Abstract
In up to one-third of patients with acute myeloid leukemia, a C-terminal frame-shift mutation results in abnormal and abundant cytoplasmic accumulation of the usually nucleoli-bound protein nucleophosmin (NPM), and this is thought to function in cancer pathogenesis. Here, we demonstrate a gain-of-function role for cytoplasmic NPM in the inhibition of caspase signaling. The NPM mutant specifically inhibits the activities of the cell-death proteases, caspase-6 and -8, through direct interaction with their cleaved, active forms, but not the immature procaspases. The cytoplasmic NPM mutant not only affords protection from death ligand-induced cell death but also suppresses caspase-6/-8–mediated myeloid differentiation. Our data hence provide a potential explanation for the myeloid-specific involvement of cytoplasmic NPM in the leukemogenesis of a large subset of acute myeloid leukemia.
Introduction
Nucleophosmin (NPM) is a multifunctional protein that resides predominantly in the nucleoli. It performs a multitude of nuclear functions, such as processing and transporting of ribosomal RNA and proteins,1 molecular chaperoning,2 and regulating the stability of tumor suppressors, such as p53,3 ARF,4 and c-Myc.5 However, its cytoplasmic role is largely unknown. In up to one-third of patients with acute myeloid leukemia (AML), a heterogeneous frame-shift mutation in the NPM1 gene results in skewed cytoplasmic accumulation of the NPM mutant protein (NPMc), and this is thought to function in cancer pathogenesis.6 As NPM1 is the most frequently mutated gene in AML with a normal karyotype, uncovering NPM's diverse cellular functions may hold the key to unraveling specific leukemogenic mechanisms involving the mutant protein.
Current AML research centers on the effect of nuclear NPM deprivation on genetic instability, derailment of cell cycle, and cancer progression. NPMc mutants were demonstrated to retain functional interactions with both their nuclear partners and wild-type NPM. Massive dislocation of the mutant protein to the cytoplasm was observed to deplete tumor suppressors and negative controllers of cell proliferation, such as p53,7 ARF,8 c-Myc-regulating Fbw7,5 Miz1,9 HEXIM1,10 and NF-κB,11 from the nucleus. The ensuing impairment in cell proliferation control was thus hypothesized to drive carcinogenesis. However, the overexpression of NPMc alone failed to demonstrate any transformation activity or enhancement of cell proliferation5,7 ; hence, these observations seemed incongruent with the aforementioned hypothesis. Furthermore, loss of p53 from the nucleus, which often results in genetic instability, is also not consistent with the fact that NPMc is exclusively associated with normal karyotypes.12 More importantly, NPMc mutations are so far identified only in AML,13 whereas ARF, c-Myc, and p53 defects are found in a multitude of cancers of diverse tissue origins.14-16 Such discrepancies argue for the existence of a hematopoietic-specific mechanism involving the mutant NPM.
Here, we identify NPMc as an inhibitor of active caspase-6 and -8, which are pivotal components of cell death signaling. Through the impediment of the caspase signaling cascade, the dislocated NPMc mutant not only raises the threshold for cell death initiation, but more interestingly, hinders the caspase-mediated myeloid differentiation process. Our data thus demonstrate a potential myeloid-specific oncogenic role for the NPMc mutant.
Methods
Reagents
Antibodies against NPM, caspase-3, -6, and -9 were purchased from Cell Signaling Technology, antibodies against cleaved caspase-3, caspase-8, poly(ADP-ribose) polymerase, and actin from Santa Cruz Biotechnology, antibody against caspase-7 from Neomarker, anti-Fas antibody from Upstate Biotechnology, and antibody against Oct-1 from Chemicon. Recombinant active caspase-3, -6, -7, -8, and -9, recombinant procaspase-3 protein, caspase-6 and -8 inhibitors, and recombinant tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) were bought from Calbiochem, and pNA-tagged caspase colorimetric substrates from Biovision. Actinomycin D, cycloheximide, and cytosine arabinoside (Ara-C) were purchased from Sigma-Aldrich.
cDNA constructs
Human wild-type NPM was cloned by polymerase chain reaction (PCR) from the OCI/AML-2 cell line. The mutant NPMc mutant was generated from the wild-type gene through PCR-based deletion and extension of the C-terminal. The glutathione-S-transferase (GST)–, His-, or GFP-tagged NPM and NPMc cDNA constructs were generated by PCR-based cloning of NPM or NPMc cDNA into pGEX-4T (GE Healthcare), pET-32 (Novagen), pcDNA3.1(−)-GFP (Invitrogen), or pLenti6.3 V5 (Invitrogen). NPM deletion mutants were generated by PCR-based cloning. cDNA for caspase-3, -6, and -8 and p53 was cloned by PCR from HeLa or HEK293T cells and subcloned into pXJ-40 plasmid, kindly provided by Dr BC Low, National University of Singapore.
Cell culture and transient transfection
For plasmid transfection in HeLa, 60% to 80% confluent cells were transfected with various amounts of plasmid DNA using Lipofectamine 2000 (Invitrogen). Suspension cells were transfected using Nucleofector Kit V (Amaxa). A total of 1.0 × 106 cells were resuspended in 100 μL of Nucleofector Solution V, together with 1 to 5 μg of the plasmids in a Nucleofector cuvette. HL-60 cells were transfected with program T-019, THP-1 with program U-013, and OCI/AML-3 with program X-001.
Lentiviral production
pLenti6.3 V5 plasmids (Invitrogen) containing gene constructs encoding GFP or GFP-tagged NPMc were cotransfected with pLP1, pLP2, and PVSV-G packaging constructs into HEK293FT cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. HEK293FT cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 10mM sodium pyruvate, 20mM l-glutamine, 1mM minimal essential medium nonessential amino acids, and 1% penicillin/streptomycin. The culture medium was collected 48 hours after transfection, cell debris was removed via centrifugation at 2500g at 4°C for 15 minutes, and the virus-containing supernatant was used for transduction.
Lentiviral transduction of human cord blood CD34+ cells
Peripheral blood samples were procured with informed consent in accordance with the Declaration of Helsinki, mainly from patients with NPMc− AML at initial diagnosis. Studies using these clinical samples were approved by the Institutional Review Board of Yong Loo School of Medicine at the National University of Singapore.
Human cord blood CD34+ cells (StemCell Technologies) were cultured in StemSpan SFEM (StemCell Technologies) supplemented with 100 ng/mL human Fms-like tyrosine kinase 3, 100 ng/mL human stem cell factor, 20 ng/mL human interleukin-3 (hIL-3), and 20 ng/mL hIL-6 (StemCell Technologies). A total of 5 × 105 cells were transferred onto 100-mm retronectin (Takara Bio)–coated culture dishes preloaded with lentivirus particles (multiplicity of infection, 50-100) according to the manufacturer's recommendations. Transduced cells were used to assay for apoptosis and differentiation 48 hours after transduction.
Immunoprecipitation
This is performed using the Seize Primary Immunoprecipitation Kit (Pierce Chemical) according to the manufacturer's instructions. Immune complexes were then eluted, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and analyzed by Western blotting.
