Abstract
Point mutations in the kinase domain of BCR-ABL are the most common mechanism of drug resistance in chronic myeloid leukemia (CML) patients treated with ABL kinase inhibitors, including imatinib. It has also been shown in vitro that mutations outside the kinase domain in the neighboring linker, SH2, SH3, and Cap domains can confer imatinib resistance. In the context of ABL, these domains have an autoinhibitory effect on kinase activity, and mutations in this region can activate the enzyme. To determine the frequency and relevance to resistance of regulatory domain mutations in CML patients on imatinib, we screened for such mutations in a cohort of consecutive CML patients with various levels of response. Regulatory domain mutations were detected in 7 of 98 patients, whereas kinase domain mutations were detected in 29. One mutation (T212R) conferred in vitro tyrosine kinase inhibitor resistance and was associated with relapse, whereas most other mutations did not affect drug sensitivity. Mechanistic studies showed that T212R increased the activity of ABL and BCR-ABL and that T212R-induced resistance may be partially the result of stabilization of an active kinase conformation. Regulatory domain mutations are uncommon but may explain resistance in some patients without mutations in the kinase domain.
Introduction
Chronic myeloid leukemia (CML) is caused by BCR-ABL, a constitutively active tyrosine kinase. The treatment of CML patients with imatinib, a small molecule inhibitor of ABL, induces durable complete cytogenetic responses in many patients in the chronic phase.1 However, the disease persists at low levels detectable only by reverse-transcribed polymerase chain reaction (RT-PCR) for BCR-ABL, and imatinib cessation usually leads to recurrence.2,3 In CML patients with progression beyond the chronic phase, imatinib responses are typically transient and development of imatinib resistance is common.4 Point mutations in the kinase domain of BCR-ABL that impair drug binding either directly or through allosteric mechanisms are the most common mechanism of drug resistance, whereas increased expression of BCR-ABL is observed in others.5-8 In a substantial fraction of patients, resistance remains mechanistically unexplained.
In vitro studies have shown that mutations in domains of BCR-ABL other than the kinase domain may also cause resistance. Specifically, mutagenesis screening has identified mutations outside the kinase domain in the neighboring linker, SH2, SH3, and Cap domains that confer imatinib resistance in the context of BCR-ABL.9 In the context of ABL, directed mutations in these domains at residues important for intramolecular interaction were shown to activate the kinase, consistent with an autoinhibitory function of the affected residues on the kinase domain.10,11 This was further elucidated by crystal structure analysis of the Cap-SH3-SH2-kinase domain fragment of ABL.12 The results from the in vitro mutagenesis screen and the crystal structures implied that residues critical for ABL autoinhibition are capable of rendering BCR-ABL resistant to imatinib in vitro and that mechanisms regulating BCR-ABL might overlap with those observed in ABL.13 To determine whether mutations in regulatory domains may contribute to clinical resistance, we screened a cohort of CML patients in various disease stages and with various levels of response to imatinib.
Methods
Patient samples
This study was approved by the Institutional Review Boards of Oregon Health & Science University (OHSU) and the University of Leipzig (Leipzig, Germany), and informed consent was provided by all patients in accordance with the Declaration of Helsinki. Peripheral blood or bone marrow mononuclear cells were isolated by density gradient centrifugation. Aliquots of cells were used for RNA extraction (RLT reagent, QIAGEN). Ninety-eight consecutive patients seen in the OHSU outpatient clinic were included, based on informed consent and the availability of high-quality sequence tracing throughout the ABL N-terminal regulatory and kinase domains. OHSU patients were grouped according to imatinib response on the basis of achievement of a complete cytogenetic response (CCyR) at any point during imatinib treatment. CCyR patients were those with absence of the Philadelphia chromosome (by standard karyotyping) of the index sample (n = 38), relapsed patients were defined by the loss of a previous CCyR (n = 22), and nonresponders were those not reaching a CCyR (n = 38). Samples from Leipzig (n = 27) were selected based on primary or acquired hematologic or cytogenetic resistance to imatinib.
Mutation analysis
RNA was isolated from all samples using the RNeasy kit (QIAGEN). Random hexamer primed cDNA was synthesized using Superscript reverse transcriptase (Invitrogen). Mutation screening was done by direct sequencing of BCR-ABL products generated by nested PCR using 2 sets of primers on BCR and ABL as described.14 All PCR reactions were set up in DNA-free enclosures. Potential mutations were cross-referenced against publicly accessible single nucleotide polymorphism (SNP) databases. All mutations were confirmed by repeat amplification and sequencing of the cDNA sample. Further, to exclude rare not previously reported SNPs, the N-terminal domains of native ABL gene were amplified using nested RT-PCR using published ABL forward primers15 in combination with the ABL reverse primers of the BCR-ABL amplification protocol.
