Abstract
The short actin filaments in the red blood cell (RBC) membrane skeleton are capped at their pointed ends by tropomodulin 1 (Tmod1) and coated with tropomyosin (TM) along their length. Tmod1-TM control of actin filament length is hypothesized to regulate spectrin-actin lattice organization and membrane stability. We used a Tmod1 knockout mouse to investigate the in vivo role of Tmod1 in the RBC membrane skeleton. Western blots of Tmod1-null RBCs confirm the absence of Tmod1 and show the presence of Tmod3, which is normally not present in RBCs. Tmod3 is present at only one-fifth levels of Tmod1 present on wild-type membranes, but levels of actin, TMs, adducins, and other membrane skeleton proteins remain unchanged. Electron microscopy shows that actin filament lengths are more variable with spectrin-actin lattices displaying abnormally large and more variable pore sizes. Tmod1-null mice display a mild anemia with features resembling hereditary spherocytic elliptocytosis, including decreased RBC mean corpuscular volume, cellular dehydration, increased osmotic fragility, reduced deformability, and heterogeneity in osmotic ektacytometry. Insufficient capping of actin filaments by Tmod3 may allow greater actin dynamics at pointed ends, resulting in filament length redistribution, leading to irregular and attenuated spectrin-actin lattice connectivity, and concomitant RBC membrane instability.
Introduction
The membrane skeleton is composed of a highly cross-linked network of spectrin, actin filaments, and accessory proteins that underlies the plasma membrane of differentiated cells. This network creates membrane domains by anchoring and restricting the long-range distribution of membrane proteins and plays an important role in determining cell shapes, membrane contours, and mechanical properties.1,2 The organization of the membrane skeleton is best known in red blood cells (RBCs), where it is organized as a quasi-hexagonal network with connecting strands formed by long, flexible spectrin molecules and vertices formed by short actin filaments.3-5 Each short actin filament forms the core of a junctional complex with 2 rod-shaped tropomyosin (TM) molecules along the filament, 2 tropomodulin 1 (Tmod1) molecules capping the pointed filament end and an α/β-adducin heterodimer capping the barbed filament end.6,7 Dematin (protein 4.9) is also associated with the short actin filaments as are β1-spectrin protein 4.1R complexes that extend from the sides of the filaments to form the extended spectrin-actin network. The network is connected to membrane macromolecular complexes by multiple linkages: from β-spectrin by ankryin to band 3, and from the junctional complex by protein 4.1, α/β-adducin and dematin to band 3, glycophorin C, the glucose transporter, and other components.8 Disruptions either of attachments of the spectrin-actin lattice to membrane macromolecular complexes (“vertical” connections), or linkages within the plane of the spectrin-actin lattice (“horizontal” connections) lead to perturbations in RBC shapes, membrane deformability, and stability, impairing RBC survival in the circulation.8-11
A particularly striking feature of the RBC actin filaments is their uniformly short length of 33 to 37 nm (15-18 subunits), which may restrict the number of spectrin molecules5-7 that bind to each filament, thereby resulting in quasi-hexagonal symmetry for the spectrin-actin lattice underlying the membrane.3-5,7 These short actin filaments are extraordinarily stable, persisting for the lifetime of the RBCs (∼ 120 days in humans and ∼ 40 days in mice). The uniform lengths and stability of RBC actin filaments contrast with rapid turnover and variable lengths of actin filaments in motile and proliferating cells.7,12 Tight capping of actin filament ends by Tmod1 and α/β-adducin is hypothesized to be a prerequisite for the connectivity and ordered geometry of the spectrin-actin network. This is because actin subunit association/dissociation from filament ends would be expected to lead to redistribution of filament lengths toward many possible lengths, thereby resulting in relaxation of stoichiometric and structural constraints for formation of the ordered geometry of the spectrin-actin lattice. Here, we have investigated the role of Tmod1 in controlling actin filament length, spectrin-actin lattice organization, and physiologic function of the RBC membrane skeleton.
Tmods are a conserved family of TM-binding and actin filament pointed end-capping proteins that regulate actin filament lengths and stability by blocking actin association and dissociation at pointed (slow growing) ends, and by strengthening binding of TMs to actin filaments.12,13 Tmod1 is expressed in mammalian RBCs, striated muscles, lens fiber cells, and neurons; Tmod2 is restricted to neurons; Tmod3 is in most cell types; and Tmod4 is restricted to skeletal muscle.12 Targeted deletion of Tmod1 in mice leads to embryonic lethality at embryonic day 9.5 (E9.5), because of inability to assemble myofibrils in cardiac myocytes and aborted cardiac development.14,15 Absence of Tmod1 also leads to mechanically unstable Tmod1-null primitive erythroid cells in E9.5 embryos, consistent with a function for Tmod1 in the membrane skeleton.15 To study the role of Tmod1 in RBC membrane skeleton structure and physiology, we rescued the embryonic lethality of Tmod1-null embryos by expressing a Tmod1 transgene in the heart under the control of the cardiac-specific α-myosin heavy chain (α-MHC) promoter.16,17 We showed previously that, except for the heart, these Tmod1−/−Tg+ embryos (stage E15.5) have no Tmod1 in any tissues, including their definitive RBCs.17 The Tmod1−/−Tg+ mice are viable and fertile, allowing study of circulating RBCs in adult mice. Here, we show that RBCs from Tmod1−/−Tg+ mice now contain low levels of Tmod3, exhibiting alterations in RBC shapes, increased osmotic fragility, and reduced cellular deformability by ektacytometry, consistent with a mild spherocytic elliptocytosis. Strikingly, Tmod1-null RBCs also show mis-regulation of actin filament lengths that can be explained by reduced capping with enhanced actin dynamics, leading to altered spectrin-actin lattice connectivity and disruptions in the membrane skeleton.
Methods
Mice and genotyping
We used a viable Tmod1 knockout mouse line that expresses a Tmod1 transgene only in the heart Tg(αMHC-Tmod1).17 These Tmod1lacZ;Tg(αMHC−Tmod1) mice are on a mixed background of 129Sv, C57BL/6, and FVB/N.18 The line was maintained as heterozygotes (Tmod1+/−Tg+) to avoid high overexpression of the αMHC-Tmod1 transgene in the heart, which leads to dilated cardiomyopathy.16 Phenotypic analyses of blood and RBCs were performed on progeny derived from crosses of Tmod1+/− mice with Tmod1+/−Tg+ mice (approximately one-eighth of progeny are null for endogenous Tmod1).17 Genotyping was performed by polymerase chain reaction as described.17 All procedures were in accordance with The Scripps Research Institute animal care guidelines and were approved by the Institutional Animal Care and Use Committee.