GST pulldown
Recombinant GST-fusion NPM or NPMc proteins bound to glutathione beads were incubated with cell lysates (HEK293T, HeLa, or primary AML cells) at 4°C for 1 hour. The beads were collected by centrifugation and washed with phosphate-buffered saline containing 0.1% Triton X-100 3 times before loading onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis for immunoblotting.
siRNA design and transient transfection
siRNA for NPM was designed based on previous studies.3 The transfection of siRNAs into OCI/AML-3 was performed using Nucleofector electroporation system (Amaxa). Transfection efficiency of OCI/AML-3 cells was calculated to be 87.4% by transfecting the cells using Nucleofector with BLOCK-iT AlexaFluor Red Fluorescent Oligo (Invitrogen).
Apoptosis assay
Apoptosis was assessed by examination of nuclear morphology. Cells were loaded with 4mM Hoechst 33342 (Sigma-Aldrich) for 10 minutes. Apoptosis was characterized by scoring condensed and fragmented highly fluorescent nuclei from images captured using an Olympus BX60 compound microscope with an attached DP70 camera.
Flow cytometry
siRNA-transfected OCI/AML-3 cells were washed in phosphate-buffered saline and resuspended in 100 μL of phosphate-buffered saline with 3% fetal bovine serum, containing saturating amounts of fluorescein isothiocyanate-conjugated-CD11b, -CD14 (both Caltag), or -CD33 (Chemicon) antibodies. GFP or GFP-tagged NPMc transduced primary blood cells were similarly labeled with phycoerythrin-conjugated CD11b or CD14 antibodies (Santa Cruz Biotechnology). Data for the OCI/AML-3 and primary blood cells were collected using Beckman Coulter Epics Altra and BD FACSAria II flow cytometers, respectively. HL-60 and THP-1 cells transfected with GFP or GFP-tagged NPMc were isolated from the nontransfected cells using BD FACs Vantage SE (BD Biosciences).
May-Grünwald-Giemsa stain
Cells were fixed in 100% methanol for 10 minutes. The cells were then stained with May-Grünwald stain (BDH; diluted 1:2 times) for 7 minutes, followed by Giemsa stain (BDH; diluted 1:9 times) for 8 minutes. The slides were then washed briefly with deionized water and mounted with DPX (BDH) and analyzed under an Olympus BX60 compound microscope at 100× magnification.
Colony-forming assay
siRNA-transfected OCI/AML-3 (2.5 × 104 cells) were plated in triplicates in each well of 6-well plates containing Methocult methylcellulose medium (Stem Cell Technologies), mixed with α-minimal essential medium supplemented with 20% fetal bovine serum, and incubated for 21 days at 37°C with 5% CO2/95% air and 95% relative humidity.
Statistical analysis
Data are mean plus or minus SD for 3 separate experiments. Differences were analyzed by Student t test. P values less than .05 were considered statistically significant. More details can be found in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Microscopy
For Figure 2E, the cells were mounted with glycerol in PBS and micrographs were obtained with an Olympus BX60 fluorescent compound microscope under a UPlanApo 100×/1.35 oil objective. For Figure 3A (middle), cells were imaged with a Zeiss Axiovert 25 CFL inverted fluorescent microscope under a CP-Achromat 10×/0.12 objective. In Figure 3A (bottom) and Figure 3C (bottom), cells were mounted in Dako Fluoromount (Dako) and viewed with an Olympus BX60 microscope under a UPlanApo 20×/0.70 NA objective. For Figure 3C (top), cells were mounted in DAPI-containing Vectashield mounting medium (Vector Labs) and images were acquired with a Zeiss LSM 510 META laser scanning confocal microscope under EC Plan-Neofluar 100×/1.3 Oil M27 objective and the supplied LSM 510 META acquisition software. For Figure 4A, B, and E, cells were mounted in DPX and images were obtained using an Olympus BX60 microscope under UPlanApo 100×/1.35 oil objective. Figure 4D was obtained with a Zeiss Axiovert 25 CFL inverted microscope under CP-Achromat 5×/0.12 objective. Except for confocal microscopy images, all other images were acquired with an Olympus DP70 camera using DP Controller (Version 3.1.1.267) acquisition software. All images were re-sized and cropped using Microsoft PowerPoint 2007.
Results
NPM specifically inhibits caspase-6 and -8
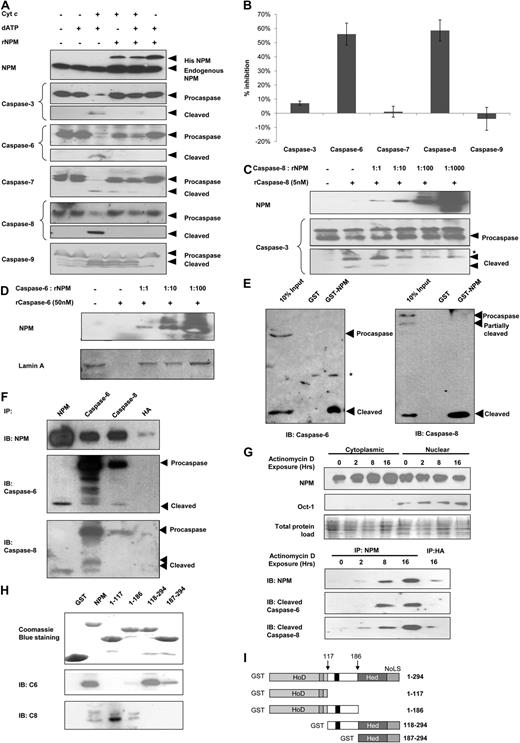
Overexpression of NPM has been shown to reduce apoptosis induced by different agents, whereas its down-regulation increased susceptibility to apoptosis in the p53-null AML cell line HL-60.17,18 To investigate how NPM acts as an antiapoptotic agent, we first examined the effects of NPM on the caspase activation cascade in vitro because apoptotic cell death is orchestrated by the activation of various caspases.19 We reconstituted the apoptotic signaling pathway using a cell-free system, in which cytochrome c was added to cytosolic extracts, with the addition of recombinant NPM. The addition of cytochrome c to the cytosol initiates an apoptotic program involving the sequential activation of the caspase-9, -3/-7, -6, and -8.20 Caspase-8 also cleaves caspase-3 and -7,21 thereby forming an amplification loop downstream of caspase-9. Addition of 5μM recombinant polyhistidine-tagged NPM to the cytochrome c–induced HEK293T cytosolic extracts inhibited the activation of caspase-3, -6, -7, and -8, but not that of caspase-9 (Figure 1A), suggesting inhibitions downstream of the initiator caspase.