Generation of mutant alleles
Mutagenesis of the native BCR-ABL cDNA in the pSRα mammalian expression vector was performed using the Quick Change mutagenesis kit (Stratagene). Correct mutagenesis was confirmed by sequence analysis covering the entire BCR-ABL sequence for each mutant.
Ba/F3 cell retroviral transduction
Infectious virus particles packaging wild-type (WT) and mutant p210BCR-ABL were produced by transient transfection of HEK293 T17 cells (ATCC) and supernatant collection after 48 hours of incubation at 37°C. Parental Ba/F3 cells were then infected with retroviral supernatants. Isolation of BCR-ABL-expressing cells was performed by 2 weeks of neomycin selection followed by withdrawal of interleukin-3.
Cell proliferation assays
Exponentially growing Ba/F3 cells expressing WT or mutant BCR-ABL were plated (in quadruplicate) at 4 × 103 per well in 96-well plates and exposed to increasing doses of imatinib, nilotinib, or dasatinib. After 72 hours, the methane thiosulfonate-based viability assay was performed and analyzed as described.16 The average of at least 4 experiments was reported from the mean inhibition of growth at each dose.
Phosphotyrosine immunoblot analysis
Ba/F3 cells expressing WT or mutant BCR-ABL were plated at 4 × 106 per well in 6-well plates and exposed to concentrations of imatinib that matched the cell proliferation experiments. Cells were incubated for 3 hours at 37°C, pelleted, and lysed directly in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer for 5 minutes at 95°C. BCR-ABL phosphorylation and expression were detected by immunoblot with mouse monoclonal phosphotyrosine antibody 4G10 (Upstate Biotechnology) and rabbit ABL antibody K12 (Santa Cruz Biotechnology), respectively.
Kinase assays
Transfection of HEK293 cells and immunoprecipitation of ABL protein and ABL in vitro kinase assay were carried out as described previously.11,17 The relative concentration of immunoprecipitated ABL protein was determined by immunoblotting (anti-ABL Ab-3; Oncogene Science) and subsequent quantification using the Li-Cor Odyssey system and normalized for ABL WT. Alternatively, hexahistidine-tagged full-length mutant and native BCR-ABL proteins were expressed in SF9 cells by baculoviral transduction and purified using nickel columns. Equal amounts of proteins were used for in vitro kinase assays, using a peptide substrate, as described.18
Quantitative RT-PCR
Quantitative RT-PCR was performed on peripheral blood or bone marrow aspirate specimens as described.19 Briefly, BCR-ABL and G6PDH transcripts were quantified using real-time quantitative PCR and fluorescent resonance energy transfer hybridization probes in a LightCycler instrument (Roche Applied Science). The BCR-ABL transcript levels are reported in percentages after conversion to the international scale, using a conversion factor derived from comparison of standard samples with a reference laboratory.20
Results
Prevalence of BCR-ABL SH3-SH2 domain mutations
Mutation screening in CML patients treated with imatinib yielded 9 novel mutations in regulatory domains, confirmed by bidirectional sequencing. These were dispersed in the Cap region (R47C), SH3 domain (K84N), SH3-SH2 connector (E123Q), SH2 domain (G144E, S154N, A196V, and T212R), and SH2-kinase linker (N231D and N231I; Table 1). Overall, regulatory domain mutations occurred in 7 of the 98 patients tested (7%), with 3 having multiple mutations: Patient 2 had 2 mutations, T212R and S154N. Patient 4 with CML in blast crisis had multiple mutations in both the SH3-SH2 domains (R47C and K84N) and the kinase domain (Q346R and H396P). Patient 5 with CML in accelerated phase had a mutation in the kinase domain (E292V) in addition to the N231I mutation in the SH2-kinase linker. Twenty-nine of the 98 patients (30%) had kinase domain mutations. Patients of all disease stages and with differing degrees of imatinib response were tested. SH3-SH2 domain mutations were found in 5 of 38 (13%) with a CCyR, 2 of 22 (9%) relapsed patients, and 0 of 38 nonresponders (Table 2). By comparison, kinase domain mutations were detected in 9 of 38 (24%) with a CCyR (consistent with our previous work21 ), 7 of 22 (32%) relapsed patients, and 13 of 38 (34%) nonresponders. A comparison of all novel mutations with publicly available SNP databases was negative. To exclude that the novel mutations reflect previously unrecognized rare polymorphisms, we sequenced ABL in all patients with available material (patients 1-5, Table 1) but found only WT sequence (data not shown). To address the question whether mutations within the SH2 and SH3 domains may occur in addition to mutations in the kinase domain, we sequenced the kinase and the regulatory domains in an additional cohort of patients with overt cytogenetic or hematologic resistance to imatinib, with (n = 14) and without (n = 13) previously detected kinase domain mutations. Sequencing of SH2 and SH3 domains was WT in 26 of 27 patients. In one patient, a 2-amino acid exchange in codon 117 was seen in 50% of amplicons that is predicted to introduce a stop codon (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These data suggest that the coexistence of kinase domain and regulatory domain mutations is rare, although such composite mutations might be detected in a larger cohort.
