Abstract
Fanconi anemia (FA) is a genetic disease characterized by congenital abnormalities, bone marrow failure, and susceptibility to leukemia and other cancers. FANCJ, one of 13 genes linked to FA, encodes a DNA helicase proposed to operate in homologous recombination repair and replicational stress response. The pathogenic FANCJ-A349P amino acid substitution resides immediately adjacent to a highly conserved cysteine of the iron-sulfur domain. Given the genetic linkage of the FANCJ-A349P allele to FA, we investigated the effect of this particular mutation on the biochemical and cellular functions of the FANCJ protein. Purified recombinant FANCJ-A349P protein had reduced iron and was defective in coupling adenosine triphosphate (ATP) hydrolysis and translocase activity to unwinding forked duplex or G-quadruplex DNA substrates or disrupting protein-DNA complexes. The FANCJ-A349P allele failed to rescue cisplatin or telomestatin sensitivity of a FA-J null cell line as detected by cell survival or γ-H2AX foci formation. Furthermore, expression of FANCJ-A349P in a wild-type background exerted a dominant-negative effect, indicating that the mutant protein interferes with normal DNA metabolism. The ability of FANCJ to use the energy from ATP hydrolysis to produce the force required to unwind DNA or destabilize protein bound to DNA is required for its role in DNA repair.
Introduction
Fanconi anemia (FA) is a recessively inherited disease characterized by congenital abnormalities, aplastic anemia, and an abnormally high risk for the development of malignant diseases, especially acute myeloid leukemia and epithelial tumors.1 Progressive bone marrow failure and late-developing myeloid malignant diseases are responsible for the majority of mortality in patients with FA. Bone marrow failure persists in children with FA because of elevated apoptosis and subsequent failure of the hematopoietic stem cell compartment. Cells from patients with FA are hypersensitive to DNA cross-linking agents such as mitomycin C and cisplatin. Among the 13 FA complementation groups, only a few of the corresponding FA proteins are predicted to have direct roles in DNA metabolism.2 The identification of FANCJ mutations in a DNA helicase gene in patients with early-onset breast cancer3,4 and patients with FA group J5-7 implicate FANCJ as a tumor suppressor caretaker that ensures genomic stability. FANCJ interacts with the tumor suppressor BRCA18 and, indeed, is a bonafide DNA helicase that catalytically unwinds duplex DNA3,9 or resolves G-quadruplex DNA structures10,11 in a reaction dependent on adenosine triphospate (ATP) hydrolysis.
Several genotyping studies have addressed the association between FANCJ mutations and FA clinical abnormalities5,6 and breast cancer risk.4,12-17 Notably, the 2533C→T nonsense mutation in exon 17, resulting in a premature stop codon (R798X), was reported in a high percentage of patients with FA,5,6 as well as in patients with breast cancer.4 The R798X mutation that truncates the protein before the seventh motif of the helicase core domain was shown to encode an ATPase-dead and helicase-dead protein (London et al10 and our unpublished data). Missense mutations in the FANCJ gene have also been identified in patients with FA complementation group J.5,6 One of the mutations identified is an alanine-to-proline mutation at residue 349.6 The Ala349 residue resides immediately adjacent to a highly conserved cysteine of the predicted iron-sulfur (Fe-S) domain of FANCJ18 (Figure 1A); however, the molecular defects of the A349P mutation or any other FA patient missense mutation have not been determined.
Clinical heterogeneity within a given complementation group (FA-J) may reflect differences in the biochemical effects of FANCJ patient mutations on the functions of the protein. Inheritance of a paternal FANCJ A349P missense allele and a maternal truncating R798X allele resulted in phenotypic abnormalities, including intrauterine growth failure and death as a stillborn fetus with a gestational age of 22 weeks.6 Because the A349P missense allele resides within a conserved Fe-S domain in the helicase core, we investigated its effect on the biochemical and cellular functions of FANCJ. Our results indicate that the A349P substitution uncoupled ATP-dependent DNA translocase activity from its ability to unwind DNA or displace proteins bound to DNA. To our knowledge, the effect of the A349P substitution on the catalytic activities of the FANCJ protein is distinct from any other helicase disease mutation reported in the literature. Importantly, these results demonstrate that the ability of FANCJ to couple DNA translocase activity to its other DNA metabolic functions is required for its roles in DNA repair. Furthermore, the FANCJ-A349P mutant allele exerted a dominant-negative effect on cellular resistance to agents that induce DNA damage or replication stress, confirming that FANCJ-A349P expression exerts deleterious effects on cellular phenotypes.
Methods
Plasmid DNA constructions, biochemical assays, immunofluorescence studies, transfection of human and chicken cell lines, and coimmunoprecipitation experiments are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Recombinant proteins
Baculovirus encoding FANCJ-WT, FANCJ-A349P, or FANCJ-K52R with a C-terminal FLAG tag was used to infect High 5 insect cells, and the recombinant FANCJ protein was purified with modifications to a protocol described previously.3 Briefly, cell pellets were resuspended in buffer A (10mM Tris HCl [pH 7.5], 130mM NaCl, 1% Triton X-100, 10mM NaF, 10mM NaPi, and 10mM NaPPi). Cells were lysed in the presence of protease inhibitors (Roche) for 45 minutes at 4°C with mild agitation and centrifuged at 21 000g for 10 minutes at 4°C. The supernatant was incubated with FLAG antibody resin (Sigma-Aldrich) for 2 hours at 4°C. The resin was then washed twice with buffer B (50mM Tris·HCl [pH 7.4], 500mM NaCl, and 0.5% Nonidet P-40), followed by buffer C (50mM Tris·HCl [pH 7.4], 150mM NaCl, and 0.5% Nonidet P-40). FANCJ was eluted with 4 μg/mL 3X FLAG peptide (Sigma-Aldrich) in buffer D (25mM Tris HCl [pH 7.4], 100mM NaCl, 10% glycerol, 0.1% Tween 20, and 5mM TCEP (Tris (2-carboxyethyl) phosphine hydrochloride; Sigma-Aldrich) for 1 hour. FLAG-tagged FANCJ protein was then dialyzed for 2 hours against buffer D with use of a dialysis tube with a 50-kD molecular weight cutoff point (Tube-O-DIALYZER), and aliquots were frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined by Nanodrop (ND-1000; Thermo Scientific) with an extinction coefficient of 135 285 M−1cm−1 determined by ProtParam program. The FANCJ-A349P recombinant protein was prepared with an identical procedure as that used for FANCJ-WT, and at least 3 different batches of proteins were tested in biochemical assays.
Fluorescence quench translocation assays
High-pressure liquid chromatography–purified, fluorescein-labeled oligonucleotide was purchased from Loftstrand Labs, with the fluorophore attached at the 3′ end (T50F, supplemental Table 1). Translocation assays were performed in 96-well F-bottom plates (Greiner Bio-One) using 1nM fluorescein-labeled probe and the specified concentrations of FANCJ protein in 50 μL of reaction mixtures using the ATPase reaction buffer described in supplemental Methods with the indicated nucleotide (2mM). Translocase assays were initiated with the addition of FANCJ protein and incubated at 30°C. Fluorescence readings were taken on a FLUOstar plate reader (BMG LabTech) with excitation and emission filters at 485 nm and 520 nm, respectively. Background fluorescence (< 10%) in reactions lacking FANCJ was subtracted from the fluorescence signals detected in FANCJ-catalyzed reactions.