Ghost preparation, sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and Western blotting
Blood was isolated by retro-orbital bleed into K3EDTA Sarstedt Microvette 200 capillary tubes or by cardiac puncture into K3EDTA Becton Dickinson Vacutainer purple top tubes, and sedimented 30 minutes at 1g through 5 volumes of 0.75% Dextran T-500 in 5mM NaHPO4, pH 7.4, 150mM NaCl at 4°C. The supernatant was aspirated, and the Dextran sedimentation was step repeated, followed by washing cells 4 times in phosphate-buffered saline (10mM NaHPO4, pH 7.4, 150mM NaCl) at 4°C. Packed RBCs were lysed by vortexing in approximately 40 volumes of ice-cold Mg++-lysis buffer (5mM NaHPO4, pH 7.4, 2mM MgCl2, 1mM dithiothreitol, and 20 μg/mL PMSF [phenylmethylsulfonyl fluoride]), and Mg++ ghosts prepared as described.19 Triton-insoluble membrane skeletons were prepared by extraction of Mg++ ghosts in approximately 10 volumes of ice-cold 1% Triton X-100 in Mg++-lysis buffer and sedimented at top speed (∼ 14 000g) in an Eppendorf microcentrifuge at 4°C. Ghosts and membrane skeleton pellets were frozen in liquid nitrogen and stored at −80°C. concentrated Gel samples were prepared by adding an equal volume of 2× concentrated sodium dodecylsulfate–sample buffer and boiled for 5 minutes, and samples were electrophoresed on 7.5% to 15% linear gradient polyacrylamide gels,19 with a running buffer of pH 8.6.20,21 For experiments comparing RBCs, cytosol, and ghosts, 4M urea was included in the 7.5% to 15% gradient running gel, and 2M urea was included in the 5% stacking gel.21 Gels were stained with Coomassie blue R250 or transferred to nitrocellulose for Western blotting.21
Rabbit polyclonal antibodies were generated against purified human RBC Tmod1 (R1749)20 and recombinant human Tmod3 (R5168).22 Cross-reactivities were removed by passing Tmod1 antisera over a Tmod3 column and Tmod3 antisera over a Tmod1 column followed by affinity purification by standard procedures, eluting with 0.2M glycine-HCl, pH 2.8. Western blotting for Tmods was as described.23 Antibodies to TMs were an affinity-purified rabbit polyclonal to purified human RBC TMs19,24 that recognizes both the upper γ-TM (hTM5, TM5NM1) and lower α-TM (TM5b) bands,25,26 and a mouse monoclonal from the Developmental Studies Hybridoma Bank (CH1 that recognizes only the upper, TM5NM1 band. Other antibodies were rabbit antisera to the cytoplasmic domain of band 327 and to full-length protein 4.1R (a gift from Cathy Korsgren, Harvard Medical School), and affinity-purified chicken antibodies against full-length recombinant α1 and β1 adducins (gifts from Diana Gilligan, Puget Sound Blood Bank). Mouse monoclonals were 17C7 to αI-spectrin (AbCAM), 3F2.3 to the β2 subunit of CapZ (Developmental Studies Hybridoma Bank), and C4 to actin (a gift from Dr J. Lessard, University of Cincinnati). Primary antibodies were detected with the use of horseradish peroxidase–conjugated: goat anti–mouse (Promega), goat anti–chicken (Aves Labs Inc), or Protein-A (Sigma-Aldrich) and visualized by enhanced chemiluminescence. Film was scanned with a Cannon CanoScan 8400F at 600 dpi and bands on blots were quantified with National Institutes of Health (NIH) ImageJ 1.41q software (http://rsb.info.nih.gov/ij). Pictures were cropped in Adobe Photoshop CS4 and figures made in Adobe Illustrator CS4.
Hematologic analysis of blood
Whole blood was collected by cardiac puncture into K3EDTA tubes (for smears, osmotic fragility, and complete blood counts), or into Acid-citrate-dextrose (for ektacytometry) as described.28 Peripheral blood smears were stained with Wright-Giemsa, or new methylene blue for reticulocytes (Sigma-Aldrich). Complete blood counts were performed with the use of an automated hematology analyzer (Advia 120 Hematology System; Siemens), using multispecies software and calibrated for C57BL/6 mouse blood. Osmotic ektacytometry was performed as described,28,29 and osmotic fragility was performed as described.30
Light and electron microscopy of RBCs
Light microscopic images of RBCs in peripheral blood smears were acquired with a 100× 1.4 oil-immersion objective using a Leica DMRXE microscope, as in Robledo et al.31 For scanning electron microscopy (SEM), whole blood was collected in K3EDTA tubes as described earlier, and RBCs were washed and fixed in 2% glutaraldehyde.31 Fixed cells were shipped overnight on ice to The Jackson Laboratory for SEM imaging.31 For transmission electron microscopy (TEM) of unspread RBC membrane skeletons, whole blood was collected by cardiac puncture in K3EDTA and shipped overnight to Harvard Medical School. Platinum/carbon replicas of Triton-permeabilized and fixed RBC membrane skeletons were prepared and visualized by TEM.32,33 Membrane skeleton pore sizes were determined with ImageJ, by manually outlining the individual fibers and measuring the areas inside the outlines. To examine actin filaments in junctional complexes, Mg++ ghosts were extracted with Triton X-100, and membrane skeletons were isolated on a sucrose gradient and expanded at low ionic strength.3 Membrane skeletons were adsorbed to a carbon-coated nitrocellulose film supported on 400-mesh copper grids and stained with 2% uranyl acetate. TEM images were obtained with the use of an FEI Tecnai T12 electron microscope, operating at 120 KeV, equipped with a TVIPS 4K× 4K CCD camera. Images were acquired at a nominal magnification of 50 000× (0.164 nm/pixel) at a nominal underfocus of −2 μm and electron dose approximately (12e/Å2 ). Actin filaments were identified according to the criteria of Shen et al,5 namely, linear rodlike structures associated with multiple radiating spectrin strands, both ends clearly demonstrable, and a thickness of 5 to 10 nm. Lengths were measured with the use of the ruler tool in Appion and Leginon software from NRAMM (http://nramm.scripps.edu/). Microscopy images were cropped in Adobe Photoshop CS4 and figures made in Adobe Illustrator CS4.
Statistical analyses
Results are means plus or minus SDs. Significance of differences in means was evaluated with the Student t test with the use of Microsoft Excel, whereas differences in variances were evaluated with the F test with the use of Prism software, Version 5 (GraphPad Software Inc).
Results
Tmod3 is associated with the membrane skeleton in the absence of Tmod1
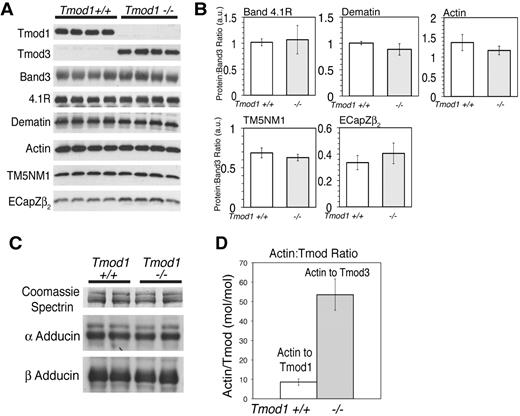
Western blotting of RBC ghosts from adult Tmod1−/−Tg+ animals with antibodies specific for Tmod1 confirms that no Tmod1 is present in these ghosts (Figure 1B left), as expected.17 However, Western blotting with antibodies specific for the widely expressed isoform, Tmod3,12 shows that RBC ghosts from Tmod1−/−Tg+ animals now contain Tmod3 associated with their membranes (Figure 1B left). No Tmod3 is present in RBC ghosts from Tmod1+/+ or Tmod1+/− animals, which contain normal levels of Tmod1, regardless of the presence of the α-MHC-Tmod1 transgene (Figure 1B right). Visual inspection of the corresponding Coomassie-stained gels and quantification by densitometry shows that levels of major RBC membrane proteins, including spectrin, band 3, band 4.1R, and actin, are not altered noticeably by the absence of Tmod1 (Figure 1A,D). To investigate whether the absence of Tmod1 might affect overall stability of the membrane skeleton, we compared Triton skeletons prepared by extraction of ghosts with Triton X-100. No differences were observed because of the absence of Tmod1 (Figure 1A,D), with the exception that Tmod3 was now associated with the Triton X-100 insoluble membrane skeleton (Figure 1B left).