Follow-up analysis of CML patients with an SH3-SH2 domain mutation
Patients in whom an SH3-SH2 domain mutation was discovered were observed for their clinical response to imatinib before and after the detection of the mutation. A graph of the quantitative RT-PCR values of BCR-ABL is shown in Figure 1A. Patient 2, harboring the T212R mutation, had a large increase in BCR-ABL transcript level beginning at 29 months after the index sample. Patient 4 with blast crisis had a very high level of BCR-ABL at the time when 4 different mutations were detected but was subsequently switched to dasatinib, resulting in a reduction in transcript to an undetectable level. Among the remaining patients with detection of a mutation in SH3-SH2, only patient 3 with the A196V mutant had an increase of BCR-ABL transcript over baseline levels. The cytogenetic and mutation analyses before and after mutation analysis are shown in Figure 1B. Patient 2 had detection of T212R at 2 follow-up time points, whereas the other mutations were not found on follow-up samples, when available. Patient 2 had cytogenetic relapse, first at 12 months and worsening at 30 months after T212R detection. Patient 4, with the R47C, K84N, Q346R, and H396P mutations, had an extremely fast response to dasatinib with undetectable BCR-ABL transcripts after 1 month. The patients with the N231D, A196V, or G144E mutations have maintained a CCyR. For patient 7 with the E123Q mutation, follow-data were not available. Thus, of the mutations detected in the SH3-SH2 domains, only T212R was closely correlated with clinical loss of imatinib response. There was no evidence for clonal cytogenetic evolution in any of the cytogenetic analyses from this patient.
Follow-up data for patients with detection of a regulatory domain mutation. (A) Available quantitative RT-PCR data for BCR-ABL in patients with a SH3-SH2 domain mutation were graphed for the period of 12 months before to 36 months after the index sample with mutation detection. (B) Clinical timelines corresponding to the quantitative RT-PCR data are shown to indicate the cytogenetic and mutation analysis during the study period. All patients were taking imatinib during the period shown, unless switched to dasatinib when noted.
Follow-up data for patients with detection of a regulatory domain mutation. (A) Available quantitative RT-PCR data for BCR-ABL in patients with a SH3-SH2 domain mutation were graphed for the period of 12 months before to 36 months after the index sample with mutation detection. (B) Clinical timelines corresponding to the quantitative RT-PCR data are shown to indicate the cytogenetic and mutation analysis during the study period. All patients were taking imatinib during the period shown, unless switched to dasatinib when noted.
Structural modeling of mutations in ABL regulatory domains
The locations of the mutations detected in the Cap, SH2, SH3, and linker domains are shown in the crystal structure of autoinhibited ABL (Figure 2A).12 The location of some mutations suggests that they may activate the ABL kinase by destabilizing intramolecular interactions. For example, A196V occurs at the interface between the SH2 domain and the N-terminal Cap that latches the SH3-SH2 domain clamp in the autoinhibited conformation. Another example is residue 231, where N231D and N231I may destabilize the packing of SH3, the SH2-kinase domain linker, and the kinase domain in the autoinhibited conformation. On the other hand, T212R is a mutation in the SH2 domain that does not have obvious consequences in the autoinhibited conformation, as this residue is solvent exposed in this structure and is not in proximity to interfaces with other domains critical for autoinhibition. In contrast, in the active structure of ABL, the T212R mutation is close to the interface of the SH2 domain with the kinase domain (see Figure 5 for a more detailed discussion).