Results
Reduced iron content in FANCJ-A349P
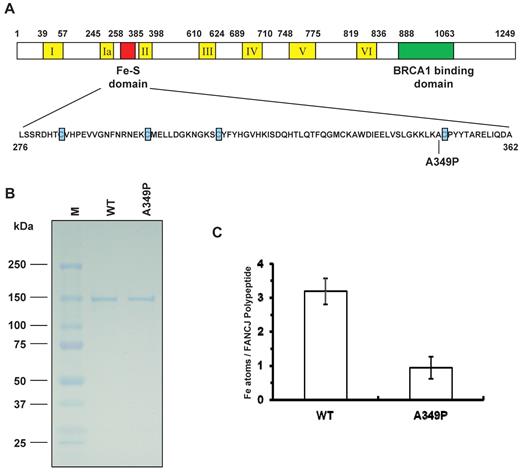
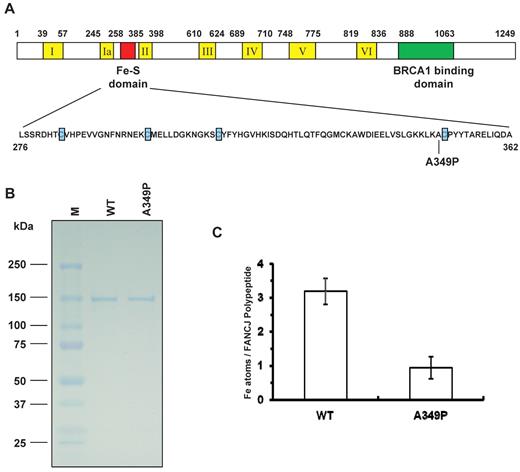
Fe-S domains found in certain DNA repair proteins have been shown to be important for their structure and biochemical function.18-20 On the basis of its sequence homology with XPD helicase, FANCJ was proposed to be an Fe-S domain protein18 ; however, this issue had not been experimentally addressed. Within the predicted Fe-S domain of FANCJ is a FA missense mutation in which an alanine immediately adjacent to one of the highly conserved 4 cysteines is mutated to proline (A349P, Figure 1A). To compare the biochemical properties and catalytic activities of FANCJ-WT and FANCJ-A349P, the recombinant human proteins were expressed using a baculovirus system and purified in an identical manner using a modified protein purification procedure that avoided the inclusion of reducing agents or divalent cation-chelating compounds that might perturb iron content in the recombinant protein during purification. FANCJ-A349P and FANCJ-WT recombinant proteins were purified to near homogeneity as demonstrated by single Coomassie-stained bands on sodium dodecyl sulfate (SDS) polyacrylamide gel (Figure 1B). Atomic absorption spectrometry was used to measure iron content in FANCJ-WT and FANCJ-A349P proteins. It was determined that multiple preparations of purified recombinant FANCJ-WT possessed 3 Fe atoms per polypeptide, whereas FANCJ-A349P only had 1 Fe atom per polypeptide (Figure 1C).
Purification and determination of iron content in FANCJ-WT and FANCJ-A349P proteins. (A) Cartoon depicting FANCJ protein with the conserved helicase core domain and position of the conserved Fe-S domain. The conserved helicase motifs are indicated by yellow boxes, and the BRCA1 binding domain is indicated by green. The Fe-S domain is expanded, and the 4 conserved cysteines are indicated with blue, with the A349P missense mutation of a FANCJ patient marked. (B) The purity of the FANCJ-WT and FANCJ-A349P proteins was evaluated by their detected migration after sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on Coomassie-stained gels according to their predicted sizes. For each protein, 2 μg was loaded. (C) Stoichiometry of iron atoms per purified recombinant FANCJ-WT or FANCJ-A349P. The determination of iron content is described in supplemental Methods. Data represent the mean of at least 3 independent experiments, with SD indicated by error bars.
Purification and determination of iron content in FANCJ-WT and FANCJ-A349P proteins. (A) Cartoon depicting FANCJ protein with the conserved helicase core domain and position of the conserved Fe-S domain. The conserved helicase motifs are indicated by yellow boxes, and the BRCA1 binding domain is indicated by green. The Fe-S domain is expanded, and the 4 conserved cysteines are indicated with blue, with the A349P missense mutation of a FANCJ patient marked. (B) The purity of the FANCJ-WT and FANCJ-A349P proteins was evaluated by their detected migration after sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on Coomassie-stained gels according to their predicted sizes. For each protein, 2 μg was loaded. (C) Stoichiometry of iron atoms per purified recombinant FANCJ-WT or FANCJ-A349P. The determination of iron content is described in supplemental Methods. Data represent the mean of at least 3 independent experiments, with SD indicated by error bars.
FANCJ-A349P substitution abolishes DNA helicase activity
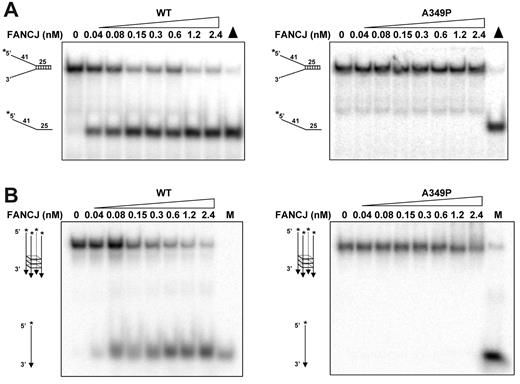
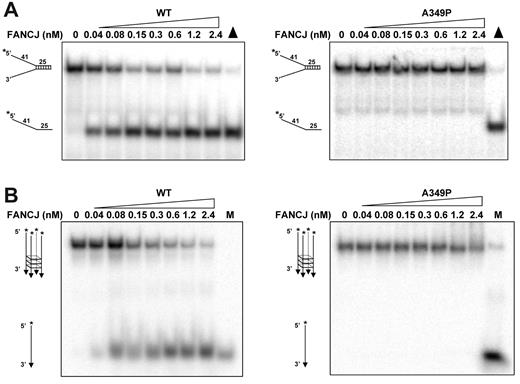
Previously, we reported that purified recombinant FANCJ protein efficiently unwound a 25-basepair (bp) duplex DNA substrate flanked by noncomplementary single-stranded DNA arms (forked duplex DNA substrate).21 Purified recombinant FANCJ-A349P protein was tested on the same 25-bp forked duplex DNA substrate and found to fail completely to unwind it under conditions that FANCJ-WT unwound the forked duplex DNA molecules to near-completion (Figure 2A). A shorter 19-bp forked duplex substrate (data not shown) or a D-loop substrate (supplemental Figure 1A) that were efficiently unwound by FANCJ-WT9 also failed to be unwound by FANCJ-A349P. Inability of FANCJ-A349P to unwind the duplex DNA substrates was observed with multiple preparations of purified recombinant protein, whereas homogeneous FANCJ-WT efficiently unwound these substrates in an ATP-dependent manner. These results contrast with another report that a recombinant form of FANCJ-WT lacks helicase activity on forked duplex substrates.10 The recombinant FANCJ-WT protein purified by London et al10 may lack an important cofactor or contain a posttranslational modification that alters the activity of the enzyme; however, this requires further study.
FANCJ-A349P mutant is inactive as a helicase on forked duplex and G-quadruplex DNA substrates that FANCJ-WT efficiently unwinds. (A) Helicase reactions (20 μL) were performed by incubating the indicated FANCJ concentration with a 0.5nM forked duplex DNA substrate at 30°C for 15 minutes as described in supplemental Methods. Triangle, heat-denatured DNA substrate control. (B) Helicase reactions were performed the same as with panel A, except that G4 DNA was used instead of forked duplex. M, radiolabeled TP-G4 49-mer oligonucleotide (supplemental Table 1) marker.
FANCJ-A349P mutant is inactive as a helicase on forked duplex and G-quadruplex DNA substrates that FANCJ-WT efficiently unwinds. (A) Helicase reactions (20 μL) were performed by incubating the indicated FANCJ concentration with a 0.5nM forked duplex DNA substrate at 30°C for 15 minutes as described in supplemental Methods. Triangle, heat-denatured DNA substrate control. (B) Helicase reactions were performed the same as with panel A, except that G4 DNA was used instead of forked duplex. M, radiolabeled TP-G4 49-mer oligonucleotide (supplemental Table 1) marker.
Recently, we reported that FANCJ-WT unwinds 5′ tailed G4 DNA substrates.11 Because G4 DNA is a very different substrate than a forked duplex, we tested FANCJ-A349P to unwind a G4 DNA molecule and found that it failed under conditions that FANCJ-WT efficiently unwound the G4 substrate (Figure 2B). Human replication protein A, which physically and functionally interacts with FANCJ,22 failed to stimulate FANCJ-A349P to unwind either the G4 or forked duplex DNA substrate, whereas FANCJ-WT helicase activity on both DNA substrates was stimulated (data not shown). Thus, FANCJ-A349P was unable to unwind forked duplex or G4 DNA substrates.