Tmod3 is associated with RBC membranes (Mg++ ghosts) and Triton-insoluble membrane skeletons from Tmod1-null mice. (A) Coomassie blue–stained sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of ghosts and Triton-insoluble membrane skeletons showing no alterations in major membrane proteins in the absence of Tmod1. (B) Western blots showing Tmod3 in ghosts from Tmod1-null (Tmod1−/−Tg+) but not wild-type (Tmod1+/+Tg+ and Tmod1+/+) or heterozygous (Tmod1−/+Tg+ and Tmod1−/+) mice. Levels of Tmod1 in ghosts from wild-type and heterozygous mice are similar regardless of the presence of the α-MHC-Tmod1 transgene (Tg+). Each lane in panels A and B is from a different individual animal. Equivalent ghost volumes were loaded to compare relative amounts of proteins. (C) Coomassie blue–stained gels and Western blots showing antibody specificities for Tmod1 or Tmod3. (D) Ratios of spectrin, band 4.1R, and actin to band 3 determined by densitometry from Coomassie blue–stained gels as in panel A. Data from wild-type and heterozygous animals were pooled (+/+) becauseTmod1 levels are the same in both genotypes (n = 9 for ghosts; n = 6 for skeletons). For Tmod1-nulls (−/−), n = 7 for ghosts; n = 6 for skeletons. Differences between +/+ and −/− datasets were not significantly different.
Tmod3 is associated with RBC membranes (Mg++ ghosts) and Triton-insoluble membrane skeletons from Tmod1-null mice. (A) Coomassie blue–stained sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of ghosts and Triton-insoluble membrane skeletons showing no alterations in major membrane proteins in the absence of Tmod1. (B) Western blots showing Tmod3 in ghosts from Tmod1-null (Tmod1−/−Tg+) but not wild-type (Tmod1+/+Tg+ and Tmod1+/+) or heterozygous (Tmod1−/+Tg+ and Tmod1−/+) mice. Levels of Tmod1 in ghosts from wild-type and heterozygous mice are similar regardless of the presence of the α-MHC-Tmod1 transgene (Tg+). Each lane in panels A and B is from a different individual animal. Equivalent ghost volumes were loaded to compare relative amounts of proteins. (C) Coomassie blue–stained gels and Western blots showing antibody specificities for Tmod1 or Tmod3. (D) Ratios of spectrin, band 4.1R, and actin to band 3 determined by densitometry from Coomassie blue–stained gels as in panel A. Data from wild-type and heterozygous animals were pooled (+/+) becauseTmod1 levels are the same in both genotypes (n = 9 for ghosts; n = 6 for skeletons). For Tmod1-nulls (−/−), n = 7 for ghosts; n = 6 for skeletons. Differences between +/+ and −/− datasets were not significantly different.
Tmod1 has 2 known binding partners in the RBC membrane skeleton, actin filament pointed ends and RBC TMs.13,21 The RBC TMs comprise TM5b, a short TM product of the α-TM gene, and TM5NM1 (hTM5), a short TM product of the γ-TM gene.19,25,26 To investigate whether substitution of Tmod1 by Tmod3 is accompanied by alterations in TMs or actin on membranes, we used Western blotting to compare levels of TM5NM1 and actin with respect to band 3 in ghosts from Tmod1+/+Tg+ and Tmod1−/−Tg+ mice (Figure 2A). Ghosts were prepared in the presence of Mg++ to assure retention of TMs on the membrane.19 This analysis showed that the ratios of TM5NM1 and actin to band 3 in ghosts or in membrane skeletons were unchanged in the absence of Tmod1 (Figure 2A-B). Therefore, substitution of Tmod3 for Tmod1 does not affect significantly the amount of either actin or TM5NM1 associated with the membrane (also see Figures 1 and 3A).
Tmod3 levels in Tmod1-null ghosts are one-fifth that of Tmod1 in ghosts from wild-type mice, but levels of actin, TM, and other actin-binding proteins are unchanged. (A) Western blots of Tmod1, Tmod3, band 3, 4.1R, dematin, actin, TM5NM1, and EcapZβ2 in Mg++ ghosts from wild-type and Tmod1-null mice. Each lane is from a different individual animal. (B) Ratios of proteins to band 3 determined by densitometry from Western blots in panel A, showing no significant differences between wild-type and Tmod1-null samples (4.1R, P < .8; dematin, actin, TM5NM1, and EcapZβ2, P < .2). (C) Western blots of α- and β-adducin in Mg++ ghosts from 2 wild-type and 2 Tmod1-null mice, using Coomassie blue staining for spectrin from a duplicate gel as a loading control. Adducin levels are unaffected by the absence of Tmod1. (D) Molar ratio of actin/Tmod1 in wild-type ghosts and actin/Tmod3 in Tmod1-null ghosts. Nanograms of actin and Tmod1 or Tmod3 were quantified per microliter of ghosts by Western blotting according to standard curves for each purified protein, as shown in supplemental Figure 1.
Tmod3 levels in Tmod1-null ghosts are one-fifth that of Tmod1 in ghosts from wild-type mice, but levels of actin, TM, and other actin-binding proteins are unchanged. (A) Western blots of Tmod1, Tmod3, band 3, 4.1R, dematin, actin, TM5NM1, and EcapZβ2 in Mg++ ghosts from wild-type and Tmod1-null mice. Each lane is from a different individual animal. (B) Ratios of proteins to band 3 determined by densitometry from Western blots in panel A, showing no significant differences between wild-type and Tmod1-null samples (4.1R, P < .8; dematin, actin, TM5NM1, and EcapZβ2, P < .2). (C) Western blots of α- and β-adducin in Mg++ ghosts from 2 wild-type and 2 Tmod1-null mice, using Coomassie blue staining for spectrin from a duplicate gel as a loading control. Adducin levels are unaffected by the absence of Tmod1. (D) Molar ratio of actin/Tmod1 in wild-type ghosts and actin/Tmod3 in Tmod1-null ghosts. Nanograms of actin and Tmod1 or Tmod3 were quantified per microliter of ghosts by Western blotting according to standard curves for each purified protein, as shown in supplemental Figure 1.