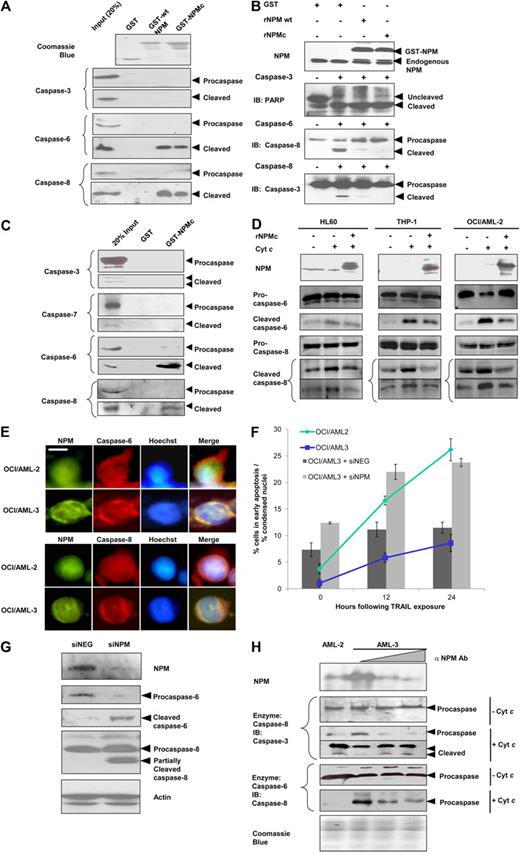
NPM inhibits and interacts with caspase-6 and -8. (A) Western blot analysis for cleaved caspases in S-100 cytosolic fractions extracted from HEK293T cells and incubated with cytochrome c (300nM) and/or deoxyadenosine triphosphate (dATP; 900nM), in the presence or absence of recombinant NPM (5μM) for 3 hours at 30°C. (B) S-100 cytosolic fractions from HEK293T were incubated with 1 unit of the various indicated recombinant active caspases (or 32 units for recombinant active caspase-8) in the presence or absence of 5μM recombinant His-tagged NPM for 1 hour at 30°C. Caspase activity was measured colorimetrically using pNA-taggd synthetic caspase substrates. (C) Western blot analysis of caspase-8 activity. Purified active recombinant caspase-8 (5nM) was incubated with 5nM to 5μM recombinant NPM and 150nM substrate recombinant procaspase-3 for 3 hours at 30°C. (D) Western blot analysis for caspase-6 activity. Purified recombinant caspase-6 (30nM) was incubated with 30nM to 3μM recombinant NPM and 625nM substrate GST-tagged lamin A for 3 hours at 30°C. (E) GST pull-down assay using equivalent amounts of GST or GST-NPM for cleaved caspase-6 and -8 in cytochrome c–treated HEK293T cytosolic extracts. (F) MN9D cells were treated with 500μM MPP for 12 hours, after which the cells were harvested and the total cell lysates extracted. Immunoprecipitation was carried out using antibodies against NPM, caspase-6, caspase-8, or hemagglutinin (HA; control). (G) Top: Western blot analysis for NPM in cytosolic or nuclear fractions of HeLa cells exposed to 0.40μM actinomycin D for 0 to 16 hours. Bottom: Cytosolic fractions collected were normalized to the same protein concentration and subjected to immunoprecipitation using antibodies against NPM or HA as a control. (H) Whole-cell lysate from apoptotically induced HEK293 cells was used in GST pull-down assay using either empty GST beads, GST-tagged full-length NPM, or its various deletion mutants. Immunoprecipitates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subjected to immunoblotting using anti-NPM, anti–caspase-6, or anti–caspase-8 antibody. (I) Domain structure of the full-length GST-NPM protein and the various deletion mutants used in the GST pull-down in panel H. Hed indicates heterodimerization domain; HoD, homodimerization domain; and NoLS, nucleolar localization signal. Arrows indicate the exact boundary of deletion in the various mutants.
NPM inhibits and interacts with caspase-6 and -8. (A) Western blot analysis for cleaved caspases in S-100 cytosolic fractions extracted from HEK293T cells and incubated with cytochrome c (300nM) and/or deoxyadenosine triphosphate (dATP; 900nM), in the presence or absence of recombinant NPM (5μM) for 3 hours at 30°C. (B) S-100 cytosolic fractions from HEK293T were incubated with 1 unit of the various indicated recombinant active caspases (or 32 units for recombinant active caspase-8) in the presence or absence of 5μM recombinant His-tagged NPM for 1 hour at 30°C. Caspase activity was measured colorimetrically using pNA-taggd synthetic caspase substrates. (C) Western blot analysis of caspase-8 activity. Purified active recombinant caspase-8 (5nM) was incubated with 5nM to 5μM recombinant NPM and 150nM substrate recombinant procaspase-3 for 3 hours at 30°C. (D) Western blot analysis for caspase-6 activity. Purified recombinant caspase-6 (30nM) was incubated with 30nM to 3μM recombinant NPM and 625nM substrate GST-tagged lamin A for 3 hours at 30°C. (E) GST pull-down assay using equivalent amounts of GST or GST-NPM for cleaved caspase-6 and -8 in cytochrome c–treated HEK293T cytosolic extracts. (F) MN9D cells were treated with 500μM MPP for 12 hours, after which the cells were harvested and the total cell lysates extracted. Immunoprecipitation was carried out using antibodies against NPM, caspase-6, caspase-8, or hemagglutinin (HA; control). (G) Top: Western blot analysis for NPM in cytosolic or nuclear fractions of HeLa cells exposed to 0.40μM actinomycin D for 0 to 16 hours. Bottom: Cytosolic fractions collected were normalized to the same protein concentration and subjected to immunoprecipitation using antibodies against NPM or HA as a control. (H) Whole-cell lysate from apoptotically induced HEK293 cells was used in GST pull-down assay using either empty GST beads, GST-tagged full-length NPM, or its various deletion mutants. Immunoprecipitates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subjected to immunoblotting using anti-NPM, anti–caspase-6, or anti–caspase-8 antibody. (I) Domain structure of the full-length GST-NPM protein and the various deletion mutants used in the GST pull-down in panel H. Hed indicates heterodimerization domain; HoD, homodimerization domain; and NoLS, nucleolar localization signal. Arrows indicate the exact boundary of deletion in the various mutants.
To determine which of the caspases were inhibited by NPM, we measured the activities of the various recombinant active caspases by monitoring the cleavage of their respective colorimetric substrates in the absence or presence of recombinant NPM. Among the 5 caspases examined, caspase-6 and -8 were most markedly inhibited (by 56.1% and 58.6%, respectively) in the presence of recombinant NPM. The activities of the other caspases were not inhibited significantly (Figure 1B). In the presence of recombinant NPM, recombinant caspase-6 and -8 added to HEK293T cell extracts also showed reduced cleavage of their physiologic targets (lamin A and procaspase-8 for caspase-6; Bid and procaspase-3 for caspase-8; supplemental Figure 1A). In contrast, reduction in the cleavage of poly(ADP-ribose) polymerase (substrate for active caspase-3 and -7) was not observed with the addition of recombinant NPM (supplemental Figure 1B). Taken together, these results suggest that NPM inhibited only caspase-6 and -8, but not caspase-3 or -7.
To further establish NPM as a caspase inhibitor, in vitro assays were reconstituted using purified recombinant caspases and saturating amount of substrates, as well as increasing amounts of recombinant polyhistidine-tagged NPM. For caspase-8, a 1:10 caspase-to-NPM stoichiometry resulted in reduction of cleavage of its downstream target procaspase-3 (Figure 1C), whereas a 1:1 stoichiometry for caspase-6 was efficient in inhibiting the cleavage of lamin A (Figure 1D). This further ascertained that NPM inhibited the proteolytic activities of caspase-6 and -8.