FANCJ-A349P binds DNA similar to FANCJ-WT
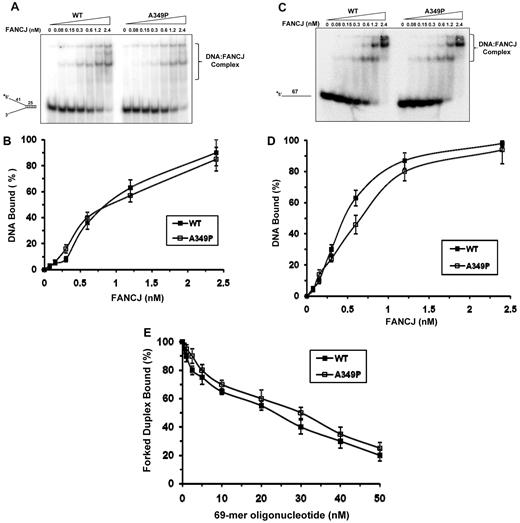
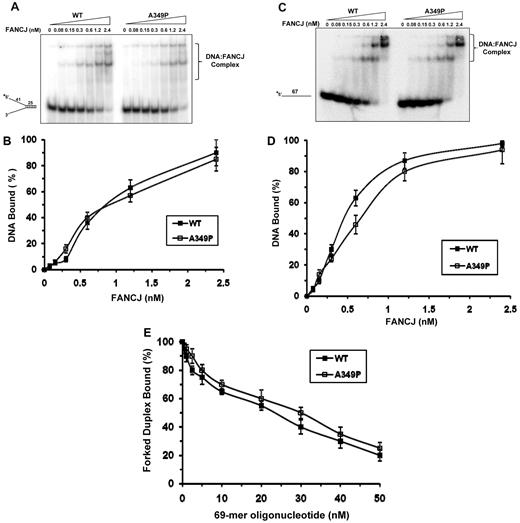
The inability of FANCJ-A349P to unwind DNA might reflect an impairment of its DNA-binding activity. To address this issue, we performed gel mobility shift assays with FANCJ-A349P and FANCJ-WT and the corresponding radiolabeled forked duplex DNA substrate or a radiolabeled single-stranded (ss) DNA oligonucleotide. Analyses of DNA binding to forked duplex (Figure 3A-B), the single-stranded 67-mer (Figure 3C-D), or a D-loop DNA structure (supplemental Figure 1B-C) demonstrated that wild-type and mutant FANCJ proteins bound the DNA molecules in a very similar protein concentration–dependent manner. FANCJ-A349P also bound these DNA molecules similar to FANCJ-WT in the presence of the poorly hydrolysable analog ATPγS or adenosine diphosphate (ADP) as well (data not shown).
DNA binding of mutant and wild-type FANCJ proteins as detected by gel mobility shift assays. (A) The indicated concentrations of FANCJ-WT or FANCJ-A349P protein were incubated with 0.5nM forked duplex DNA substrate at 25°C for 30 minutes, as described in supplemental Methods. The DNA-protein complexes were resolved on native 5% polyacrylamide gels. (B) Quantitative analyses of DNA gel-shift experiments as performed in (A). (C) Gel mobility shift experiments with radiolabeled single-stranded 67-mer oligonucleotide. (D) Quantitative analyses of DNA gel-shift experiments as performed in (C). (E) 2.4nM FANCJ-WT or FANCJ-A349P was incubated with 0.5nM radiolabeled forked duplex DNA at 25°C for 15 minutes, and the indicated concentration of 67-mer ssDNA was subsequently added and incubated for an additional 15 minutes at 25°C. DNA-protein complexes were resolved on native 5% polyacrylamide gels. Quantitative analyses of DNA-binding data from DNA competition experiments are shown with SD indicated by error bars.
DNA binding of mutant and wild-type FANCJ proteins as detected by gel mobility shift assays. (A) The indicated concentrations of FANCJ-WT or FANCJ-A349P protein were incubated with 0.5nM forked duplex DNA substrate at 25°C for 30 minutes, as described in supplemental Methods. The DNA-protein complexes were resolved on native 5% polyacrylamide gels. (B) Quantitative analyses of DNA gel-shift experiments as performed in (A). (C) Gel mobility shift experiments with radiolabeled single-stranded 67-mer oligonucleotide. (D) Quantitative analyses of DNA gel-shift experiments as performed in (C). (E) 2.4nM FANCJ-WT or FANCJ-A349P was incubated with 0.5nM radiolabeled forked duplex DNA at 25°C for 15 minutes, and the indicated concentration of 67-mer ssDNA was subsequently added and incubated for an additional 15 minutes at 25°C. DNA-protein complexes were resolved on native 5% polyacrylamide gels. Quantitative analyses of DNA-binding data from DNA competition experiments are shown with SD indicated by error bars.
To further examine a potential DNA-binding defect of FANCJ-A349P, we performed forked duplex–binding experiments in the presence of increasing concentrations of ssDNA competitor molecules. Quantification of the reduction in FANCJ-forked duplex gel-shift species as a function of ss 69-mer competitor demonstrated a very similar profile between FANCJ-WT and FANCJ-A349P (Figure 3E), suggesting that FANCJ-A349P binds DNA in a manner similar to FANCJ-WT. Similar binding of FANCJ-A349P and FANCJ-WT was also observed for ssDNA molecules containing a thymine glycol or 8-oxoguanine base damage (supplemental Figure 2) or the G4 substrate (data not shown).
FANCJ-A349P retains ATPase activity comparable with FANCJ-WT
Next, we examined the DNA-dependent ATPase activity of FANCJ-A349P and FANCJ-WT (Table 1). Using covalently closed M13 ssDNA as the effector molecule, we determined Km values for ATP hydrolysis for the mutant and wild-type enzymes. FANCJ-WT displayed a Km of 0.88mM, a value close to the Km value of 0.73mM determined for FANCJ-A349P. Using an ATP concentration (8.5mM) approximately 10-fold greater than the Km, ATPase assays with the FANCJ-WT and FANCJ-A349P proteins showed that FANCJ-A349P had a slightly greater (1.7-fold) kcat for ATP hydrolysis than FANCJ-WT. As a negative control, the FANCJ-K52R Walker A box mutant protein was tested and found to have a kcat for ATP hydrolysis significantly reduced versus FANCJ-WT or FANCJ-A349P (Table 1). The Keff values for ATP hydrolysis using M13 ssDNA were similar between FANCJ-WT and FANCJ-A349P (Table 1). We also examined the kinetic parameters Km and kcat for ATP hydrolysis for the FANCJ-WT and FANCJ-A349P proteins using forked duplex DNA, a preferred substrate of FANCJ helicase activity, and found that FANCJ-A349P retained similar ATPase kinetic rate constants comparable with FANCJ-WT (data not shown).
The observation that the FANCJ-A349P mutant exhibited a Km for ATP similar to wild-type FANCJ suggested that the mutant was able to favorably interact with ATP, at least in the presence of DNA. To determine whether the mutation affected the ability of the FANCJ protein to form a binary complex with ATP, [α32P]-ATP binding by FANCJ-WT and FANCJ-A349P was measured using a gel filtration assay. The results from these experiments demonstrated that ATP binding by FANCJ-A349P was comparable with that of FANCJ-WT (supplemental Figure 3).