Locations of patient regulatory domain mutations in the autoinhibited conformations of ABL. The crystal structure of autoinhibited ABL in complex with the kinase inhibitor PD166326 (PDB entry 2FO0)12 displayed with the locations of the mutated residue side chains shown in green stick format. The mutations are dispersed across the SH2 (green), SH3 (yellow), and linker domains (red, magenta). Stick format atoms are color-coded for nitrogen (dark blue), oxygen (red), and sulfur (orange). The figure was created using PyMol.22
Locations of patient regulatory domain mutations in the autoinhibited conformations of ABL. The crystal structure of autoinhibited ABL in complex with the kinase inhibitor PD166326 (PDB entry 2FO0)12 displayed with the locations of the mutated residue side chains shown in green stick format. The mutations are dispersed across the SH2 (green), SH3 (yellow), and linker domains (red, magenta). Stick format atoms are color-coded for nitrogen (dark blue), oxygen (red), and sulfur (orange). The figure was created using PyMol.22
Kinase inhibitor sensitivity of SH3-SH2 domain mutants
To measure whether the mutations we discovered in patients confer imatinib resistance in vitro, each was introduced into BCR-ABL, stably expressed in Ba/F3 cells, and tested in cell proliferation assays with graded concentrations of kinase inhibitors. The T212R mutation decreased imatinib sensitivity by 2.4-fold compared with native BCR-ABL (Table 3; supplemental Figure 1). The E123Q mutation also caused a subtle but reproducible decrease in sensitivity by 1.3-fold. The other mutations did not significantly alter sensitivity to imatinib (Table 3). In a second set of experiments, the autophosphorylation of each BCR-ABL mutant was analyzed by immunoblot. Once again, the T212R mutant was uniquely resistant among those tested (Figure 3). For comparison, T224A, the most resistant SH3-SH2 mutant from an in vitro screen,9 was also tested. This mutant was found to be imatinib resistant to a level comparable with E123Q (Table 3; supplemental Figure 2). Thus, among the SH3-SH2 mutations, T212R conferred the highest degree of resistance.
Imatinib sensitivity of regulatory domain mutations in BCR-ABL. SH3-SH2 mutations were introduced into BCR-ABL and stably expressed in Ba/F3 cells. These cell lines were exposed to graded concentrations of imatinib for 4 hours and lysed directly into sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Immunoblots were performed for BCR-ABL autophosphorylation, and expression. T212R was the only mutant to display detectable resistance of BCR-ABL autophosphorylation to imatinib.
Imatinib sensitivity of regulatory domain mutations in BCR-ABL. SH3-SH2 mutations were introduced into BCR-ABL and stably expressed in Ba/F3 cells. These cell lines were exposed to graded concentrations of imatinib for 4 hours and lysed directly into sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Immunoblots were performed for BCR-ABL autophosphorylation, and expression. T212R was the only mutant to display detectable resistance of BCR-ABL autophosphorylation to imatinib.
SH3-SH2 mutations can have positive or negative effects on the kinase activity of ABL
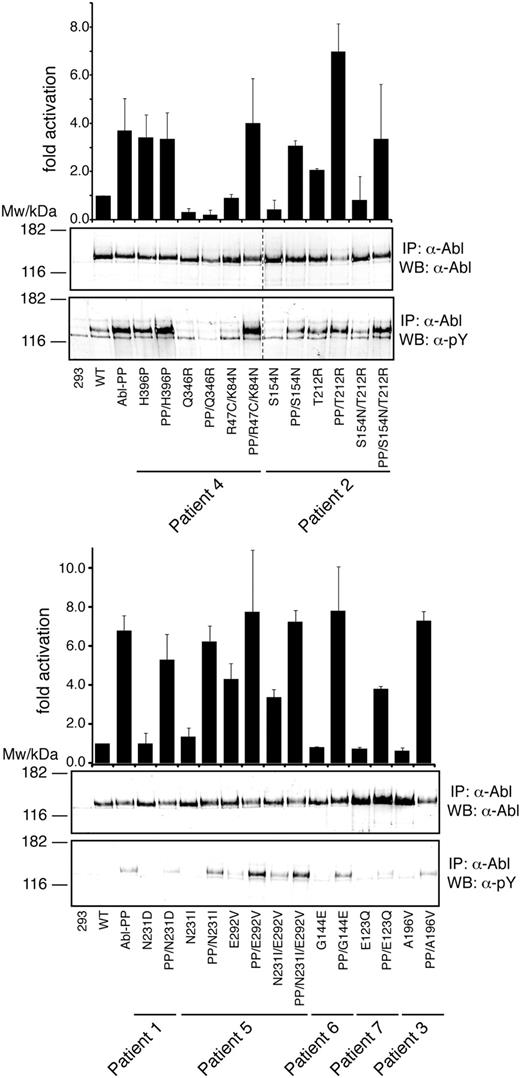
Given the location of the mutations in autoregulatory domains of ABL, we hypothesized that they may activate the kinase. Thus, we tested the effects of the regulatory domain mutations (and the concomitant E292V, Q346R, and H396P mutations) within the context of native and constitutively active ABL (Figure 4). Constitutive activation was achieved by introducing the P223E/P230E double mutation (ABL PP), as described.10 We found that the H396P, T212R, and E292V mutations led to a strong increase of kinase activity. The H396P mutation is located in the activation loop of the kinase domain and was found to confer resistance by stabilizing the active kinase conformation to which imatinib cannot bind.23 For H396P, no further increase in kinase activity was observed on additional introduction of the PP mutation (Figure 4). In contrast, T212R caused less pronounced activation of ABL than H396P but clearly “overactivated” ABL PP (Figure 4). A second group of mutations, including R47C/K84N, N231I, and G144E, had no consistent effects on ABL kinase activity alone or in combination with the PP mutation. Unexpectedly, a third group of mutations showed an intermediate (S154N) or very strong (Q346R) negative effect on kinase activity (Figure 4). For both mutations, the negative effect on kinase activity appeared to dominate over the activating mutations detected in the same patient. For example, even the strongly activating H396P mutation causes only a minor rescue of kinase activity in combination with the Q346R mutation. Likewise, S154N abrogated the activation conferred by T212R. These data reveal that point mutations in the kinase and regulatory domains of ABL exert complex and sometimes opposing effects on the kinase activity. However, all 3 mutations that conferred a significant degree of imatinib resistance (T212R, E292V, and H396P) consistently activated ABL.