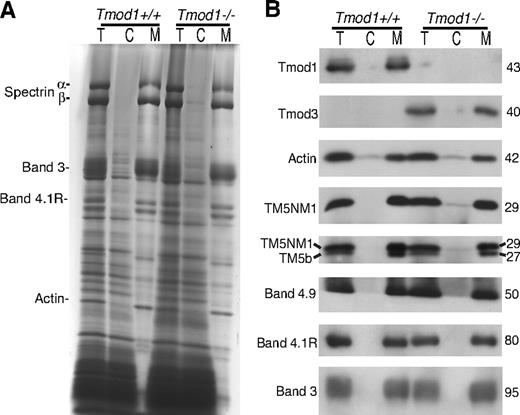
Comparison of cytosol/membrane levels for Tmods and other actin-binding proteins in wild-type or Tmod1-null RBCs. (A) Coomassie blue–stained sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of total RBC extracts (T), RBC cytosol (C), and washed membranes (M) prepared by hypotonic lysis of RBCs isolated from wild-type and Tmod1-null mice. (B) Western blots of Tmods, actin, TMs, dematin, 4.1R, and band 3 in total RBC extracts, cytosol, and washed membranes. Tmod1 and Tmod3 are predominantly associated with membranes, and associations of actin, TMs, dematin, and 4.1R with membranes are not altered in the absence of Tmod1. A small amount of the upper TM5NM1 but not the lower TM5b band is detected in RBC cytosol from Tmod1-null mice.
Comparison of cytosol/membrane levels for Tmods and other actin-binding proteins in wild-type or Tmod1-null RBCs. (A) Coomassie blue–stained sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of total RBC extracts (T), RBC cytosol (C), and washed membranes (M) prepared by hypotonic lysis of RBCs isolated from wild-type and Tmod1-null mice. (B) Western blots of Tmods, actin, TMs, dematin, 4.1R, and band 3 in total RBC extracts, cytosol, and washed membranes. Tmod1 and Tmod3 are predominantly associated with membranes, and associations of actin, TMs, dematin, and 4.1R with membranes are not altered in the absence of Tmod1. A small amount of the upper TM5NM1 but not the lower TM5b band is detected in RBC cytosol from Tmod1-null mice.
In addition to TM, RBC actin filaments are associated with several accessory actin-binding proteins, α and β adducins and dematin. Western blotting of Mg++ ghosts for these proteins shows that their levels are unchanged in the absence of Tmod1 (Figure 2A-B). We also saw no changes in levels of the EcapZ β2 subunit (Figure 2A), unlike α- or β-adducin–null ghosts in which EcapZ levels are increased several fold.31,34-36
Next, we performed quantitative Western blotting to determine the molar ratio of actin to Tmod1 or Tmod3 in RBC ghosts from Tmod1+/+Tg+ or Tmod1−/−Tg+ mice, respectively. For this experiment, the nanograms of Tmods per microliter of packed ghosts were determined according to standard curves of the purified proteins loaded on the same gels (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In ghosts from Tmod1+/+Tg+ mice, the ratio of actin to Tmod1 was calculated to be approximately 8 actin subunits to 1 Tmod1 (Figure 2C), corresponding to 2 Tmod1 molecules per short actin filament of approximately 16 subunits long, as shown previously for human RBC ghosts.20,21 In contrast, in RBC ghosts from Tmod1−/−Tg+ mice the ratio of actin to Tmod3 was calculated to be much greater, at more than 50 actin subunits to 1 Tmod3 molecule (Figure 2C). Because the ratio of actin to Tmod is increased approximately 5- to 6-fold (Figure 2C), while total actin levels in ghosts are unchanged (Figure 2A), the absolute level of Tmod3 in ghosts from Tmod1−/−Tg+ mice is less than one-fifth the normal level of Tmod1 in ghosts from Tmod1+/+Tg+ mice.
Tmod3 is not in RBC cytosol and membrane/cytosolic proportions of actin and TMs are unaffected by absence of Tmod1
One possible explanation for the presence of Tmod3 in ghosts from Tmod1−/−Tg+ mice could be that Tmod3 is normally present in the cytosol of RBCs and that, in the absence of Tmod1, Tmod3 can now bind to the actin filament pointed ends, compensating for Tmod1's capping function. However, comparison of Tmod levels in RBCs, cytosol, and Mg++ ghosts showed that no Tmods are in the cytosol of Tmod1+/+Tg+ RBCs and that Tmod3 is predominantly associated with membranes prepared from RBCs from Tmod1−/−Tg+ mice, as is Tmod1 in RBCs from Tmod1+/+Tg+ mice21 (Figure 3; data not shown). Thus, the presence of Tmod3 in Mg++ ghosts cannot be explained by Tmod3 translocation to the membrane from a cytosolic pool.
Comparison of whole RBCs, cytosol, and membranes by Coomassie and Western blotting also shows that associations of α- and β-spectrins, 4.1R, and dematin with the membrane are stable in the absence of Tmod1 (Figure 3). The proportion of actin in cytosol and on membranes is also similar for either Tmod1- or Tmod3-containing RBCs (Figure 3B). (Note that the presence of a small amount of cytosolic actin is consistent with previous observations on human RBCs.37 ) Thus, normal levels of membrane-associated actin in Tmod1-null ghosts (Figure 2A) are not due to compensatory polymerization from a larger cytosolic pool. However, a small amount of the upper TM5NM1 band is detected in cytosol of RBCs from Tmod1−/−Tg+ mice but not Tmod1+/+Tg+ mice, using either the CH1 monoclonal or our rabbit polyclonal that detects both TM bands (Figure 3B). The lower TM5b band was not detected in the cytosol. Because the amounts of both TM bands associated with membranes appear similar with or without Tmod1 (Figure 3B), as shown previously in Figure 2A, the presence of cytosolic TM5NM1 could be due to a small increase in the amount of total TM5NM1 in Tmod1-null RBCs. The increased TM could reflect increased numbers of reticulocytes in anemic Tmod1−/−Tg+ mice (see Tables 1 and 2), because reticulocytes contain more TM than mature RBCs.38 In conclusion, all of the Tmod1 and Tmod3 are associated with RBC membranes, and substitution of Tmod3 for Tmod1 does not affect significantly the association of actin or TMs with the membrane.
Hematologic analysis of Tmod1+/+Tg+ and Tmod1−/− Tg+ adult mice
| . | Tmod1+/+Tg+ (n = 9) . | Tmod1−/−Tg+ (n = 9) . |
|---|---|---|
| RBC count, ×106 cells/μL | 7.82 ± 0.55 | 6.90 ± 1.45 |
| Hgb level, g/dL | 12.13 ± 0.079 | 10.54 ± 2.09* |
| Hct, % | 41.64 ± 2.69 | 35.06 ± 7.93† |
| MCV, fL | 53.31 ± 2.44 | 50.67 ± 1.32† |
| MCH, pg | 15.52 ± 0.72 | 15.33 ± 0.52† |
| MCHC, g/dL | 29.1 ± 0.65 | 30.29 ± 1.09† |
| RDW, % | 13.61 ± 0.50 | 13.98 ± 0.51 |
| HDW, g/dL | 1.73 ± 0.11 | 1.88 ± 0.11† |
| Retic, % | 2.31 ± 0.93 | 2.01 ± 0.53 |
| WBC count, ×103 cells/μL | 5.39 ± 1.38 | 4.90 ± 1.28 |
| PLT count, ×103 cells/μL | 1241.88 ± 372.31 | 935.11 ± 402.20 |
| Spleen weight, % BW | 0.29 ± 0.12 | 0.29 ± 0.05 |
| . | Tmod1+/+Tg+ (n = 9) . | Tmod1−/−Tg+ (n = 9) . |
|---|---|---|
| RBC count, ×106 cells/μL | 7.82 ± 0.55 | 6.90 ± 1.45 |
| Hgb level, g/dL | 12.13 ± 0.079 | 10.54 ± 2.09* |
| Hct, % | 41.64 ± 2.69 | 35.06 ± 7.93† |
| MCV, fL | 53.31 ± 2.44 | 50.67 ± 1.32† |
| MCH, pg | 15.52 ± 0.72 | 15.33 ± 0.52† |
| MCHC, g/dL | 29.1 ± 0.65 | 30.29 ± 1.09† |
| RDW, % | 13.61 ± 0.50 | 13.98 ± 0.51 |
| HDW, g/dL | 1.73 ± 0.11 | 1.88 ± 0.11† |
| Retic, % | 2.31 ± 0.93 | 2.01 ± 0.53 |
| WBC count, ×103 cells/μL | 5.39 ± 1.38 | 4.90 ± 1.28 |
| PLT count, ×103 cells/μL | 1241.88 ± 372.31 | 935.11 ± 402.20 |
| Spleen weight, % BW | 0.29 ± 0.12 | 0.29 ± 0.05 |
All mice were 1 year old. Values are ± SD.