FANCJ-A349P can translocate on ssDNA
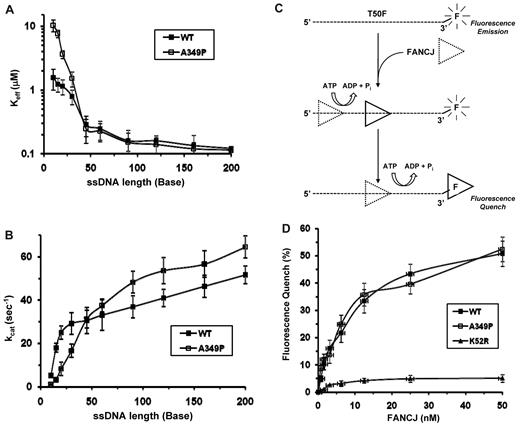
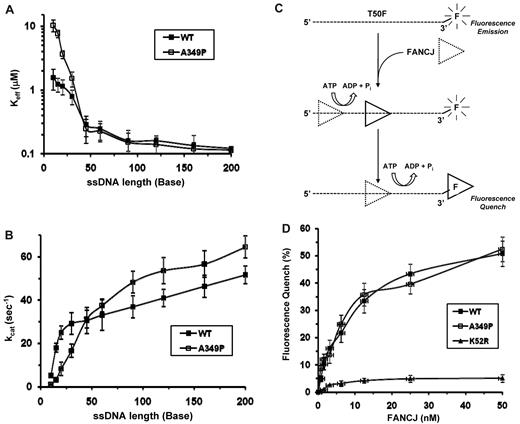
Next, we wanted to ask if FANCJ-A349P was defective in translocating on DNA because this would prevent it from unwinding duplex or quadruplex DNA substrates. Moreover, a helicase that translocates less processively over longer ssDNA tracts may only be able to partially unwind structured DNA molecules, which can rehybridize if the helicase dissociates. To study the effect of the A349P substitution on FANCJ DNA translocase activity, we performed 2 types of experiments with purified FANCJ-WT and FANCJ-A349P. In the FANCJ-WT, we measured DNA-dependent ATPase activity as a function of oligonucleotide (dT) length. A processive translocase driven by its ATP hydrolysis would display increasing ATPase activity as a function of oligonucleotide length because the protein would have to dissociate and rebind a shorter ssDNA effector more frequently than a longer ssDNA effector.23 To compare the kinetics of ATP hydrolysis between the wild-type and mutant FANCJ proteins, we determined both Keff and kcat values for increasing lengths of dT oligonucleotides (range, 5-200 nt). As expected, the Keff values for FANCJ-WT decreased with increasing lengths of oligonucleotides dT, whereas kcat values increased (Figure 4A-B). For FANCJ-A349P, the Keff values also decreased, and kcat values increased with increasing oligonucleotide dT lengths (Figure 4A-B), suggesting that it behaves as an ATP-driven ssDNA translocase. A noticeable difference in Keff values between FANCJ-WT and FANCJ-A349P was observed using the shorter oligonucleotides (< 45-mer), suggesting that the mutant protein displays a reduced ability to use oligonucleotides of 10, 15, 20, or 30 nt as ssDNA effectors for DNA-dependent ATPase activity compared with FANCJ-WT; consequently, the kcat values for dT tracts of 10 to 30 nt were reduced for FANCJ-A349P.
FANCJ-A349P translocates on single-stranded DNA. The Keff-ssDNA values (A) and kcat values (B) for ATP hydrolysis catalyzed by FANCJ-WT and FANCJ-A349P proteins were determined as a function of oligonucleotide dT length, as described in supplemental Methods. Error bars represent the SD of the fit to the Michaelis-Menton equation. (C) Cartoon depicting fluorescence quenching of fluorescein covalently attached to 3′ end of ssDNA molecule by ATP-driven translocation of FANCJ to a position near the fluorophore. Binding site size or the assembly state of FANCJ is not known. For simplicity, cartoon depicts initial ATP-dependent translocation of FANCJ followed by second translocation step placing FANCJ in the vicinity of fluorescein where it quenches the fluorophore; however, physical or kinetic step size is not known. (D) Fluorescence quenching experiments, performed as described in “Fluorescence quench translocation assays,” with use of FANCJ-WT, FANCJ-A349P, and FANCJ-K52R proteins.
FANCJ-A349P translocates on single-stranded DNA. The Keff-ssDNA values (A) and kcat values (B) for ATP hydrolysis catalyzed by FANCJ-WT and FANCJ-A349P proteins were determined as a function of oligonucleotide dT length, as described in supplemental Methods. Error bars represent the SD of the fit to the Michaelis-Menton equation. (C) Cartoon depicting fluorescence quenching of fluorescein covalently attached to 3′ end of ssDNA molecule by ATP-driven translocation of FANCJ to a position near the fluorophore. Binding site size or the assembly state of FANCJ is not known. For simplicity, cartoon depicts initial ATP-dependent translocation of FANCJ followed by second translocation step placing FANCJ in the vicinity of fluorescein where it quenches the fluorophore; however, physical or kinetic step size is not known. (D) Fluorescence quenching experiments, performed as described in “Fluorescence quench translocation assays,” with use of FANCJ-WT, FANCJ-A349P, and FANCJ-K52R proteins.
FANCJ interaction with a 3-stranded D-loop DNA molecule would be an example of a more complex structure that might represent a physiologic substrate. Using the D-loop structures with a 10- or 20-nt tailed invading strand as the effector in the ATPase reaction, we observed that FANCJ-A349P displayed a Keff value greater than that of FANCJ-WT (data not shown), reflecting a difference that was comparable with that observed for 10- and 20-nt dT oligonucleotides. Moreover, FANCJ-A349P failed to unwind either D-loop substrate (supplemental Figure 1).
To directly test the ability of FANCJ-A349P translocation on ssDNA, we used a fluorescence-quenching assay. The principle of this assay is that when a 5′ to 3′ helicase translocates on ssDNA in an ATP-dependent manner and reaches the position of the fluorescein covalently attached to the 3′ end of the oligonucleotide, the fluorescence intensity emitted on excitation of the fluorophore covalently bonded to the DNA molecule is changed by the close proximity of helicase to fluorescein.24,25 For our purpose, we compared fluorescence quench as a function of FANCJ-A349P and FANCJ-WT concentration with a 3′-fluorescein–conjugated 50-mer oligonucleotide (Figure 4C), because FANCJ is presumed to translocate 5′ to 3′ based on results from helicase assays with a DNA directionality substrate3 or duplex DNA substrates with a 5′ ssDNA overhang.9 These experiments demonstrated very similar increases in fluorescence quench as a function of FANCJ-WT or FANCJ-A349P concentrations (Figure 4D). Importantly, the fluorescence quench for either FANCJ-WT or FANCJ-A349P was dependent on intrinsic ATPase activity, because there was only a very small increase in fluorescence quench when an ATPase-inactive FANCJ-K52R mutant3,9 was incubated with the 3′-fluorescein-conjugated oligonucleotide and ATP (Figure 4D). The FANCJ-K52R protein retained its ability to bind the fluorescein-conjugated oligonucleotide used for the quenching assays similar to FANCJ-WT or FANCJ-A349P (supplemental Figure 4), indicating that the change in fluorescence quenching signal is not simply the result of ssDNA binding by the protein. In the absence of ATP or in the presence of ATPγS, the increase in fluorescence quench was very small (∼ 5%) at the highest concentration of FANCJ-WT or FANCJ-A349P (50nM; data not shown), indicating that ssDNA translocation by either the A349P mutant or wild-type protein was dependent on hydrolysable ATP.