NPM interacts with cleaved caspase-6 and -8 but not their zymogens
X-linked inhibitor of apoptosis (XIAP) was previously shown to inhibit the activities of caspase-3 and -7 through direct interactions with the active fragments but not their inactive zymogens.22 NPM was likewise noted to directly bind both active caspase-6 and -8, but not their zymogens. GST-tagged recombinant NPM was shown to pull down cleaved forms of both caspase-6 and -8 from cytochrome c–induced HEK293T cell extracts, but not their respective zymogens (Figure 1E). Meanwhile, other components of the apoptotic signaling apparatus (cytochrome c, Apaf-1, or any forms of caspase-3, -7, and -9) showed no interaction with GST-tagged NPM (supplemental Figure 1D). In addition, antiibodies against endogenous NPM coprecipitated endogenous cleaved caspase-6 and -8 but not their procaspase forms, from apoptotically induced MN9D cells (Figure 1F). In this case, the neuronal MN9D cells were used, as we have previously demonstrated the activation of caspase-6 and -8, in response to the neurotoxin multipotent progenitor+,23 as well as the accelerated activation of both caspases after immunodepletion of NPM from MN9D lysates (supplemental Figure 1C). Notably, NPM knock-down increased neuronal cell apoptosis in mouse models.24 Finally, reciprocal immunoprecipitations were performed and antibodies against either caspase-6 or -8 coprecipitated NPM as well (Figure 1F), further confirming their interactions.
The spatial difference between the subcellular localizations of the nucleoli-bound NPM and the cytoplasmic apoptotic apparatus raises the question as to how NPM can inhibit the caspases in vivo. Here, immunoblotting analysis demonstrated that, with increasing duration of exposure to increasing dosage of the transcriptional inhibitor actinomycin D, more NPM translocated from the nucleus to the cytoplasm, as shown by the increase in cytoplasmic NPM and the concomitant decrease in nuclear NPM (Figure 1G top). When immunoprecipitation was performed using the aforementioned cytoplasmic extracts, normalized to the same protein concentration, with increasing exposure to actinomycin D, the increasing amounts of cytoplasmic NPM expectedly coprecipitated increasingly more cleaved caspase-6 and -8 (Figure 1G bottom). This demonstrates that, in response to apoptotic stimuli, NPM translocates to the cytoplasm to arrest the protease activities of capsase-6 and -8.
In addition, to identify the NPM determinants involved in active caspase-6 and -8 binding, several deletion mutants of NPM fused to GST were produced. These were tested for their ability to bind active caspase-6 and -8 in in vitro pull-down assays (Figure 1H-I). The deletion mutants were designed based on the functionally relevant NPM domains (the homo- and heterodimerization domains) (Figure 1I). We found that the NPM heterodimerization domain (residues 186-259) is essential for the interaction between NPM and active caspase-8. Coincidentally, this region is also crucial for the interaction of NPM with p53.3,25 In contrast, the N-terminal homodimerization domain is more important for interaction with cleaved caspase-6 (Figure 1H).
Cytoplasmic NPM mitigates cytotoxin-induced cell death
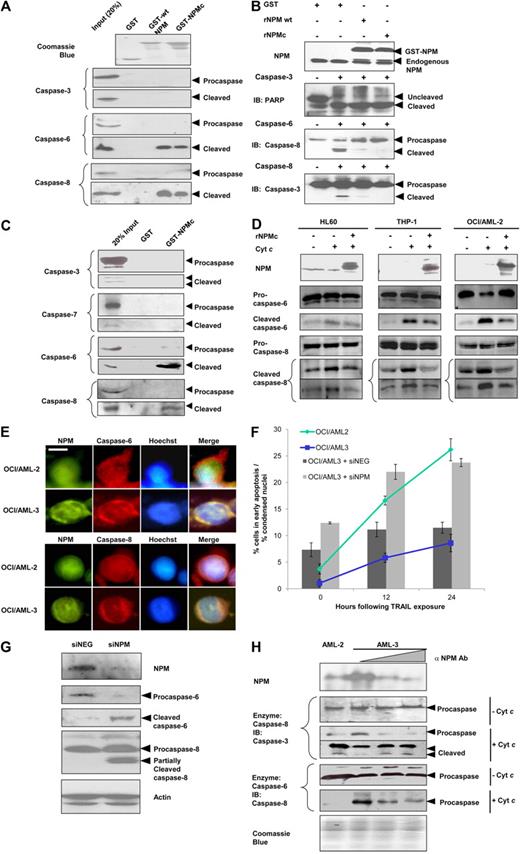
Given the ability of NPM to bind cleaved caspase-6 and -8, cytotoxin-induced cytoplasmic translocation of NPM may serve to attenuate a full-fledged apoptotic response until when cell death is inevitable. We thus hypothesize that aberrant and massive localization of NPM in the cytoplasm in AML may excessively inhibit the activities of the cleaved caspases and increase the threshold for the initiation of apoptosis. Like its wild-type counterpart, NPMc retained the ability to interact with both cleaved caspase-6 and -8 (Figure 2A) and inhibit the proteolytic activities of recombinant active caspase-6 and caspase-8 on their downstream substrates (procaspase-8 and procaspase-3, respectively, Figure 2B), despite harboring the mutation to its extreme C-terminal. Similarly, as with wild-type NPM, the mutant form of NPM showed no interaction with any form of caspase-3 and did not inhibit the proteolytic activity of added recombinant caspase-3 on endogenous poly(ADP-ribose) polymerase in the HeLa lysate. Moreover, the overexpression of NPMc rescued cell death induced by caspase-6 or -8 cotransfection, but not cotransfection of caspase-3 (supplemental Figure 2A).
Cytoplasmic dislocation of NPM impairs caspase signaling and protects against TRAIL-induced cell death. (A) GST, GST-NPM wild-type, or GST-NPMc immobilized on glutathione-Sepharose were assayed by immunoblotting for binding to caspase-6, -8, and -3 in cytochrome c–induced HeLa S-100 cytosolic extracts. As a control, lysates (30-50 μg) were run directly in gels. (B) Western blot analysis for caspase activities on physiologic substrates. S-100 cytosolic fractions from HeLa cells incubated separately with either recombinant active caspase-3, -6, or -8 in the absence or presence of 5μM recombinant GST tag alone, GST-tagged NPM wild-type, or NPMc for 1 hour at 30°C. Cleavage of the respective caspase substrates were assayed in immunoblots. (C) GST or GST-NPMc immobilized on glutathione-Sepharose were assayed by immunoblotting for binding to caspase-3, -7, -6, or -8 in cytochrome c–treated protein extracts of blast cells from peripheral blood of AML patients. (D) Western blot analysis for caspase activation. S-100 cytosolic fractions from 3 hematopoietic cell lines treated with cytochrome c (300nM), dATP (900nM), in the presence or absence of 5μM recombinant NPMc at 30°C for 30 minutes. (E) Subcellular localization of NPM, caspase-6 and -8 in OCI/AML-2 or -3 cells. Scale bar represents 5 μm. (F) Apoptosis was scored by analyzing condensed nuclei in Hoechst-stained OCI/AML-2 or OCI/AML-3 cells subjected to 50 ng/mL TRAIL treatment for up to 24 hours at 37°C (line graph). In addition, OCI/AML-3 cells transfected with 6nM specific (siNPM) or control (siNEG) siRNAs and subjected to 50 ng/mL TRAIL treatment for up to 24 hours, before flow cytometric analysis by annexin V staining with propidium iodide counterstain (bar graph). Apoptotic cells are defined as cells that stain positively for annexin V but not propidium iodide. (G) Western blot analysis for caspase activation in OCI/AML-3 cells transfected with siNPM or siNEG for 48 hours. (H) Western blot analysis of caspase activity on physiologic substrates. Cytoplasmic fractions from OCI/AML-2, or OCI/AML-3 cells treated separately with either recombinant caspase-6 or -8. Portions of the extract from OCI/AML-3 cells were immunodepleted of NPM.