Kinase activation of regulatory domain mutations in ABL. Immunoprecipitated ABL proteins expressing patient SH3-SH2 domain mutations alone or together with the ABL-activating PP (P223E/P230E) mutations were assayed for activity by in vitro kinase assays with an optimal substrate peptide. The graphs show catalytic activity relative to ABL for 4 experiments (from 2 independent transfections) done in duplicate (mean ± SD). Representative immunoblots showing equal Abl protein levels (top panel) and degree of Abl autophosphorylation (bottom panel) are shown below the bar graphs. Note that, in the left panel, 2 lanes with unrelated samples were cropped between lanes 8 and 9 (indicated by dashed line).
Kinase activation of regulatory domain mutations in ABL. Immunoprecipitated ABL proteins expressing patient SH3-SH2 domain mutations alone or together with the ABL-activating PP (P223E/P230E) mutations were assayed for activity by in vitro kinase assays with an optimal substrate peptide. The graphs show catalytic activity relative to ABL for 4 experiments (from 2 independent transfections) done in duplicate (mean ± SD). Representative immunoblots showing equal Abl protein levels (top panel) and degree of Abl autophosphorylation (bottom panel) are shown below the bar graphs. Note that, in the left panel, 2 lanes with unrelated samples were cropped between lanes 8 and 9 (indicated by dashed line).
To test the converse, we introduced several activating ABL regulatory domain mutations (PP, K51A [Cap region], W99A [SH3 domain], and Y139D [SH2 domain]) into BCR-ABL. These mutations disrupt intramolecular contacts within the autoinhibited conformation of ABL.10-12 The mutants were expressed in BaF3 cells and the cells tested for their sensitivity to imatinib. No significant differences in sensitivity were seen compared with cells expressing native BCR-ABL (Table 3, supplemental Figure 3), suggesting that there is no tight correlation between constitutive activation in ABL and imatinib resistance in BCR-ABL.
Mechanistic analysis of T212R-induced resistance
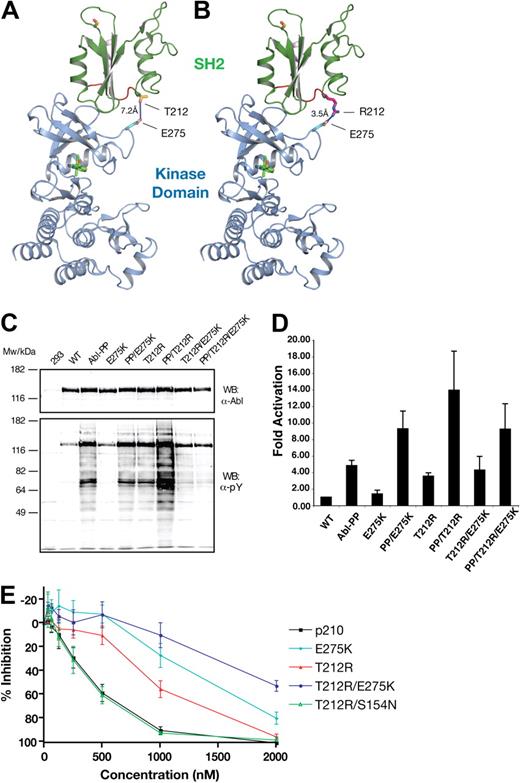
Because the T212R mutant was correlated with clinical loss of imatinib response, was resistant to imatinib in vitro, and showed a strong activation in kinase activity assays, we decided to analyze the structural and mechanistic basis for T212R-mediated resistance in more detail. Given that T212 is distant from the imatinib-binding site and is not located in a region that is known to be critical to maintain ABL autoinhibition, we reasoned that this mutation may confer resistance by a different mechanism. We modeled T212R on the active conformation of ABL, in which the SH2 domain changes its position substantially with respect to the kinase domain, binding its N-terminal lobe (Figure 5A).24,25 This binding of the SH2 domain on top of the N-lobe was shown to be critical for efficient catalytic activity in ABL.25 In this conformation, T212R localizes near the interface formed between SH2 and N-lobe of the kinase domain (Figure 5A). The threonine side chain at this position does not extend far enough to interact with the kinase domain, whereas arginine (in the T212R mutation) appears to be able to form a new electrostatic interaction with E275 (Figure 5B). We hypothesized that this may result in the stabilization of the active kinase conformation, to which imatinib binds less effectively.