A349P mutation uncouples FANCJ ATP hydrolysis from its ability to disrupt protein-DNA interactions
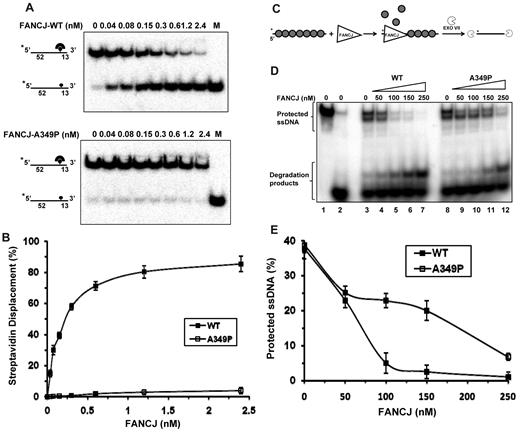
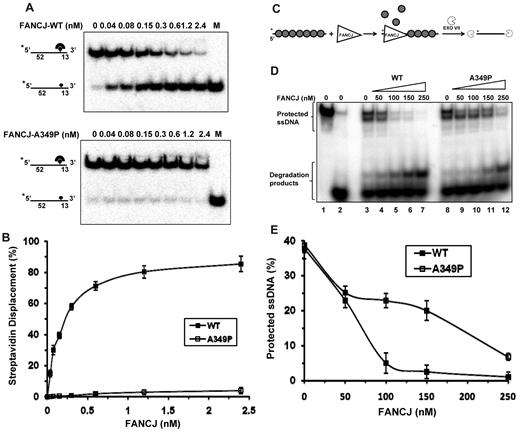
Recently, we demonstrated that FANCJ can catalytically disrupt the high-affinity interaction of streptavidin bound to a biotinylated ssDNA molecule, and inhibit RAD51 strand exchange by destabilizing the RAD51 nucleoprotein filament.26 The ability of FANCJ-A349P to catalyze ATP hydrolysis but fail to unwind duplex or quadruplex DNA substrates posed the question if the mutant helicase might be able to use its motor ATPase function to disrupt protein-DNA interactions. To address this issue, we tested FANCJ-A349P to displace streptavidin bound to a biotinylated oligonucleotide. FANCJ-A349P failed to disrupt the streptavidin-biotinylated oligonucleotide interaction under conditions that FANCJ-WT accomplished more than 80% streptavidin displacement in a kinetic manner (Figure 5A-B).
FANCJ-A349P is compromised in its ability to displace DNA-bound protein. (A) The indicated concentrations of FANCJ-WT (upper) or FANCJ-A349P (lower) were incubated with 2mM ATP and DNA substrate (0.5nM) that had streptavidin bound to the covalently linked biotin moiety residing 52 nt from the 5′ end of the radiolabeled oligonucleotide (supplemental Table 1). (B) Quantitative analyses of FANCJ streptavidin displacement assays as shown in panel A. Data represent the mean of at least 3 independent experiments with SD indicated by error bars. (C) Experimental scheme to examine FANCJ displacement of RAD51 protein filament on ssDNA. (D) The degradation of the intact 32P-labeled ssDNA fragment by exonuclease VII was analyzed in a 10% polyacrylamide gel. The preformed RAD51-ssDNA complex was incubated with FANCJ-WT (lanes 4-7) or FANCJ-A349P (lanes 9-12) followed by the addition of exonuclease VII. Protein concentrations are indicated at the top of the gel. The ssDNA fragment before and after the treatment with exonuclease VII is shown in lanes 1 and 2, respectively. (E) The data from panel D represented as a graph.
FANCJ-A349P is compromised in its ability to displace DNA-bound protein. (A) The indicated concentrations of FANCJ-WT (upper) or FANCJ-A349P (lower) were incubated with 2mM ATP and DNA substrate (0.5nM) that had streptavidin bound to the covalently linked biotin moiety residing 52 nt from the 5′ end of the radiolabeled oligonucleotide (supplemental Table 1). (B) Quantitative analyses of FANCJ streptavidin displacement assays as shown in panel A. Data represent the mean of at least 3 independent experiments with SD indicated by error bars. (C) Experimental scheme to examine FANCJ displacement of RAD51 protein filament on ssDNA. (D) The degradation of the intact 32P-labeled ssDNA fragment by exonuclease VII was analyzed in a 10% polyacrylamide gel. The preformed RAD51-ssDNA complex was incubated with FANCJ-WT (lanes 4-7) or FANCJ-A349P (lanes 9-12) followed by the addition of exonuclease VII. Protein concentrations are indicated at the top of the gel. The ssDNA fragment before and after the treatment with exonuclease VII is shown in lanes 1 and 2, respectively. (E) The data from panel D represented as a graph.
Next, we compared the activity of FANCJ-A349P with FANCJ-WT to destabilize the RAD51 protein filament bound to ssDNA using an exonuclease protection assay (Figure 5C). In this assay, the ssDNA precoated with RAD51 is protected from exonuclease VII digestion (Figure 5D, compare lane 3 with lane 2). Several cleavage fragments generated by exonuclease VII are consistent with the RAD51 polar polymerization and dissociation on ssDNA, which leaves the ssDNA ends uncovered by the protein. As shown in Figure 5D, 100nM of FANCJ-WT was required for almost complete removal of the RAD51-dependent protection of ssDNA. This amount of FANCJ compares well with those of other proteins known to disrupt the RAD51 filament on ssDNA.27-30 An increasing amount of exonuclease VII digestion product was detected with greater amounts of FANCJ-WT in the reaction (Figure 5D lanes 4-7). In contrast, more RAD51-coated ssDNA was protected from exonucleaseVII digestion in reactions containing FANCJ-A349P compared with FANCJ-WT (Figure 5D-E). Thus, FANCJ-A349P failed to destabilize protein bound to DNA as efficiently as FANCJ-WT.
FANCJ-A349P fails to genetically complement mitomycin C sensitivity of FA-J cells
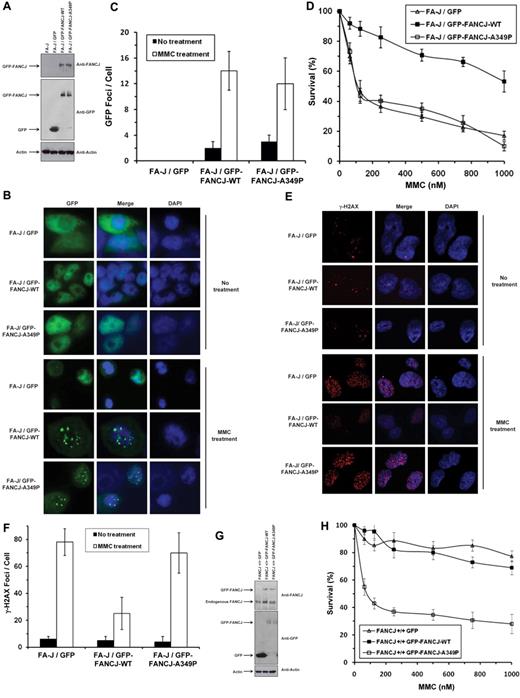
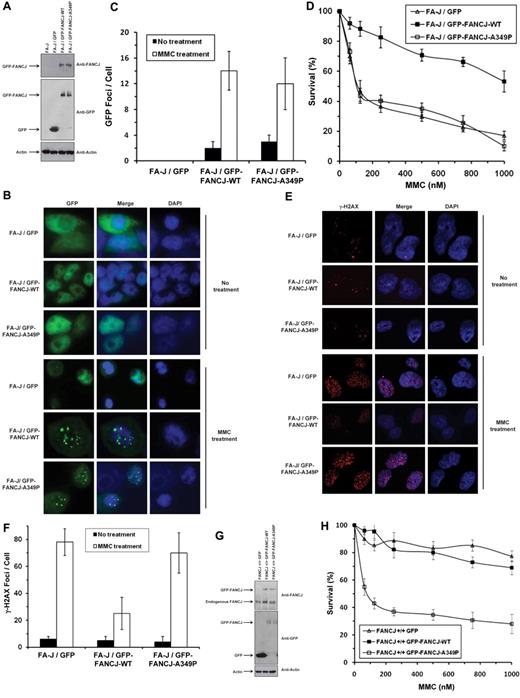
Biochemical analyses of the purified recombinant FANCJ-A349P demonstrated that the missense mutation in the Fe-S domain of FANCJ abolished its helicase activity on duplex and G4 DNA substrates, yet the mutant retained its ability to hydrolyze ATP and translocate on ssDNA. To determine whether FANCJ-A349P could function in vivo, we performed a series of genetic complementation experiments using a human FA-J cell line (EUFA30/hTert/SV40)5 with both FANCJ mutant alleles encoding the truncated FANCJ-R798X gene product. Genetic assays were performed with the FA-J cells that had been transfected with plasmid encoding green fluorescence protein (GFP)–tagged human FANCJ-WT or FANCJ-A349P proteins. Western blot analysis demonstrated that GFP-tagged FANCJ-A349P was expressed at a similar level as GFP-tagged FANCJ-WT (Figure 6A). In addition, GFP-tagged FANCJ-A349P protein was found to be associated with previously identified FANCJ protein partners (BRCA18 and MLH131 ), as demonstrated by coimmunoprecipitation experiments (supplemental Figure 5). Both GFP-tagged FANCJ-WT and GFP-tagged FANCJ-A349P formed DNA damage–inducible foci when the corresponding transfected cells were exposed to a 100nM concentration of the DNA cross-linking agent mitomycin C (MMC), as detected by immunofluorescence microscopy (Figure 6B-C).