RBC indicates red blood cell; Hgb, hemoglobin; Hct, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red cell distribution width; HDW, hemoglobin distribution width; Retic, reticulocytes; WBC, white blood cell; PLT, platelet; and BW, body weight.
P < .1.
P < .05.
Percentage of reticulocytes and spleen weights in young Tmod1+/+Tg+ and Tmod1−/−Tg+ mice
| . | Reticulocytes, %* . | Spleen weight, % BW† . |
|---|---|---|
| Tmod1+/+Tg+ | 1.36 ± 0.54 (5) | 0.32 ± 0.037 (7) |
| Tmod1+/−Tg+ | 1.20 ± 0.32 (5) | |
| Tmod1−/−Tg+ | 2.20 ± 0.70 (7)‡ | 0.29 ± 0.020 (10) |
| . | Reticulocytes, %* . | Spleen weight, % BW† . |
|---|---|---|
| Tmod1+/+Tg+ | 1.36 ± 0.54 (5) | 0.32 ± 0.037 (7) |
| Tmod1+/−Tg+ | 1.20 ± 0.32 (5) | |
| Tmod1−/−Tg+ | 2.20 ± 0.70 (7)‡ | 0.29 ± 0.020 (10) |
Values are ± SD. Numbers in parentheses indicate numbers of animals from each genotype.
BW indicates body weight.
Mice were 2 to 3 months old. Reticulocytes were counted in smears with the use of new methylene blue staining. Between 3000 and 4000 total cells were counted for each mouse.
Mice were 2 to 5 months old. Spleen weights are shown for males only.
P < .05 for comparison of percentage of reticulocytes in Tmod1−/−Tg+ mice with Tmod1+/+Tg+ mice. Other comparisons were not statistically significant.
Membrane skeleton organization is disrupted and actin filament lengths are mis-regulated in absence of Tmod1
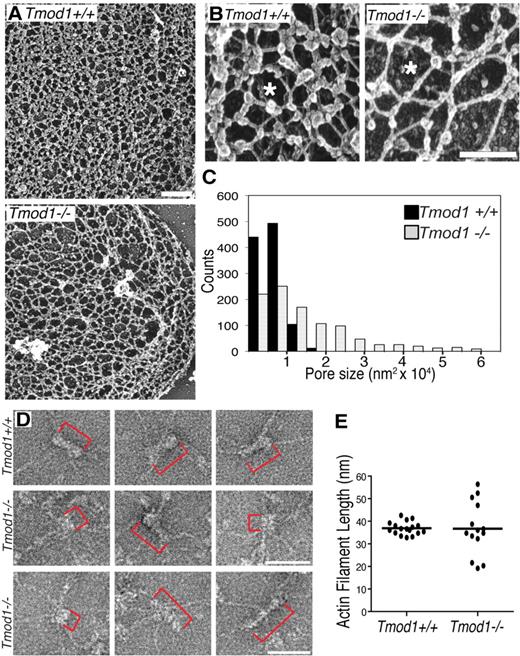
To investigate the organization of the membrane skeleton, we used electron microscopy to visualize unspread membrane skeletons of Mg++ ghosts attached to poly-l-lysine–coated grids, rapidly frozen, freeze-dried, and platinum shadowed.32,33 The unspread membrane skeleton of ghosts from Tmod1+/+Tg+ mice is visualized as a complex network of filaments intersecting at multiple points. Previous work has shown that some of these intersections represent connections of spectrin strands to junctional complexes, whereas others represent convergence of spectrin strands to form lateral associations along a portion of their length, resulting in a filament intersection.39,40 Comparison of wild-type membrane skeletons with those of Tmod1-null ghosts shows a notable increase in sizes of lattice fenestrations (pores) in the absence of Tmod1 (Figure 4A). High magnification images show further that connecting strands appear longer in the absence of Tmod1 (Figure 4B). Quantification of lattice pore sizes by tracing their perimeters with the use of Image J shows that, in wild-type membrane skeletons, lattice pore sizes are distributed in a narrow range between approximately 5000 to 10 000 nm2 with a mean of 6310 nm2 (± 2820 nm2; Figure 4C). In contrast, in Tmod1-null membrane skeletons, pore sizes span over a 10-fold size range from 5000 to 60 000 nm2, with a mean of 13 352 nm2 (± 12 355 nm2), significantly different from pore sizes in wild-type (P < .001; Figure 4C). Thus, the absence of Tmod1 leads to significant structural changes in the global organization of the membrane skeleton lattice.
The spectrin-actin lattice displays larger and more variable pore sizes, and actin filament lengths are more variable in the absence of Tmod1. (A-B) Electron micrographs of platinum replicas of unspread RBC membrane skeletons from Tmod1+/+Tg+ mice or Tmod1−/−Tg+ mice. Asterisks indicate pores in the lattice that appear larger in the absence of Tmod1. Bars, (A) 400 nm; (B) 200 nm. (C) Histogram of pore sizes shows 10-fold more size variability and a 2-fold increase in average sizes in the absence of Tmod1; n = 5 skeletons per genotype, with 200 adjacent pores per micrograph. Average pore sizes: 6310 nm2 (± 2820 nm2) for wild-type and 13 352 nm2 (± 12 355 nm2) for Tmod1-null RBCs (P < .001). (D) Electron micrographs of negatively stained junctional complexes in expanded membrane skeletons from wild-type (top) and Tmod1-null mice (middle and bottom). Red brackets indicate actin filaments. Bars, 50 nm. Actin filament lengths are uniform in wild-type junctional complexes but appear shorter or longer in Tmod1-null junctional complexes. (E) Measurements of actin filament lengths in junctional complexes of membrane skeletons from wild-type or Tmod1-null mice. Each dot represents 1 measurement. Average filament lengths: 36.90 nm (± 2.85 nm; n = 17) for wild-type and 36.74 nm (± 11.70 nm; n = 14) for Tmod1-null. Range of filament lengths: 32.80 to 42.64 nm for wild-type and 19.18 to 56.42 nm for Tmod1-null. Differences in average filament lengths between genotypes are not significant, but differences in the range of filament lengths are significant based on an F test for variance (P < .001).