Compensatory mutation analysis in the T212R mutant. (A) The threonine of residue 212 (green) is located near the interface between SH2 and the kinase domain's N-terminal lobe in the active conformation of ABL (PDB entry 1OPL, molecule B).24 (B) In the T212R mutant, arginine is in position to form an electrostatic interaction with E275 in the kinase domain (gray). The kinase domain is shown in gray with bound inhibitor in yellow. Stick format atoms are color-coded for nitrogen (dark blue), oxygen (red), and sulfur (orange). The figure was created using PyMol.22 (C) Single-, double-, and triple-mutant ABL expression constructs with T212R, ABL PP (P223E/P230E), and mutations at the E275 position were transiently transfected in HEK293 cells, lysed 40 hours later, and total protein extracts analyzed by anti-ABL and antiphosphotyrosine immunoblotting. (D) These ABL mutant proteins were next immunoprecipitated and assayed for activity by in vitro kinase assays with an optimal substrate peptide. The graphs show catalytic activity relative to ABL for 2 experiments done in duplicate (mean ± SD). (E) The E275K and S154N mutants were tested in native and T212R BCR-ABL by Ba/F3 cell proliferation assays, as before. E275K displayed substantial imatinib resistance alone, and this effect was additive to that of T212R in the double mutants. T212R/E275A had the same sensitivity to that of the T212R single mutant. In the T212R/S154N double mutant, the sensitivity of imatinib was reverted back to that of WT BCR-ABL.
Compensatory mutation analysis in the T212R mutant. (A) The threonine of residue 212 (green) is located near the interface between SH2 and the kinase domain's N-terminal lobe in the active conformation of ABL (PDB entry 1OPL, molecule B).24 (B) In the T212R mutant, arginine is in position to form an electrostatic interaction with E275 in the kinase domain (gray). The kinase domain is shown in gray with bound inhibitor in yellow. Stick format atoms are color-coded for nitrogen (dark blue), oxygen (red), and sulfur (orange). The figure was created using PyMol.22 (C) Single-, double-, and triple-mutant ABL expression constructs with T212R, ABL PP (P223E/P230E), and mutations at the E275 position were transiently transfected in HEK293 cells, lysed 40 hours later, and total protein extracts analyzed by anti-ABL and antiphosphotyrosine immunoblotting. (D) These ABL mutant proteins were next immunoprecipitated and assayed for activity by in vitro kinase assays with an optimal substrate peptide. The graphs show catalytic activity relative to ABL for 2 experiments done in duplicate (mean ± SD). (E) The E275K and S154N mutants were tested in native and T212R BCR-ABL by Ba/F3 cell proliferation assays, as before. E275K displayed substantial imatinib resistance alone, and this effect was additive to that of T212R in the double mutants. T212R/E275A had the same sensitivity to that of the T212R single mutant. In the T212R/S154N double mutant, the sensitivity of imatinib was reverted back to that of WT BCR-ABL.
To test for the proposed interaction of T212R with E275, a series of double mutants were made in an attempt to revert the observed effects of T212R. We hypothesized that E275K would prevent the formation of the electrostatic interaction and create a repulsive interaction between the 2 positively charged side chains. Thus, we introduced the T212R/E275K double mutant into ABL and ABL PP and tested the effects on total cellular tyrosine phosphorylation and in vitro kinase activity. As described in Figure 4, T212R led to an “overactivation” of ABL PP (Figure 5C lanes 3 and 7; and Figure 5D). Although the E275K mutation alone also “overactivated” ABL PP (Figure 5D) in combination with the T212R mutant, it reverted the observed increased cellular tyrosine phosphorylation and kinase activity (Figure 5C lanes 7 and 9; and Figure 4). In addition, we have introduced the T212R and T212R/E275K mutations into another activating ABL mutant (Y139D) and observed similar changes in in vitro kinase activity and total cellular tyrosine phosphorylation as for ABL PP (supplemental Figure 4; data not shown). In contrast, the T212R mutation had no activating effect in the context of ABL G2A that is activated by a different structural mechanism (supplemental Figure 4). In conclusion, these observations strongly argue for the formation of the hypothesized ionic interaction between the T212R mutant residue and E275 leading to stabilization of the active conformation of Abl. We cannot completely exclude the possibility that T212R may interfere with the binding of a negative regulator protein; but given its remoteness from the phosphotyrosine binding groove of the SH2 domain and any other known protein-protein binding surface, this is very unlikely.