Cytoplasmic dislocation of NPM impairs caspase signaling and protects against TRAIL-induced cell death. (A) GST, GST-NPM wild-type, or GST-NPMc immobilized on glutathione-Sepharose were assayed by immunoblotting for binding to caspase-6, -8, and -3 in cytochrome c–induced HeLa S-100 cytosolic extracts. As a control, lysates (30-50 μg) were run directly in gels. (B) Western blot analysis for caspase activities on physiologic substrates. S-100 cytosolic fractions from HeLa cells incubated separately with either recombinant active caspase-3, -6, or -8 in the absence or presence of 5μM recombinant GST tag alone, GST-tagged NPM wild-type, or NPMc for 1 hour at 30°C. Cleavage of the respective caspase substrates were assayed in immunoblots. (C) GST or GST-NPMc immobilized on glutathione-Sepharose were assayed by immunoblotting for binding to caspase-3, -7, -6, or -8 in cytochrome c–treated protein extracts of blast cells from peripheral blood of AML patients. (D) Western blot analysis for caspase activation. S-100 cytosolic fractions from 3 hematopoietic cell lines treated with cytochrome c (300nM), dATP (900nM), in the presence or absence of 5μM recombinant NPMc at 30°C for 30 minutes. (E) Subcellular localization of NPM, caspase-6 and -8 in OCI/AML-2 or -3 cells. Scale bar represents 5 μm. (F) Apoptosis was scored by analyzing condensed nuclei in Hoechst-stained OCI/AML-2 or OCI/AML-3 cells subjected to 50 ng/mL TRAIL treatment for up to 24 hours at 37°C (line graph). In addition, OCI/AML-3 cells transfected with 6nM specific (siNPM) or control (siNEG) siRNAs and subjected to 50 ng/mL TRAIL treatment for up to 24 hours, before flow cytometric analysis by annexin V staining with propidium iodide counterstain (bar graph). Apoptotic cells are defined as cells that stain positively for annexin V but not propidium iodide. (G) Western blot analysis for caspase activation in OCI/AML-3 cells transfected with siNPM or siNEG for 48 hours. (H) Western blot analysis of caspase activity on physiologic substrates. Cytoplasmic fractions from OCI/AML-2, or OCI/AML-3 cells treated separately with either recombinant caspase-6 or -8. Portions of the extract from OCI/AML-3 cells were immunodepleted of NPM.
To demonstrate that NPMc has similar characteristics in vivo, blast cell lysates harvested from the peripheral blood of AML patients, were obtained, and interactions between NPMc and cleaved caspase-6 and -8, but not any forms of caspase-3 or -7, were observed (Figure 2C). As in nonhematopoietic cell lysates, addition of untagged recombinant NPMc retarded cytochrome c-induced caspase-6 and -8 activation in the cytosolic extracts of the leukemic cell lines HL-60, THP-1, and OCI/AML-2 (Figure 2D). To further establish that NPMc retards the apoptotic response in cells, NPMc overexpression was found to protect against death ligand-induced cell death in HeLa and THP-1 cells (supplemental Figure 2B-C).
To further our investigation, we used a pair of AML cell lines with different subcellular localizations of NPM for comparison. The myelogenous OCI/AML-3 leukemic cells showed significantly cytoplasmic NPMc localization,26 whereas its counterpart, OCI/AML-2, which do not harbor the NPM mutation, showed nucleolar NPM localization. The cytoplasmic NPM mutant was further shown to colocalize with procaspase-6 and -8 in OCI/AML-3 (Figure 2E). This close proximity of NPMc to cytoplasmic procaspase-6 and -8 may allow NPMc to swiftly arrest caspase activation after death stimulation. Expectedly, in response to treatment with the death-receptor ligand TRAIL, which exerts its cytotoxicity via caspase-8, OCI/AML-2 cells demonstrated a greater extent of cell death compared with OCI/AML-3 cells (Figure 2F line graph). Conversely, siRNA-mediated knock-down of total NPM (inclusive of both wild-type NPM and NPMc because NPMc was reported to heterodimerize wild-type-NPM; hence, “pulling” along the wild-type form out to the cytoplasm27 ) from the OCI/AML-3 cells resulted in enhanced sensitivity to TRAIL-induced cell death, further indicating a role for NPMc in apoptotic inhibition in myeloid cells (Figure 2F bar chart).
The importance of NPM in maintaining the inhibition of caspases was exemplified by the enhanced cleavage of caspase-6 and -8 that was observed when NPM was knocked down in OCI/AML-3 cells, even in the absence of TRAIL treatment (Figure 2G). In vitro testing revealed that the relatively greater abundance of NPMc in OCI/AML-3 cytosolic extract, compared with that of OCI/AML-2, coincided with reduced cleavage of downstream procaspases after the addition of recombinant active caspase-6 or -8. Moreover, the removal of cytoplasmic NPM from OCI/AML-3 cytosolic extracts with immunodepletion allowed processing of downstream procaspases to resume (Figure 2H). Taken together, the results demonstrate how the excessive presence of NPMc in the cytoplasm may tamper with caspase-mediated signal flow and alter the cell death response.
Overexpression of cytoplasmic NPM blocks myeloid differentiation
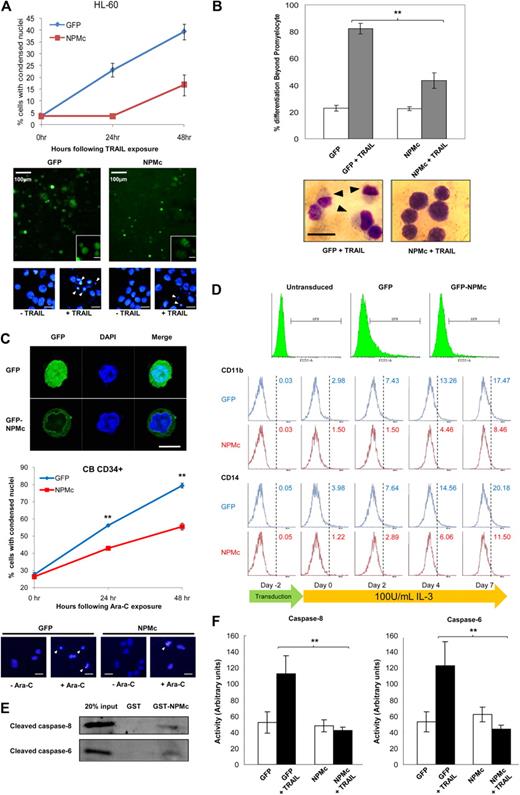
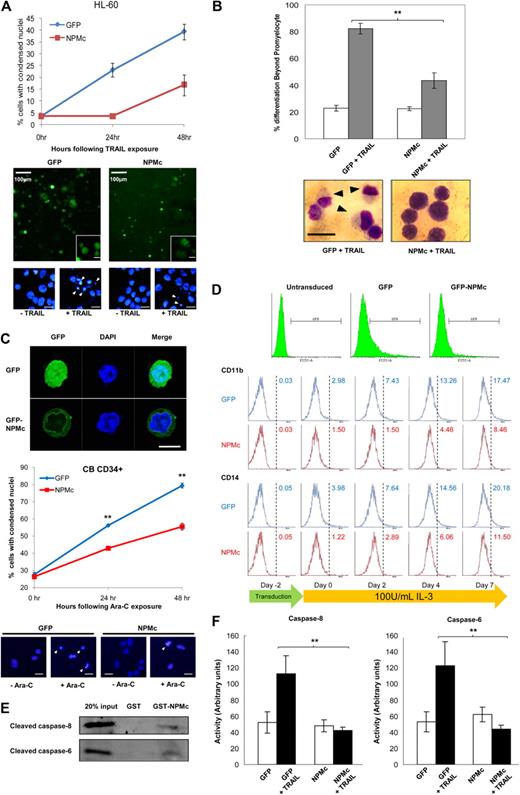
Treatment of the human leukemic HL-60 cell line with TRAIL was previously reported to result not only in rapid cytotoxicity, but also progressive maturation of the surviving cells along the monocytic lineage. Such dichotomous effect of TRAIL on HL-60 was also shown to be mediated by caspase-8.28 Here, we further demonstrated that NPMc overexpression in HL-60 cells not only markedly alleviated the cytotoxicity induced by TRAIL treatment (Figure 3A) but also significantly reduced the proportion of surviving cells with postpromyelocytic differentiation (Figure 3B). Similar results were obtained when GFP-tagged NPMc was overexpressed in cord blood-derived CD34+ cells, where cells overexpressing NPMc were both more resistant to Ara-C–induced cytotoxicity and IL-3–mediated myeloid differentiation (Figure 3C and Figure 3D, respectively). Differences in the extent of differentiation started to appear after transduction, possibly because of the cytokines supplemented in the culture media during transduction. These cytokines, although added to minimize differentiation and increase viability, inadvertently resulted in slight differentiation. Even so, primary cells overexpressing NPMc showed less differentiation compared with GFP-overexpressing cells.
