for each protein. Neither the isolated α-spectrin EF domain nor the β-spectrin actin-binding domain binds to actin at these concentrations, whereas the assembled minispectrin binds robustly.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260612/4/m_zh89991058100006.jpeg?Expires=1769144926&Signature=sxet4aM~x2CCxQqKtxswxjLvYyAOPcxHwmo1129Q32yNSXjxFvzCLYdLbGQLUEf-nbSGYjzuy6eGACBpxT-DD36vpF6qNt7b~1a346sXkbGmNqmvnxGCPZQ~lBjEXYyqfHBO65Iq6fU1qk3canCWlWCVjD0~O-2N05jFt~ynXeORy1Zt90-GKTvoJZ-ksGELkTf2xHKvpOXsPnfY24zfgDKoh09vmVTOiNL9NJ7FoAYHkzivcXJwlgd-Z-ignIJgAAKJHg9Bx3y4gY6HJoC1O0QiuJq3ztxmauMVFpjGKBs9neeAilRIYdQIWfWgzS1Xz34yWv90e36pz1nUvPvmAA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)