Expression of FANCJ-A349P fails to rescue mitomycin C sensitivity of FA-J cells and exerts a dominant-negative effect on mitomycin C resistance of wild-type cells. (A) Western blot analysis of human FA-J (EUFA30/hTert/SV40) fibroblasts stably transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Protein was detected with antibody against FANCJ (Sigma-Aldrich), GFP (Clontech) or actin (loading control, 10% loaded). (B) MMC-induced GFP-tagged FANCJ foci formation. GFP fluorescence detection was performed on FA-J cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. FA-J cells were treated with or without 100nM MMC for 2 hours. (C) Quantitative analyses of GFP-tagged FANCJ foci shown in panel B. (D) MMC sensitivity of stably transfected human FA-J fibroblasts with indicated genotypes was evaluated by cell survival assay. (E) γ-H2AX immunofluorescence staining of FA-J cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Stably transfected FA-J cells were treated with or without 100nM MMC for 2 hours, washed with phosphate-buffered saline (PBS), allowed to recover for 6 hours, and analyzed by immunofluorescence detection as described in supplemental Methods. (F) Quantitative analyses of γ-H2AX foci shown in panel E. Data represent mean of > 100 cells; SD indicated by error bars. (G) Western blot analysis of human wild-type (VH10/hTert) fibroblasts transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-FANCJ-A349P. Protein was detected with antibody against FANCJ (resolved on 6% SDS-PAGE), GFP, or actin (resolved on 4%-12% gradient SDS-PAGE). (H) MMC sensitivity of wild-type fibroblasts with indicated genotypes was evaluated by survival assay. (I) γ-H2AX immunofluorescence staining of wild-type fibroblasts transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Wild-type cells were either untreated or exposed to 100nM MMC for 2 hours, washed with PBS, allowed to recover for 6 hours, and analyzed by immunofluorescence detection. (J) Quantitative analyses of γ-H2AX foci in the corresponding transfected wild-type cell lines as shown in panel I.
Expression of FANCJ-A349P fails to rescue mitomycin C sensitivity of FA-J cells and exerts a dominant-negative effect on mitomycin C resistance of wild-type cells. (A) Western blot analysis of human FA-J (EUFA30/hTert/SV40) fibroblasts stably transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Protein was detected with antibody against FANCJ (Sigma-Aldrich), GFP (Clontech) or actin (loading control, 10% loaded). (B) MMC-induced GFP-tagged FANCJ foci formation. GFP fluorescence detection was performed on FA-J cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. FA-J cells were treated with or without 100nM MMC for 2 hours. (C) Quantitative analyses of GFP-tagged FANCJ foci shown in panel B. (D) MMC sensitivity of stably transfected human FA-J fibroblasts with indicated genotypes was evaluated by cell survival assay. (E) γ-H2AX immunofluorescence staining of FA-J cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Stably transfected FA-J cells were treated with or without 100nM MMC for 2 hours, washed with phosphate-buffered saline (PBS), allowed to recover for 6 hours, and analyzed by immunofluorescence detection as described in supplemental Methods. (F) Quantitative analyses of γ-H2AX foci shown in panel E. Data represent mean of > 100 cells; SD indicated by error bars. (G) Western blot analysis of human wild-type (VH10/hTert) fibroblasts transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-FANCJ-A349P. Protein was detected with antibody against FANCJ (resolved on 6% SDS-PAGE), GFP, or actin (resolved on 4%-12% gradient SDS-PAGE). (H) MMC sensitivity of wild-type fibroblasts with indicated genotypes was evaluated by survival assay. (I) γ-H2AX immunofluorescence staining of wild-type fibroblasts transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Wild-type cells were either untreated or exposed to 100nM MMC for 2 hours, washed with PBS, allowed to recover for 6 hours, and analyzed by immunofluorescence detection. (J) Quantitative analyses of γ-H2AX foci in the corresponding transfected wild-type cell lines as shown in panel I.
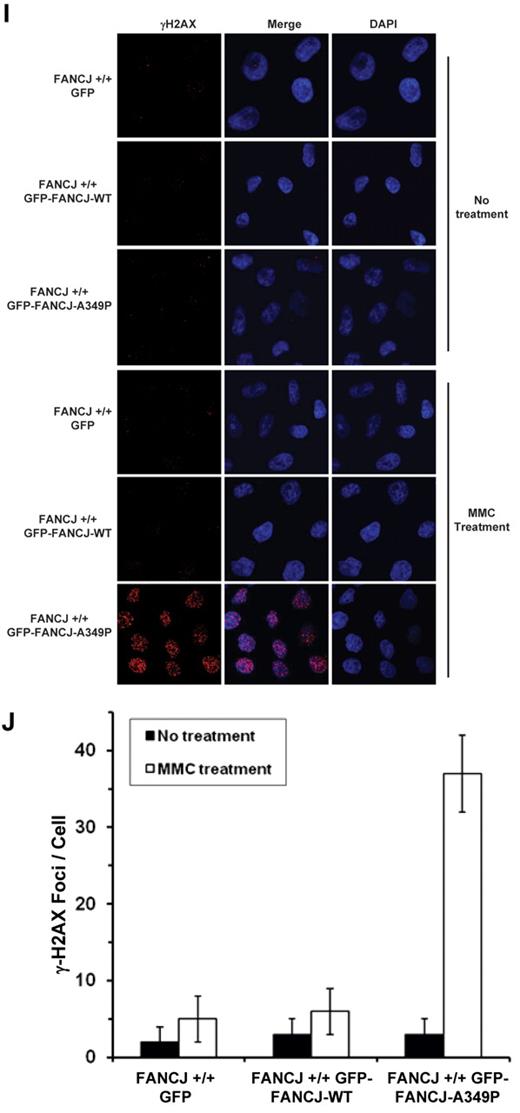
The results of MMC survival assays demonstrated that the FA-J/GFP-tagged FANCJ-A349P cells were as sensitive to MMC as the FA-J/vector cells (Figure 6D). Compared with FA-J/GFP-tagged FANCJ-WT cells, FA-J cells transfected with either empty vector or GFP-tagged FANCJ-A349P showed reduced survival as a function of MMC dose. γH2AX foci, a marker of double-strand breaks, were elevated by 3-fold in MMC-treated FA-J/GFP–tagged FANCJ-A349P cells compared with the isogenic FA-J/GFP–tagged FANCJ-WT cells (Figure 6E-F). Together with the cell survival and immunofluorescence data, the results indicate that the GFP-tagged FANCJ-A349P mutant protein can form MMC-induced foci but fails to render the cells resistant to the effects of the cross-linking agent, as measured by cell survival or accumulation of DNA damage. In genetic complementation experiments using a chicken fancj knockout cell line (supplemental Figure 6), FANCJ-A349P expressed at a level similar to FANCJ-WT (supplemental Figure 7A) failed to rescue cisplatin sensitivity by either colony survival assays (supplemental Figure 7B) or resistance to γH2AX foci formation (supplemental Figure 7C-D).