The spectrin-actin lattice displays larger and more variable pore sizes, and actin filament lengths are more variable in the absence of Tmod1. (A-B) Electron micrographs of platinum replicas of unspread RBC membrane skeletons from Tmod1+/+Tg+ mice or Tmod1−/−Tg+ mice. Asterisks indicate pores in the lattice that appear larger in the absence of Tmod1. Bars, (A) 400 nm; (B) 200 nm. (C) Histogram of pore sizes shows 10-fold more size variability and a 2-fold increase in average sizes in the absence of Tmod1; n = 5 skeletons per genotype, with 200 adjacent pores per micrograph. Average pore sizes: 6310 nm2 (± 2820 nm2) for wild-type and 13 352 nm2 (± 12 355 nm2) for Tmod1-null RBCs (P < .001). (D) Electron micrographs of negatively stained junctional complexes in expanded membrane skeletons from wild-type (top) and Tmod1-null mice (middle and bottom). Red brackets indicate actin filaments. Bars, 50 nm. Actin filament lengths are uniform in wild-type junctional complexes but appear shorter or longer in Tmod1-null junctional complexes. (E) Measurements of actin filament lengths in junctional complexes of membrane skeletons from wild-type or Tmod1-null mice. Each dot represents 1 measurement. Average filament lengths: 36.90 nm (± 2.85 nm; n = 17) for wild-type and 36.74 nm (± 11.70 nm; n = 14) for Tmod1-null. Range of filament lengths: 32.80 to 42.64 nm for wild-type and 19.18 to 56.42 nm for Tmod1-null. Differences in average filament lengths between genotypes are not significant, but differences in the range of filament lengths are significant based on an F test for variance (P < .001).
To investigate whether actin filament lengths were altered in the absence of Tmod1, we visualized junctional complexes by negative-staining electron microscopy of Triton X-100–extracted membrane skeletons expanded at low ionic strength.3 As expected, junctional complexes with short actin filaments and radiating spectrin strands can be identified in images of expanded membrane skeletons from Tmod1+/+Tg+ mice (Figure 4D top row). Measurements indicate that average actin filament lengths are similar to lengths of human RBC actin filaments (36.90 ± 2.85 nm; Figure 4D-E).4,5 Junctional complexes with radiating spectrin strands can also be identified in Tmod1-null membrane skeletons, but actin filament lengths do not appear to be uniform (Figure 4 middle and bottom rows, red brackets). Although average filament lengths in the absence of Tmod1 (36.74 ± 11.70 nm) are similar to wild type, they span a greater length range from 19.18 to 56.42 nm, compared with a relatively narrow length range for wild type of 32.80 to 42.64 nm (Figure 4E). Although our sample size is admittedly small because of technical difficulties in obtaining undamaged lattices from either genotype, differences in the variance of filament lengths between wild-type and Tmod1-null are statistically significant (P < .001). Interestingly, filament lengths in Tmod1-null membrane skeletons appear to fall into 3 populations, with one similar to wild type, one shorter, and one longer; although this will require a greater sample size to substantiate. These observations indicate that spectrin and actin assemble into junctional complexes but that actin filament lengths in these complexes are more variable in the absence of Tmod1.
Tmod1−/−Tg+ mice exhibit a mild sphero-elliptocytic anemia with osmotically fragile RBCs
Analysis of peripheral blood shows a mild anemia with significant reductions in hematocrit and hemoglobin (Hgb) concentration for Tmod1−/−Tg+ mice compared with Tmod1+/+Tg+ mice (Table 1). In addition, the mean corpuscular volume is reduced in the absence of Tmod1, indicating a population of microcytic cells. The mean corpuscular Hgb is also reduced, albeit less so, whereas the mean corpuscular Hbg concentration and Hgb distribution width are both elevated significantly in Tmod1−/−Tg+ mice. These data suggest a population of cells with increased Hgb concentration. In separate control experiments, we found that hematologic values in Tmod1+/+Tg+ mice did not differ significantly from nontransgenic Tmod1+/+ mice (data not shown), consistent with the normal levels of Tmod1 observed in ghosts from transgenic and nontransgenic mice (Figure 1).
Examination of cells in Wright-Giemsa–stained blood smears and by SEM shows considerable heterogeneity in cell sizes with numerous smaller and oddly shaped cells (Figure 5A-B). Strikingly, in addition to populations of microcytic and spherocytic cells, numerous rounded (“fat”) elliptocytic cells are also evident (Figure 5A-B). Despite the mild anemia and abnormalities in RBC shapes, the percentage of circulating reticulocytes is not increased (Table 1), but this is probably because of a hematopoiesis-restricted anemia that often occurs with age in many normal inbred strains of mice of 1 year of age or older.41 To confirm this supposition, we examined reticulocyte percentages in younger animals between 2 and 3 months of age and observed a small but significant increase in the percentage of circulating reticulocytes in Tmod1−/−Tg+ mice compared with Tmod1+/−Tg+ or Tmod1+/+Tg+ mice (Table 2). Despite the increased percentage of reticulocytes in the absence of Tmod1, spleen weights did not differ significantly between genotypes.
Tmod1-null RBC shapes are heterogeneous with rounded elliptocytes, spherocytes, and microcytes, resembling spherocytic elliptocytosis. (A) Peripheral blood smears from Tmod1−/−Tg+ mice show significant populations of rounded elliptocytes (arrowheads), as well as spherocytes and a few microcytes (arrows). (B) Scanning electron microscopy (SEM) of RBCs from a Tmod1+/+Tg+ mouse and a Tmod1−/−Tg+ mouse show reduction of the central biconcavity as well as oval-shaped, spherical, and some smaller cells in the absence of Tmod1. Bars, 10 μm.
Tmod1-null RBC shapes are heterogeneous with rounded elliptocytes, spherocytes, and microcytes, resembling spherocytic elliptocytosis. (A) Peripheral blood smears from Tmod1−/−Tg+ mice show significant populations of rounded elliptocytes (arrowheads), as well as spherocytes and a few microcytes (arrows). (B) Scanning electron microscopy (SEM) of RBCs from a Tmod1+/+Tg+ mouse and a Tmod1−/−Tg+ mouse show reduction of the central biconcavity as well as oval-shaped, spherical, and some smaller cells in the absence of Tmod1. Bars, 10 μm.
Osmotic gradient ektacytometry shows that RBCs from Tmod1−/−Tg+ mice exhibited significantly decreased maximum deformability (DImax), indicative of decreased surface area to volume and more rigid cells (Figure 6A-C).29 Tmod1-null RBCs also exhibited an increased osmolarity at which the deformability was at a minimum, also consistent with decreased surface area to volume (Figure 6D). This conclusion is supported by osmotic fragility curves that show an increased osmolarity at which 50% of cells are lysed (Figure 6F-G). Unexpectedly, the Tmod1-null RBCs also exhibited an increase in osmolarity for the DI along the hypertonic arm of the curve (Figure 6E). However, the broad, double peak for the DImax in the Tmod1-null RBCs is indicative of considerable heterogeneity in cell water content, with an increased subpopulation of more dehydrated cells. This double peak profile in osmotic ektacytometry is characteristic of RBCs in human hereditary elliptocytosis (HE), indicating 2 cell populations with different deformabilities.42,43 Overall, the ektacytometry is consistent with the presence of rounded elliptocytes, microcytes, and spherocytes with a reduced surface area to volume for RBCs from Tmod1−/−Tg+ mice, as shown in the smears and hematology analyses above.