Next, the T212R, E275K, and composite T212R/E275K mutations were introduced into BCR-ABL and the mutants stably expressed in Ba/F3 cells and tested for imatinib sensitivity in cell proliferation assays. Contrary to expectations, the T212R/E275K was more resistant than the T212R mutant (Figure 5E). The reason might be that the E275K single mutation was not neutral with respect to imatinib sensitivity but conferred 3.3-fold resistance on its own. We therefore tested 2 additional mutations (E275V and E275A) for their ability to abolish resistance in conjunction with T212R. However, compared with E275K, imatinib sensitivity was either unchanged (T212R/E275A) or slightly reduced (T212R/E275V), the latter probably reflecting reduced imatinib sensitivity of the E275V single mutant (data not shown). Thus, although the notion that T212R interacts with E275 is supported by the ABL kinase activity data, the situation is complicated by the fact that mutations in E275 alone affect imatinib sensitivity.
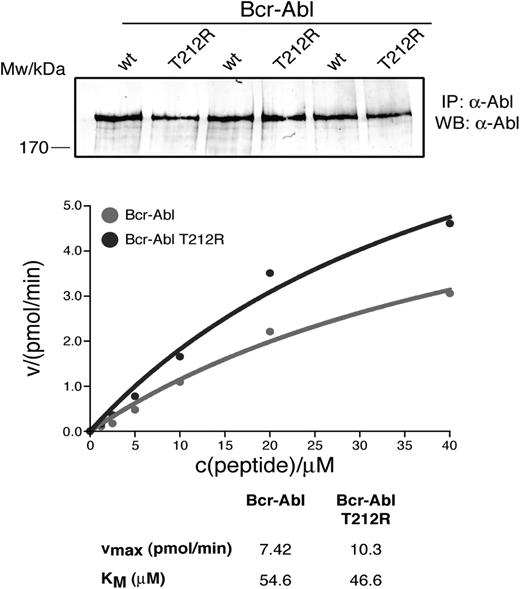
To test whether introduction of T212R into BCR-ABL would increase kinase activity over the native protein, similar to what we and others have reported for some kinase domain mutations, we compared the activity of the purified proteins in an in vitro kinase assay, using a peptide substrate.18,26 We observed a small, but reproducible, increase in Vmax of the T212R mutant over native BCR-ABL, suggesting that the mutation causes “overactivation” of BCR-ABL (Figure 6).
Kinase activity of full-length T212R BCR-ABL. Native BCR-ABL and T212R mutant BCR-ABL were overexpressed in HEK293 cells and immunoprecipitated in triplicate. Kinase assays were performed in triplicate at the indicated substrate concentrations, and the enzyme velocity was plotted as a function of peptide concentration. BCR-ABL levels were quantified from anti-ABL immunoblots of immunoprecipitated native and T212R mutant BCR-ABL and used to normalize levels of protein used in the kinase assay.
Kinase activity of full-length T212R BCR-ABL. Native BCR-ABL and T212R mutant BCR-ABL were overexpressed in HEK293 cells and immunoprecipitated in triplicate. Kinase assays were performed in triplicate at the indicated substrate concentrations, and the enzyme velocity was plotted as a function of peptide concentration. BCR-ABL levels were quantified from anti-ABL immunoblots of immunoprecipitated native and T212R mutant BCR-ABL and used to normalize levels of protein used in the kinase assay.
Discussion
We describe the occurrence of mutations in the Cap, SH3, SH2, and SH2-kinase linker domains of BCR-ABL in 7 of 98 CML patients on imatinib therapy. Point mutations upstream of the SH1 domain of BCR-ABL were also described by Talpaz et al in a study of dasatinib in patients with resistance to imatinib27 ; but to the best of our knowledge, we provide the first systematic analysis describing mutations outside of the kinase domain in CML patients treated with imatinib. Importantly, none of these mutations had previously been observed in our laboratory, and they were all confirmed in a second round of amplification and sequencing. Thus, although impossible to exclude with absolute certainty, PCR artifacts or contamination are very unlikely explanations for our findings. However, when characterized for imatinib sensitivity, only the T212R mutant proved substantially resistant and only the patient with the T212R (patient 2) subsequently relapsed. An important practical consequence is that biochemical validation of resistance mutations is mandatory, before assuming a causal relation between the presence of a mutation and resistance. This may also apply to some of the uncharacterized kinase domain mutations reported in patients with resistance. Conversely, most other SH3-SH2 domain mutations occurred in patients with CCyR and were not detected in subsequent samples, suggesting that they may have occurred in transiently amplifying cell clones, similar to the kinase domain mutant clones that have been reported in some CCyR patients.21 Thus, such mutations may either reflect bystander mutations without immediate phenotypic consequences, or they may affect disease biology in an as yet unknown fashion. We considered the possibility that regulatory domain mutations may occur in conjunction with kinase domain mutations, enhancing their resistance. However, sequencing of 14 patients with overt resistance to imatinib and documented kinase domain mutations failed to detect any such mutations, suggesting that this is not the case (supplemental Table 1). However, it remains possible that they might confer resistance in conjunction with mutations in the ABL C-terminus, which was not subjected to sequence analysis. In addition, their ability to confer drug resistance may be limited to the context of a primary CML cell, for example, if they interfered with the binding of a regulatory protein that is absent from the BaF/3 cells used in this study. Differences between primary cells and cell lines may also explain why we failed to detect any of the regulatory domain mutations previously identified by Azam et al in their in vitro screening experiment.9 Alternatively, more of these mutations may be detected in a larger cohort of patients.