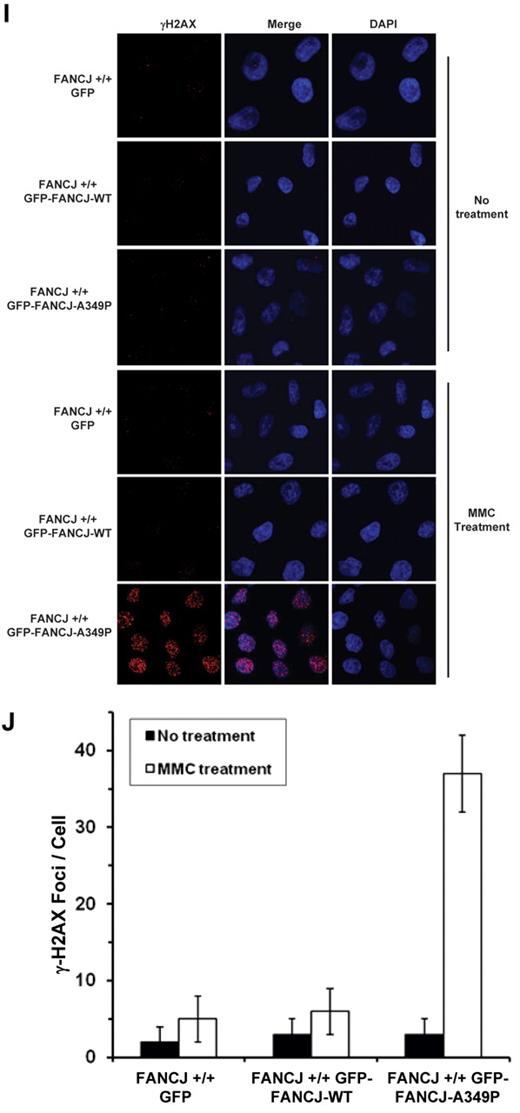
We also examined whether the FANCJ-A349P mutant allele exerted a dominant-negative effect when expressed in a wild-type FANCJ background. Because exogenously expressed FANCJ protein contained a GFP tag, we were able to detect its migration from that of endogenous wild-type FANCJ protein (Figure 6G). GFP-tagged FANCJ-WT and FANCJ-A349P proteins were expressed at similar levels in the wild-type cells, approximately 3-fold less than that of endogenous FANCJ. Compared with the wild-type cells expressing GFP-tagged FANCJ-WT or transfected with empty vector, wild-type cells expressing GFP-tagged FANCJ-A349P showed significantly reduced survival as a function of MMC dose (Figure 6H). γH2AX foci were elevated by 6-fold in MMC-treated wild-type cells expressing GFP-tagged FANCJ-A349P compared with the same cells expressing GFP-tagged FANCJ-WT (Figure 6I-J). In genetic experiments using the chicken DT40 fancj+/+ cell line, FANCJ-A349P exerted a dominant-negative effect on cisplatin sensitivity, as measured by either colony survival assays (supplemental Figure 7E) or resistance to γH2AX foci formation (supplemental Figure 7F-G). We conclude that expression of the FANCJ-A349P protein encoded by the mutant allele exerts a dominant- negative effect on cell survival or DNA damage accumulation after treatment with a DNA cross-linking agent.
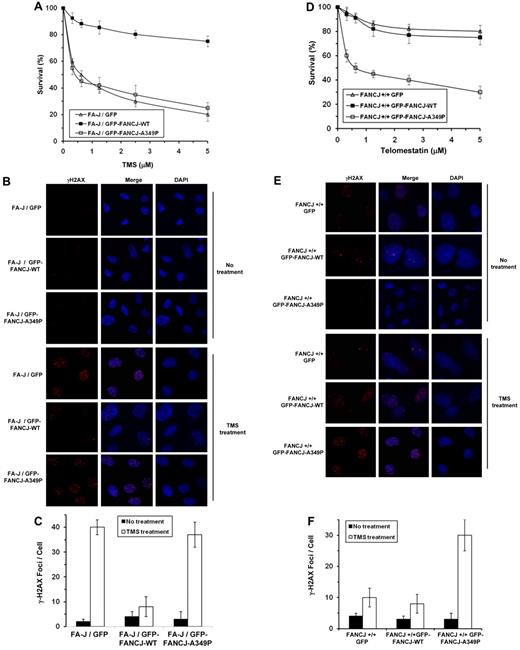
FANCJ-A349P fails to rescue the sensitivity of FA-J cells to the G4 binding compound telomestatin
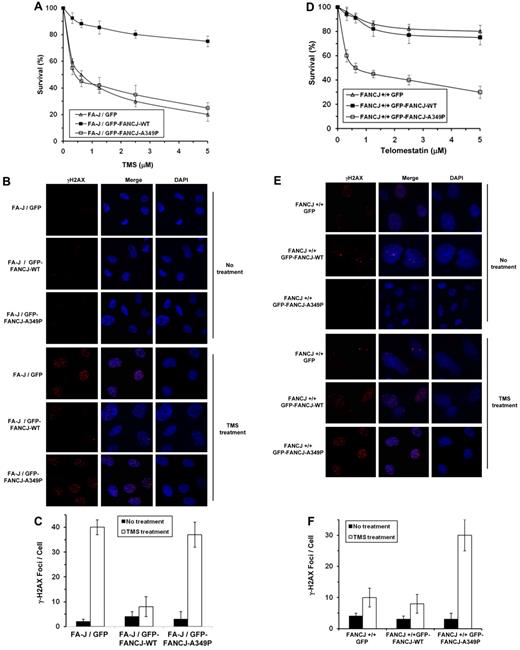
Previously we reported that FANCJ-depleted cells treated with the G4-interactive compound telomestatin (TMS) displayed impaired proliferation and elevated levels of apoptosis and DNA damage compared with control cells, providing evidence that G4 DNA is a physiologic substrate of FANCJ.11 To assess the ability of FANCJ-A349P to function in the cellular response to TMS, we performed genetic complementation assays with the FANCJ-transfected cell lines used for the MMC resistance assays. Both GFP-tagged FANCJ-WT and GFP-tagged FANCJ-A349P formed DNA damage–inducible foci when the corresponding transfected cells were exposed to 5μM TMS, as detected by immunofluorescence microscopy (data not shown). Results from survival assays demonstrated that the FA-J/GFP–tagged FANCJ-A349P cells were as sensitive to TMS as the FA-J/vector cells (Figure 7A). γH2AX foci were significantly elevated by approximately 5-fold in TMS-treated FA-J/GFP–tagged FANCJ-A349P cells compared with the isogenic FA-J/GFP–tagged FANCJ-WT cells (Figure 7B-C), indicating the accumulation of double-strand breaks in cells expressing the FANCJ-A349P mutant protein. FANCJ-A349P expressed in the transfected chicken fancj−/− cells also failed to rescue TMS sensitivity by either colony survival assays (supplemental Figure 8A) or resistance to γH2AX foci formation (supplemental Figure 8B-C).
Expression of FANCJ-A349P fails to rescue sensitivity of FA-J cells to the G-quadruplex binding compound telomestatin and exerts a dominant-negative effect on telomestatin resistance of wild-type cells. (A) TMS sensitivity of stably transfected human FA-J (EUFA30/hTert/SV40) fibroblasts with the indicated genotypes was evaluated by survival assay. (B) γ-H2AX immunofluorescence staining of the corresponding FA-J cell lines stably transfected with plasmids encoding GFP, GFP-FANCJ-WT, or GFP-FANCJ-A349P. FA-J cells were untreated or treated with 5μM TMS for 4 hours, washed with PBS, allowed to recover for 4 hours, and analyzed by immunofluorescence detection. (C) Quantitative analyses of γ-H2AX foci after 5μM TMS exposure of the corresponding transfected FA-J cell lines shown in panel B. Data represent the mean of at least 100 cells counted, with SD indicated by error bars. (D) TMS sensitivity of stably transfected human wild-type (VH10/hTert) fibroblasts with the indicated genotypes was evaluated by survival assay. (E) γ-H2AX immunofluorescence staining of wild-type cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Wild-type cells were either untreated or exposed to 5μM TMS for 4 hours, washed with PBS, allowed to recover for 4 hours, and analyzed by immunofluorescence detection. (F) Quantitative analyses of γ-H2AX foci in the corresponding transfected wild-type cell lines as shown in panel E.