Osmotic deformability and fragility curves for RBCs from Tmod1-null mice are consistent with mild spherocytic elliptocytosis. (A) Representative osmotic deformability curves measured by ektacytometry for RBCs from Tmod1+/+Tg+ mice and (B) from Tmod1−/−Tg+ mice, showing decreased deformability with a double-humped peak characteristic of elliptocytosis.43 (C) The maximum deformability, DImax, a direct measure of the ability of the cells to deform under isotonic conditions, is reduced approximately 17% for Tmod1-null RBCs, from 0.481 to 0.402. (D) The osmolality at which cells are least deformable under hypotonic conditions, Omin, is a measure of the surface area-to-volume ratio and corresponds to the osmolality at which 50% of cells lyse in a standard osmotic fragility test; Omin is increased in Tmod1-null RBCs. (E) The osmolality at which cells exhibit half-maximal deformability under hypertonic conditions is affected by both mechanical properties of the membrane and hemoglobin concentration.29 The slight increase for Tmod1-null RBCs, with an increase in mean corpuscular Hbg concentration (Table 1) is consistent with modestly weakened mechanical properties of the membrane. (F) The osmotic fragility curve shows a small rightward shift for Tmod1-null RBCs, confirming the shift in Omin. (G) The osmolality at which 50% of RBCs are lysed is increased significantly for Tmod1-null RBCs. Nine (A-E) and 6 (F-G) mice of each genotype (Tmod1+/+Tg+ and Tmod1−/−Tg+). ***P < .001; **P < .01; *P < .05.
Osmotic deformability and fragility curves for RBCs from Tmod1-null mice are consistent with mild spherocytic elliptocytosis. (A) Representative osmotic deformability curves measured by ektacytometry for RBCs from Tmod1+/+Tg+ mice and (B) from Tmod1−/−Tg+ mice, showing decreased deformability with a double-humped peak characteristic of elliptocytosis.43 (C) The maximum deformability, DImax, a direct measure of the ability of the cells to deform under isotonic conditions, is reduced approximately 17% for Tmod1-null RBCs, from 0.481 to 0.402. (D) The osmolality at which cells are least deformable under hypotonic conditions, Omin, is a measure of the surface area-to-volume ratio and corresponds to the osmolality at which 50% of cells lyse in a standard osmotic fragility test; Omin is increased in Tmod1-null RBCs. (E) The osmolality at which cells exhibit half-maximal deformability under hypertonic conditions is affected by both mechanical properties of the membrane and hemoglobin concentration.29 The slight increase for Tmod1-null RBCs, with an increase in mean corpuscular Hbg concentration (Table 1) is consistent with modestly weakened mechanical properties of the membrane. (F) The osmotic fragility curve shows a small rightward shift for Tmod1-null RBCs, confirming the shift in Omin. (G) The osmolality at which 50% of RBCs are lysed is increased significantly for Tmod1-null RBCs. Nine (A-E) and 6 (F-G) mice of each genotype (Tmod1+/+Tg+ and Tmod1−/−Tg+). ***P < .001; **P < .01; *P < .05.
Discussion
The short actin filaments capped by Tmod1 at their pointed ends and α/β-adducin at their barbed ends are key structural features of the spectrin-actin lattice in the RBC membrane skeleton. In this study, we show that Tmod1-null mice exhibit a mild anemia with features resembling human spherocytic HE, including rounded elliptocytic as well as spherocytic shapes, reduced surface area to volume, dehydration, reduced cellular deformability, and increased osmotic fragility.9,11 The sphero-elliptocytic phenotype in the Tmod1-null mouse is consistent with membrane instability and reduced deformability as a consequence of disruptions in horizontal interactions in the membrane skeleton,1,11 because of actin filament length mis-regulation and disturbances in network connectivity (Figure 4). The attenuated spectrin-actin lattice may be due to altered numbers of spectrins attached to shorter or longer actin filaments, or to weakening of spectrin interactions at junctional complexes, as described for the 4.1R-deficient spectrin-actin lattice,33,44 or both. Absence of Tmod1 may also contribute indirectly to disruptions in membrane protein linkages at junctional complexes by mis-localization or disappearance of binding sites along longer or shorter actin filaments, but this will require further investigation. The mild elliptocytic phenotype is probably a consequence of the relatively small changes in actin filament lengths and network connectivity, because of molecular compensation by Tmod3 in the absence of Tmod1.
Comparison of the Tmod1-null RBC phenotype to the α- and β-adducin-null phenotypes is instructive. The overall RBC phenotype of adducin-null mice appears to resemble HS, with stiffer, smaller, and dehydrated RBCs with loss of membrane surface area, leading to reduced survival compensated by increased RBC production.30,31,35,45 Although RBCs in peripheral blood smears from β-adducin–null mice show rounded elliptocytes in addition to spherocytes,30,45 osmotic ektacytometry profiles30 do not show the broad double peak for the DImax as we observe in RBCs from Tmod1-null mice (Figure 6). Investigation of membrane skeleton structure in RBCs from β-adducin–null mice with the use of atomic force microscopy shows some disorganization, but it is difficult to compare with our study because individual molecular features are difficult to discern in the atomic force microscopic images.46 The α-adducin–null mouse, with RBCs also lacking β- and γ-adducin, contains predominantly spherocytic and microcytic cells in peripheral blood smears.31,35 In both the β-adducin–null and the α-adducin–null RBCs, actin levels are unchanged, but levels of the barbed end capping protein EcapZ are greatly increased,31,34,35 probably compensating for adducin's function in capping actin filament barbed ends. This may ameliorate changes in actin filament length that would otherwise occur, preventing severe disruptions of spectrin-actin lattice organization (ie, disrupted horizontal interactions), but this has not been examined directly. On the basis of this reasoning, the predominant HS phenotype may arise according to the loss of adducin's linkage to band 3, thus abrogating an important junctional complex linkage to the membrane.8
A striking finding of our study is that Tmod1-null RBCs now contain Tmod3 associated with the membrane skeleton. This is not due to replacement of Tmod1 on the membrane with Tmod3 that has translocated from the cytosol, because wild-type mouse RBCs do not contain Tmod3 (Figure 3). Because Tmod3 mRNA is expressed ubiquitously in most cells and tissues,12,47 we speculate that Tmod3 protein may be produced in erythroblast progenitors but only be recruited into the membrane skeleton to cap actin filament pointed ends when Tmod1 is lacking. Insufficient levels or altered properties of Tmod348,49 as the membrane skeleton assembles during late stages of erythroblast terminal maturation may lead to aberrant actin filament assembly, resulting in abnormal spectrin-actin lattice organization in mature RBCs. Alternatively, actin filaments may assemble normally into the membrane skeleton during differentiation in the absence of Tmod1 but undergo length redistribution during shear stress in the circulation because of insufficient capping by low levels of Tmod3. This would be consistent with prevailing models to explain disorganization of the membrane skeleton lattice in human HE disorders.8,9,11,50 Additional studies will be required to address the timing of Tmod1 and Tmod3 message and protein expression and their relationship to actin and membrane skeleton assembly during terminal differentiation of erythroblasts into RBCs.