The T212R resistance mutation is unique because of its location distant to the active site where imatinib binds. Because of the positioning of the SH2 domain on the N-lobe of the kinase domain in active ABL conformations that was shown recently to be essential for an efficient catalytic output of cytoplasmic tyrosine kinases,25 we hypothesized that the resistance may result from stabilization of a more active, less imatinib accessible conformation.28 Consistent with this prediction, T212R further increased the activity of the already activated ABL variants ABL PP (Figure 2C) and Y139D, but not G2A, which activates ABL through a different mechanism (supplemental Figure 1). Structural modeling suggested that T212R may form an electrostatic interaction with E275 that would stabilize the active, disinhibited conformation. This hypothesis is supported by the fact that exchange of glutamic acid with lysine (E275K) reduced the T212R-induced overactivation of ABL PP and ABL Y139D but, as predicted, did not reduce the activity of the ABL T212R/G2A double mutant. However, E275K failed to abrogate imatinib resistance in the context of BCR-ABL. This may reflect the fact that the E275K mutation by itself confers imatinib resistance, which may obscure the reversion of imatinib resistance that could be caused by disrupting the electrostatic interaction of T212R and E275. Indeed, mutations in the Beta3-Helix-C loop have been detected in some patients with imatinib resistance. Regardless, our data show that the relationship between intrinsic kinase activity and imatinib resistance is complex and that extrapolating from ABL to BCR-ABL is not straightforward. For example, several mutations with a strong activating effect on ABL (PP, K51A, W99A, and Y139D) failed to confer appreciable imatinib resistance to BCR-ABL. One potentially important difference between these mutations and T212R is that the former mutations may primarily destabilize the inactive kinase conformation but still allow for imatinib binding. In contrast, T212R may promote an active kinase conformation, from which imatinib is excluded. This would be similar to H396P, which is also imatinib resistant, activates ABL, “overactivates” BCR-ABL,28 and has been shown to stabilize an active kinase conformation.22 On the other hand, the fact that T212R confers some resistance to dasatinib, which binds an active kinase conformation,29 suggests the involvement of additional mechanism, the clarification of which will require further structural analysis.
In conclusion, we show that mutations in the regulatory domains of BCR-ABL occur in a subset of patients on imatinib. Most of these mutations do not alter imatinib sensitivity to an appreciable degree, suggesting that they may either represent “passenger” rather than “driver” mutations or influence the biology of BCR-ABL without changing the sensitivity to imatinib. However, T212R and, to a lesser degree, E123Q reduce imatinib sensitivity and may explain clinical resistance in some patients without detectable kinase domain mutations. Given that both mutants are sensitive to clinically achievable concentrations of dasatinib and nilotinib, second-line inhibitors should be considered in such patients. The clinical significance of mutations that do not appreciably change imatinib sensitivity remains to be determined. Although not directly related to drug resistance, they could still represent a marker of genetic instability. Determining their prognostic impact will require performing SH3-SH2 sequencing on a larger group of patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Heart, Lung, and Blood Institute (grant HL082978-01; M.W.D.), Doris Duke Charitable Foundation (B.J.D.), and the Leukemia & Lymphoma Society (B.J.D., M.W.D.).
National Institutes of Health
Howard Hughes Funding
Authorship
Contribution: D.W.S. designed and performed research and wrote the paper; O.H. designed and performed research and helped write the paper; I.K., S.W., T.B., L.P.T. T.L., and K.-H.D. performed research; R.D.P. contributed vital reagents; B.J.D. and G.S.-F. helped design research; and M.W.D. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael W. Deininger, Hematology and Medical Oncology, L592, Oregon Health & Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail: deininge@ohsu.edu.
References
Author notes
D.W.S. and O.H. contributed equally to this study.
Presented in abstract form at the 48th annual meeting of the American Society of Hematology, Orlando, FL, December 10, 2006.