NPMc protects against dichotomous effects of TRAIL treatment in HL-60 and Ara-C treatment in CD34+ primary blood cells. (A) Top: Apoptosis scored with condensed nuclei in Hoechst-stained HL-60 cells transfected with GFP or GFP-tagged NPMc and exposed to 100 ng/mL TRAIL for up to 48 hours. Bottom: Representative micrographs of HL-60 cells transfected with GFP and GFP-NPMc, with insets of micrographs at higher magnifications, and of nuclear staining with Hoechst 33342 showing condensed nuclei (white arrowheads). Unless otherwise stated, scale bars represent 10 μm. (B) Top: Number of cells with differentiation beyond the promyeloid stage was scored in May-Grünwald-Giemsa–stained HL-60 cells transfected with GFP or GFP-tagged NPMc, with or without 100 ng/mL TRAIL treatment. At least 150 cells were scored for each set of experiments. Bottom: Representative micrographs of May-Grünwald-Giemsa staining of TRAIL-treated GFP- or GFP-tagged NPMc-transfected cells. Black arrows represent differentiated cells; scale bar represents 15 μm. (C) Top: Confocal microscopy image of primary blood cells showing entire cell and cytoplasmic localization of GFP and GFP-tagged NPMc, respectively. Middle: Apoptosis scored with condensed nuclei in Hoechst-stained cord blood-derived CD34+ cells transduced with GFP or GFP-tagged NPMc and exposed to 5μM Ara-C for up to 48 hours. Bottom: Representative micrographs of nuclear staining of CD34+ cells showing condensed nuclei (white arrowheads). Scale bars represent 10 μm. (D) Flow cytometric analysis of GFP or GFP-tagged NPMc-transduced CD34+ cells. Top: GFP+ cells were gated and analyzed for immunostaining of differentiation markers CD11b and CD14 before transduction (day −2), 2 days after transduction (day 0), and days 2, 4, and 7 days after stimulation with 100 U/mL IL-3 (days 2, 4, and 7). (E) Cleaved caspase-6 and -8 were pulled down with glutathione-Sepharose–immobilized GST or GST-NPM from cytosolic extracts from HL-60 cells that were apoptotically challenged with cytochrome c and dATP as in Figure 1A. (F) Cells from panel A were lysed, and caspase-6 and -8 activities were measured using pNA-tagged caspase substrates. **P < .01. Data were obtained from 3 sets of experiments and presented as mean ± SEM.
NPMc protects against dichotomous effects of TRAIL treatment in HL-60 and Ara-C treatment in CD34+ primary blood cells. (A) Top: Apoptosis scored with condensed nuclei in Hoechst-stained HL-60 cells transfected with GFP or GFP-tagged NPMc and exposed to 100 ng/mL TRAIL for up to 48 hours. Bottom: Representative micrographs of HL-60 cells transfected with GFP and GFP-NPMc, with insets of micrographs at higher magnifications, and of nuclear staining with Hoechst 33342 showing condensed nuclei (white arrowheads). Unless otherwise stated, scale bars represent 10 μm. (B) Top: Number of cells with differentiation beyond the promyeloid stage was scored in May-Grünwald-Giemsa–stained HL-60 cells transfected with GFP or GFP-tagged NPMc, with or without 100 ng/mL TRAIL treatment. At least 150 cells were scored for each set of experiments. Bottom: Representative micrographs of May-Grünwald-Giemsa staining of TRAIL-treated GFP- or GFP-tagged NPMc-transfected cells. Black arrows represent differentiated cells; scale bar represents 15 μm. (C) Top: Confocal microscopy image of primary blood cells showing entire cell and cytoplasmic localization of GFP and GFP-tagged NPMc, respectively. Middle: Apoptosis scored with condensed nuclei in Hoechst-stained cord blood-derived CD34+ cells transduced with GFP or GFP-tagged NPMc and exposed to 5μM Ara-C for up to 48 hours. Bottom: Representative micrographs of nuclear staining of CD34+ cells showing condensed nuclei (white arrowheads). Scale bars represent 10 μm. (D) Flow cytometric analysis of GFP or GFP-tagged NPMc-transduced CD34+ cells. Top: GFP+ cells were gated and analyzed for immunostaining of differentiation markers CD11b and CD14 before transduction (day −2), 2 days after transduction (day 0), and days 2, 4, and 7 days after stimulation with 100 U/mL IL-3 (days 2, 4, and 7). (E) Cleaved caspase-6 and -8 were pulled down with glutathione-Sepharose–immobilized GST or GST-NPM from cytosolic extracts from HL-60 cells that were apoptotically challenged with cytochrome c and dATP as in Figure 1A. (F) Cells from panel A were lysed, and caspase-6 and -8 activities were measured using pNA-tagged caspase substrates. **P < .01. Data were obtained from 3 sets of experiments and presented as mean ± SEM.
As in the HEK293T and HeLa cells, glutathione-Sepharose immobilized GST-NPMc pulled down cleaved forms of both caspase-6 and -8 in HL-60 (Figure 3E). The NPMc overexpression also coincided with a significant reduction in caspase-6 and -8 activities with TRAIL treatment (Figure 3F). Together with the findings by Secchiero et al,28 our data indicate a role for the caspases in mediating the myeloid differentiation process, and that NPMc may regulate the process through the inhibition of these caspases.