Tmods stabilize TM associations with actin filament pointed ends in vitro12,13 and in vivo in the membrane skeleton.18,51 Absence of Tmod1 in lens fiber cells of the Tmod1-null mouse18 or depletion of Tmod3 by short hairpin RNA in polarized intestinal epithelial Caco2 cells51 leads to reduction of F-actin and TM levels on lateral membranes along with a disrupted spectrin membrane skeleton. Thus, we were surprised to find no significant changes in TMs associated with the membranes of Tmod1-null RBCs, considering the fact that Tmod3 is present at only one-fifth of normal levels. One possibility is that the associations of TMs with RBC actin are stabilized by binding to other proteins in the junctional complex, such as dematin, adducin, or protein 4.1R. A role for adducins in TM stabilization is suggested by studies on adducin knockout mice which show that the amount of TM associated with RBC membranes is reduced by greater than 60% in β-adducin–null mice,34,35 and by greater than 80% in α-adducin–null mice.31
Despite the lack of global changes in either TMs or actin in the membrane skeleton, Tmod1-null RBCs exhibit mis-regulation of actin filament lengths and larger network pore sizes as demarcated by the spectrin strands (Figure 4). How might these structural changes be explained? Tmod1-null RBCs contain Tmod3 at approximately one-fifth of normal levels of Tmod1, well below that necessary to cap all the short actin filaments in the membrane skeleton. Because total actin levels in the membrane skeleton are unchanged, the fraction of capped filaments and their lengths must be altered. If some uncapped actin filaments were to depolymerize and others to elongate from their pointed ends, then rearrangement of spectrin-4.1R attachments from shorter filaments (with fewer binding sites) to longer filaments (with more binding sites) could plausibly account for increses in lattice pore size (Figure 7). Once depolymerization has shortened an uncapped filament past the end of TM, the TM would dissociate and the filament would tend to shorten down to a minimum length that may be stabilized by α/β-adducin (Figure 7). Conversely, an elongating filament would tend to elongate until it can accommodate 2 TMs, self-associating end to end.13,20 Longer filaments with TM could then be capped by Tmod3, which is expected to bind with higher affinity to TM-actin ends than to pure actin ends (Figure 7).12,13 The model satisfies the properties of TMs and actin, and our observations that α/β-adducins, actin and TM levels in the membrane skeleton are unchanged in the absence of Tmod1. In effect, actin dissociation from some uncapped ends is compensated for by actin reassociation at other uncapped ends, leading to no net changes in actin filament polymer or in numbers of junctional complexes. A rigorous test of this mechanism will require determination of the numbers of spectrin strands attached to each junctional complex with respect to actin filament lengths, as well as the relative numbers of junctional complexes with unchanged, shorter, or longer actin filaments. Tmod1-null mouse RBCs may provide a new model to investigate the role of actin filament lengths and spectrin-actin lattice organization in control of RBC shape and mechanical properties with relevance to human spherocytic HE.
Hypothetical mechanism explaining how actin filament length changes can lead to rearrangements of the spectrin-actin lattice in the absence of Tmod1. (Top left) Model depicting the short actin filaments in expanded views of the spectrin-actin lattice of wild-type RBCs. The actin filaments are all the same length (16 subunits long), capped by Tmod1 at their pointed ends, α/β-adducin at their barbed ends, and with 2 TMs (TM5NM1 and TM5b) bound along their sides. On average, 6 spectrin-4.1R complexes are attached to each filament, creating the hexagonal lattice structure. (For simplicity, dematin is not included). (Top right) Hypothetical model depicting actin filament length redistributions in the absence of Tmod1. Substitution of Tmod3 for Tmod1 results in some filaments with normal lengths as in wild type, but insufficient levels of Tmod3 result in some uncapped filaments that either depolymerize or elongate, resulting in shorter or longer actin filaments. These shorter or longer filaments might have variable numbers of spectrin-4.1R attachments, resulting in irregular network organization and a more open lattice with larger pore sizes (Figure 4A-B). RBC TMs span along 6 actin filament subunits, requiring that actin filaments must be at least 12 subunits long for TM binding. Thus, TMs will dissociate from depolymerizing RBC filaments and associate with elongating filaments. TMs can be stabilized on longer actin filaments by virtue of TM head-to-tail self-association20 and TM-actin capping by Tmod3,12 accounting for no net changes in TMs in the absence of Tmod1.
Hypothetical mechanism explaining how actin filament length changes can lead to rearrangements of the spectrin-actin lattice in the absence of Tmod1. (Top left) Model depicting the short actin filaments in expanded views of the spectrin-actin lattice of wild-type RBCs. The actin filaments are all the same length (16 subunits long), capped by Tmod1 at their pointed ends, α/β-adducin at their barbed ends, and with 2 TMs (TM5NM1 and TM5b) bound along their sides. On average, 6 spectrin-4.1R complexes are attached to each filament, creating the hexagonal lattice structure. (For simplicity, dematin is not included). (Top right) Hypothetical model depicting actin filament length redistributions in the absence of Tmod1. Substitution of Tmod3 for Tmod1 results in some filaments with normal lengths as in wild type, but insufficient levels of Tmod3 result in some uncapped filaments that either depolymerize or elongate, resulting in shorter or longer actin filaments. These shorter or longer filaments might have variable numbers of spectrin-4.1R attachments, resulting in irregular network organization and a more open lattice with larger pore sizes (Figure 4A-B). RBC TMs span along 6 actin filament subunits, requiring that actin filaments must be at least 12 subunits long for TM binding. Thus, TMs will dissociate from depolymerizing RBC filaments and associate with elongating filaments. TMs can be stabilized on longer actin filaments by virtue of TM head-to-tail self-association20 and TM-actin capping by Tmod3,12 accounting for no net changes in TMs in the absence of Tmod1.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jawon Lee for help with genotyping mice and osmotic fragility measurements, Carmela Ferreira-Mota for reticulocyte counts, and Renee Chow for help with the diagram of the spectrin-actin network. We are particularly indebted to Marie Lecomte for noticing the sphero-elliptocytic shape of Tmod1-deficient red blood cells on a smear at a Red Cell Gordon Conference. We thank Erica L. Jacovetty for negative staining electron microscopy of membrane skeletons, performed at the National Resource for Automated Molecular Microscopy (NRAMM).
This research was supported by the NIH (grant HL083464, V.M.F.; grant HL092535, F.A.K.; grant HL088468, L.L.P; and grants HL59561 and HL56949, J.H.), and by the National Center for Research Resources P41 program (RR17573) to NRAMM.
National Institutes of Health
Authorship
Contribution: J.D.M. purified recombinant Tmods and antibodies, made RBC ghosts, and performed sodium dodecylsulfate–polyacrylamide gel electrophoresis, Western blotting, osmotic fragility, and blood smears; R.B.N. supervised the mouse colony, collected blood, performed osmotic fragility, measured spleen weights and actin filament lengths, analyzed data, and made figures and tables; N.E.K. performed Western blotting and data analysis; J.H. performed electron microscopy of unspread membrane skeletons; S.K.L and F.A.K. performed Advia hematologic analyses and osmotic ektacytometry; L.L.P. interpreted and photographed blood smears and performed SEM; and V.M.F. designed experiments, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Velia M. Fowler, Department of Cell Biology, CB163, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: velia@scripps.edu.
References
Author notes
J.D.M. and R.B.N. contributed equally to this study.