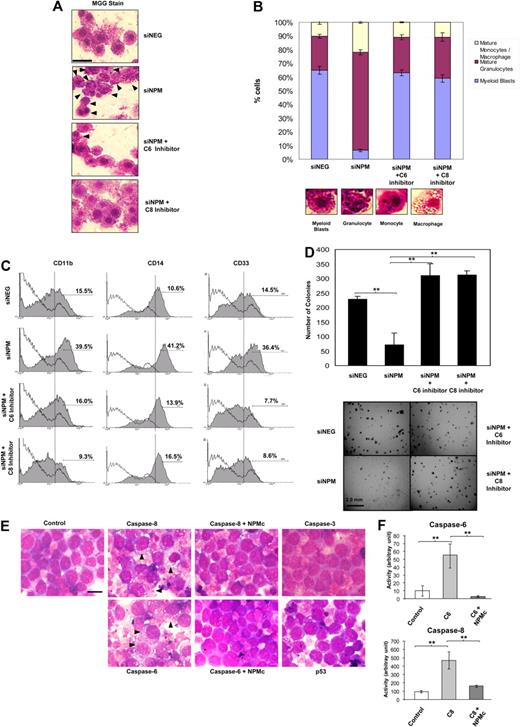
As a reciprocal experiment, we knocked down NPM in OCI/AML-3 and investigated changes in differentiation. Examination of these AML blasts revealed interesting changes in the morphology of the cells, which are suggestive of the involvement of NPMc in preventing terminal differentiation through caspase inhibition. A substantial proportion (65.1%) of the mock (siNEG) transfected cells showed morphology typical of myeloid blasts with round nuclei. Depletion of NPM substantially reduced the proportion of immature blasts to a mere 6.4%, with the majority showing granulocytic (71.1%) or monocytic (21.9%) morphology (Figure 4B). The increase in differentiated cells with NPM knockdown coincided with increased caspase cleavage (Figure 2G), enhanced expression of the myeloid differentiation markers CD11b, CD14, and CD33 (Figure 4C), and reduced colony-forming ability (Figure 4D). Treatment of the siNPM-transfected cells with caspase-6 or caspase-8 inhibitors, however, counteracted the effects of NPM depletion, resulting in the restoration of a blast-majority cell population (63.0% for caspase-6 inhibition, 59.0% for caspase-8 inhibition; Figure 4A and Figure 4B, respectively), reduced expression of the myeloid CD markers (Figure 4C), and resumption of colony-forming capability (Figure 4D).
Caspase-6– and -8–mediated myeloid differentiation is abrogated by AML-associated cytoplasmic NPM mutant. OCI/AML-3 cells were transfected with either specific (siNPM) or control (siNEG) siRNA with or without exposure to 10μM caspase-6 or caspase-8 inhibitors. (A) Forty-eight hours after transfection, the cells were subjected to May-Grünwald-Giemsa stain and observed under light microscope (original magnification ×100). Black arrowheads indicate cells differentiated past the promyelocytic stage. Scale bar represents 20 μm. (B) Top: Cells from panel A were examined, and the number of myeloid blasts, granulocytes, and monocytes/macrophages were scored. At least 200 cells were scored for each set of experiments. Bottom: Representative micrographs of a myeloid blast, mature granulocyte, monocyte, and macrophage. (C) Cells from panel A were also analyzed for expression of cell surface markers for myeloid differentiation (CD11b, CD14, and CD33). (D) Transformation-like phenotype of cells treated as in panel A. Cells 3 days after siRNA transfection were plated in methylcellulose medium and incubated at 37°C for 21 days. Top: Quantification of the colonies for each experimental condition. Bottom: Growth of the transfected cells in methylcellulose. Scale bar represents 2.0 mm. (E) HL-60 cells were transfected with caspase-6, -8, -3, or p53, and GFP. In the case of caspase-6 or -8 transfections, a portion of cells were cotransfected with GFP-tagged NPMc instead of GFP. Forty-eight hours later, cells were subjected to May-Grünwald-Giemsa stain and observed. Black arrowheads indicate cells differentiated beyond the promyelocytic stage. Scale bar represents 10 μm. (F) Caspase activities were measured in caspase-6– or -8–transfected cells from panel E. **P < .01. Data were obtained from 3 sets of experiments and presented as mean ± SEM.
Caspase-6– and -8–mediated myeloid differentiation is abrogated by AML-associated cytoplasmic NPM mutant. OCI/AML-3 cells were transfected with either specific (siNPM) or control (siNEG) siRNA with or without exposure to 10μM caspase-6 or caspase-8 inhibitors. (A) Forty-eight hours after transfection, the cells were subjected to May-Grünwald-Giemsa stain and observed under light microscope (original magnification ×100). Black arrowheads indicate cells differentiated past the promyelocytic stage. Scale bar represents 20 μm. (B) Top: Cells from panel A were examined, and the number of myeloid blasts, granulocytes, and monocytes/macrophages were scored. At least 200 cells were scored for each set of experiments. Bottom: Representative micrographs of a myeloid blast, mature granulocyte, monocyte, and macrophage. (C) Cells from panel A were also analyzed for expression of cell surface markers for myeloid differentiation (CD11b, CD14, and CD33). (D) Transformation-like phenotype of cells treated as in panel A. Cells 3 days after siRNA transfection were plated in methylcellulose medium and incubated at 37°C for 21 days. Top: Quantification of the colonies for each experimental condition. Bottom: Growth of the transfected cells in methylcellulose. Scale bar represents 2.0 mm. (E) HL-60 cells were transfected with caspase-6, -8, -3, or p53, and GFP. In the case of caspase-6 or -8 transfections, a portion of cells were cotransfected with GFP-tagged NPMc instead of GFP. Forty-eight hours later, cells were subjected to May-Grünwald-Giemsa stain and observed. Black arrowheads indicate cells differentiated beyond the promyelocytic stage. Scale bar represents 10 μm. (F) Caspase activities were measured in caspase-6– or -8–transfected cells from panel E. **P < .01. Data were obtained from 3 sets of experiments and presented as mean ± SEM.
We further showed that the overexpression of caspase-6 and -8, but not caspase-3 or p53, resulted in differentiation of the myeloid blasts in HL-60, which was characterized by widespread appearance of cells with increased cytoplasmic-to-nuclear ratio and phagocytic vesicles. Coexpression of NPMc with caspase-6 or -8, however, reverted the cells back to blast-like appearance with little cytoplasm and no vacuoles observed (Figure 4E), and this again coincided with a reduction in measured caspase-6 and -8 activities (Figure 4F). The use of the p53-null HL-60 cells at this juncture has been very useful, as it eliminated the influence of p53, whose stability was reported to be regulated by the subcellular localization of NPM mutation. Our overall data thus indicate that caspase-6– and -8–mediated signaling is essential for myeloid differentiation of the blasts. By inhibiting caspase-6 and -8, NPMc may retard terminal blast differentiation and contribute to blast transformation and accumulation during leukemogenesis.
Discussion
In this study, we have identified NPM as a novel caspase inhibitor. However, unlike the inhibitors of apoptosis and heat shock proteins (HSPs) that are collectively known to directly inhibit caspase-3, -7, and -9, NPM specifically inhibits caspase-6 and -8 through direct interactions. Whereas caspase-3, -7, and -9 are key players in the mitochondrial (intrinsic) pathway, caspase-8 is known to be part of the death receptor (extrinsic) pathway relaying signals from death-inducing signaling complex to downstream effectors. Notably, caspase-8 was proposed to trigger a feedback amplification loop through the activation of the intrinsic apoptotic pathways, resulting in the activation of caspase-9, -3, followed by -6 and -8, in that order. The cleavage of Bid29 by caspase-8 to produce tBid facilitates the release of cytochrome c from the mitochondria to initiate the intrinsic pathway. Although not the main effector caspase in the intrinsic apoptotic pathway, caspase-8 is also known to contribute to the caspase activation cascade downstream of apoptosome formation. As such, the inhibitions of caspase-8 and its upstream activator caspase-6 in this feedback loop by NPM under stress conditions may represent a parsimonious strategy in attaining effective silencing of the intricate and interconnected death-signaling network.
To date, no known nonviral enzymatic inhibitor of caspase-6 or -8 has ever been reported. c-FLIP, which inhibits death receptor pathway signaling, prevents activation of caspase-8 but is not known to be a direct enzymatic inhibitor. c-FLIP has high sequence homology and structural similarity to caspase-8 because it contains 2 death effector domains and a caspase-like domain.30 NPM, on the other hand, shows no significant protein sequence homology to caspase-8. The mechanism of inhibition of caspase-8 by NPM is probably different from that effected by c-FLIP, and possibly novel. Whereas c-FLIP prevents the recruitment of caspase-8 and FADD for proximity-induced cleavage of caspase-8, NPM appears to bind directly to cleaved caspase-8 and inhibit its proteolytic activity. Binding of the cleaved forms, but not pro-forms, of caspase-6 and -8 by NPM is reminiscent of the interaction between caspases in the intrinsic pathway and their inhibitors. Examples include c-IAP1, c-IAP2, XIAP, and survivin, which have been reported to bind to and inhibit the active forms of the terminal caspase-3 and -7.22,31,32 In addition, the small heat shock protein αB-crystallin was shown to bind to the p24 partially processed subunit of caspase-3 and inhibit its autoproteolytic maturation.33
XIAP is known to use its second baculovirus inhibitor of apoptosis repeat domain (BIR2) and an N-terminal linker segment to both interact with and inhibit caspase-3 and -7.34 Interestingly, our results demonstrate that NPM uses different domains to interact with either caspase-6 or -8, the C-terminal heterodimerization domain of NPM interacts with caspase-6, whereas the N-terminal homodimerization domain of NPM interacts with caspase-8 (Figure 1H). The mechanism of inhibition of caspase-3/-7 by XIAP is the result of steric blockade prohibitive of substrate binding, as revealed by x-ray crystallography.35,36 Whether the same mechanism underlies NPM-mediated inhibition of caspase-6 and -8 will require investigation using x-ray crystallography or nuclear magnetic resonance. Because NPM uses 2 separate domains to interact with the 2 caspases, it would be interesting to embark on further studies to determine whether NPM forms a multimeric inhibitory complex and whether NPM can interact with both caspases simultaneously. It is also interesting to note that inhibition of caspase-6 takes place at a 1:1 stoichometric ratio of caspase-6 to NPM, whereas the inhibition of caspase-8 requires a ratio of 1:10 or higher (Figure 1C-D). A possible explanation is that the interaction between NPM and caspase-8 requires the formation of oligomeric NPM, or that homodimerization of NPM competes with cleaved caspase-8 for interaction. A comprehensive structural analysis of their interactions would be required to answer this question in detail.
The recent discovery of a cytoplasmic NPM mutant in AML6 has led to our speculation that NPM may possess a cytoplasmic function that may be distinct from its nuclear or nucleolar functions. Our results suggest that abnormal impairment of caspase-6 and -8-mediated signaling by the aberrantly localized NPMc mutant could deliver a 2-pronged insult to hematopoietic cells and underlie leukemogenesis. Caspase-8 is often the apical caspase activated in the death receptor systems,37,38 which serves to eliminate unwanted hematopoietic cells through apoptosis.39 As active caspase-6 is a potent activator of procaspase-840 in cross-talks between the apoptotic pathways, inhibition of caspase-6 by cytoplasmic NPM mutant may further contribute to the suppression of caspase-8 activation and, hence, cell death signaling. Anomalies in cell death control have been known to contribute to tumorigenesis, as in the case of Bcl-2 overexpression in B-cell lymphoma, where survival, but not proliferation, is enhanced.41,42 Perturbations in the caspase signaling pathway by NPMc can likewise promote leukemic cell survival, even though NPMc does not enhance proliferation.5,7 By acquiring resistance toward cell death, leukemogenic mutations that would otherwise induce apoptotic responses can be allowed to accumulate.
Meanwhile, caspases are also known to possess nonapoptotic functions. Treatment of leukemic cells with the caspase inhibitor Z-VAD-fmk, or overexpression of the CrmA caspase-8 inhibitor prevents apoptosis and monocytic differentiation.43 Specifically, caspase-8 deficiency was shown to underlie defects in lymphocyte activation in autoimmune lymphoproliferative syndrome.44 More importantly, caspase-8 deletion has been shown to accelerate oncogenic transformation, through a yet unknown mechanism,45 and to arrest the differentiation of myelomonocytic cells in the bone marrow of mice.46 Such blockages in myeloid differentiation characterize AML. NPMc-mediated blockade of the terminal differentiation of myeloid cells, coupled to a heightened threshold for caspase-mediated apoptotic signaling, may thus lead to the accumulation of immature blasts and expansion of the transformed leukemic stem cells. It should also be noted that, besides caspase inhibition, there are other means by which inhibition of differentiation leads to leukemia. For instance, the down-regulation of the granulocytic differentiation factor C/EBPα by the fusion protein AML1-ETO results in the blockage of granulocytic differentiation and is implicated in t(8;21) myeloid leukemia.47 Events that arrest differentiation appear to be a common theme in leukemogenesis. Clearly, therapeutic strategies could be formulated to bring about the relief of such inhibitions.
The “cytoplasmic gain-of-function” hypothesis, as proposed herein, inhibition in apoptosis signaling pathways and caspase-mediated myeloid differentiation, helps to explain the tissue specificity of the NPMc mutation. Furthermore, NPMc could cooperate with other oncogenes to bring about leukemia. On that note, it was shown that NPMc cooperates specifically with adenovirus E1A to transform primary mouse embryonic fibroblasts in soft agar.48 One such potential oncogene is c-Myc, which has roles in both apoptosis and proliferation49 and was shown to be in stabilized in NPMc-expressing cells.5 c-Myc could confer a proliferative advantage in NPMc-expressing hematopoietic cells, whereas NPMc blunts the c-Myc-mediated apoptotic response. Our findings may thus complement prevalent hypotheses to provide a more comprehensive mechanistic insight into the oncogenic prowess of NPMc.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Dorothy Teo of National University Hospital for her aid in MGG staining, and D. P. Lane, B. C. Low, L. L. Chew, Y. P. Tan, L. H. Ong, B. L. Tay, C. H. Yap, and C. Wang for their kind assistance and fruitful discussion of this project.
This work was supported by Agency for Science Technology and Research/Biomedical Research Council (grant) and the Ministry of Education (AcRF grant T208B3112).
Authorship
Contribution: T.M.L., S.M.L., Y.-K.M., and L.Y.C. designed the experiments; S.M.L., B.B.A., B.X.T., T.Y., S.T.A., K.G.T., and Y.-K.M. performed the research and analyzed the data; L.P.K., A.E.J.Y., and E.S.-C.K. contributed cell lines and patient samples; and T.M.L., S.M.L., and B.X.T. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tit Meng Lim, Department of Biologic Sciences, 14 Science Dr 4, Block S1A #05-05, National University of Singapore, Singapore 117543; e-mail: dbsltm@nus.edu.sg.
References
Author notes
S.M.L., B.X.T., and B.B.A. contributed equally to this study and are considered first coauthors.