Expression of FANCJ-A349P fails to rescue sensitivity of FA-J cells to the G-quadruplex binding compound telomestatin and exerts a dominant-negative effect on telomestatin resistance of wild-type cells. (A) TMS sensitivity of stably transfected human FA-J (EUFA30/hTert/SV40) fibroblasts with the indicated genotypes was evaluated by survival assay. (B) γ-H2AX immunofluorescence staining of the corresponding FA-J cell lines stably transfected with plasmids encoding GFP, GFP-FANCJ-WT, or GFP-FANCJ-A349P. FA-J cells were untreated or treated with 5μM TMS for 4 hours, washed with PBS, allowed to recover for 4 hours, and analyzed by immunofluorescence detection. (C) Quantitative analyses of γ-H2AX foci after 5μM TMS exposure of the corresponding transfected FA-J cell lines shown in panel B. Data represent the mean of at least 100 cells counted, with SD indicated by error bars. (D) TMS sensitivity of stably transfected human wild-type (VH10/hTert) fibroblasts with the indicated genotypes was evaluated by survival assay. (E) γ-H2AX immunofluorescence staining of wild-type cells transfected with plasmids encoding GFP, GFP-tagged FANCJ-WT, or GFP-tagged FANCJ-A349P. Wild-type cells were either untreated or exposed to 5μM TMS for 4 hours, washed with PBS, allowed to recover for 4 hours, and analyzed by immunofluorescence detection. (F) Quantitative analyses of γ-H2AX foci in the corresponding transfected wild-type cell lines as shown in panel E.
We tested whether the FANCJ-A349P mutant allele exerted a dominant-negative effect on TMS resistance when expressed in a wild-type FANCJ background. Compared with the wild-type cells expressing GFP-tagged FANCJ-WT, fancj+/+FANCJ-A349P cells showed significantly reduced survival as a function of TMS dose (Figure 7D). γH2AX foci were elevated by 4-fold in TMS-treated wild-type cells expressing GFP-tagged FANCJ-A349P compared with the same cells expressing GFP-tagged FANCJ-WT (Figure 7E-F). FANCJ-A349P expressed in the transfected chicken fancj+/+ cells exerted a dominant-negative effect on TMS sensitivity as measured by either colony survival assays (supplemental Figure 8D) or resistance to γH2AX foci formation (supplemental Figure 8E-F). We conclude that the mutant allele encoding FANCJ-A349P exerts a dominant-negative effect on cell survival or the accumulation of DNA damage after TMS treatment, similar to what was observed for exposure to a DNA cross-linking agent.
Discussion
In this study, we characterized the effects of the FANCJ-A349P mutation on the biochemical and cellular functions of the protein. Biochemical characterization of the purified recombinant protein demonstrated that the A349P substitution disrupts the ability of FANCJ to couple its ATPase and DNA translocase functions to unwinding of duplex or G4 DNA substrates. Importantly, this demonstration is the first to show that a FANCJ missense mutation genetically linked to FA impairs the ability of FANCJ to transduce the energy from ATP hydrolysis to the force production required for either unwinding structured nucleic acids or displacing proteins bound to DNA. Genetic analyses demonstrated that the A349P mutation abolishes the function of FANCJ in cellular resistance to a DNA cross-linking agent or a G4-binding molecule. Thus, the catalytic functions (DNA unwinding and/or protein stripping) appear to be critical for the roles of FANCJ in vivo to function in DNA repair and defend genomic stability.
The importance of the integrity of the conserved Fe-S domain in FANCJ is attested to by the dramatic effects of the A349P mutation on the biochemical functions of the protein. Based on the crystal structure of XPD homologs, the Fe-S domain is proposed to play a structural role to stabilize the enzyme and/or to serve as a wedge to physically separate the DNA duplex strands.32-34 It was suggested that the Fe-S cluster in RAD3 (XPD) helicase from Ferroplasma acidarmanus (FacRad3) is required for the proper folding and structural stability of the auxiliary domain and is important for coupling ATP hydrolysis to unidirectional translocation of the helicase. A FacRad3 Fe-S domain mutant was suggested to be defective in ssDNA translocation based on its inability to displace streptavidin from a biotinylated oligonucleotide19 ; however, translocation by the FacRad3 mutant was not directly tested. More recently, wild-type recombinant FacXPD was shown to translocate on RPA-coated ssDNA.35 From our studies of the FANCJ-A349P mutant, we determined that this particular mutation interferes with a critical step downstream of DNA translocation that is required for disruption of standard Watson-Crick hydrogen bonds between base pairs of B-form DNA as well as alternate Hoogsteen hydrogen bonds between guanine residues of the G-tetrad stack. From steady-state measurements, we determined that the mutant protein can hydrolyze ATP and translocate along the ssDNA lattice but was impaired in its ability to destabilize protein-DNA interactions. This finding suggests that the motor ATPase and translocase functions of the FANCJ mutation can also be biochemically separated from the task of dislodging proteins bound to DNA. A helicase core domain mutation in SF1 PcrA helicase uncoupled ssDNA translocation from helicase activity36 ; however, the uncoupling of DNA translocation from a putative protein–stripping function was not examined. Soultanas et al (2000) concluded that PcrA unwinds DNA by an active mechanism and that the ATP-dependent processes of DNA translocation and destabilization of the DNA duplex ahead of the enzyme are distinct. From our studies, we propose that FANCJ also unwinds DNA by an active mechanism and suggest that the Fe-S domain plays an important role in coupling DNA translocation to either helicase activity or displacing proteins bound to DNA. Integrity of the Fe-S domain may influence DNA-motor interactions or the force-generating ability of FANCJ in its capacity to unwind structured nucleic acids or strip DNA-bound proteins.
Protein instability of FANCJ has been reported in patients with FA-J,5,6 as well as in patients with breast cancer,37 which might cause dysfunction of certain FANCJ mutant proteins in vivo. Recombinant FANCJ-A349P mutant protein expressed in insect cells was soluble, and its purity and integrity after purification were comparable with recombinant FANCJ-WT. Moreover, FANCJ-A349P retained DNA and ATP-binding activity, DNA-dependent ATPase activity, and DNA translocase activity, suggesting that the missense mutation does not globally disrupt protein structure. Indeed, the pattern of cleavage products from partial digestion of FANCJ-A349P and FANCJ-WT recombinant proteins was similar (data not shown). FANCJ-A349P protein was expressed in human cells and formed damage-inducible foci, suggesting that FANCJ-A349P is likely to be expressed in the cells of the patient.6
This study provides new insight to the importance of FANCJ catalytic activities for its role in DNA repair and G4 DNA metabolism. FANCJ is unique among the proteins genetically linked to the DNA cross-link disorder FA in that it is the only bonafide DNA helicase. Our work implicates the DNA unwinding and/or protein displacement functions of FANCJ as being critically important for its cellular role. Although FANCM is a DNA-dependent ATPase and can promote ATP-dependent branch point migration, this protein lacks detectable helicase activity; consequently, it has functions biochemically and genetically separable from FANCJ and has roles distinct from FANCJ.38 Further studies of clinically relevant mutations in FA proteins will be important to understand the molecular pathologies of the disease.
Partial loss of function in a full-length FANCJ protein, such as the helicase defective FANCJ-A349P mutant, may exert biologic effects that are distinguishable from truncated or unstable FANCJ proteins that behave as true nulls. The mutational spectrum in FA may be relevant to the question if FA heterozygotes are at an increased risk for cancer. For example, carrier grandmothers of FANCC mutations were found to be at the highest risk for breast cancer.39 The A349P mutant allele exerted a dominant-negative effect on cell survival or double-strand break formation after cellular exposure to MMC or TMS. Formation of FANCJ-A349P foci after MMC or TMS treatment raises the possibility that the mutant protein disrupts the accumulation or activity of other DNA repair/checkpoint factors at stalled replication forks. Previously, the engineered Walker A box K52R mutation that inactivated FANCJ ATPase activity was shown to exert a dominant-negative effect on sensitivity to interstrand cross-linking agents31 or IR,8,40 lead to a delayed entry into the S phase, and activate DNA damage checkpoint machinery.40 However, the K52R site-directed mutation is distinct from the A349P patient mutation because the A349P mutation uncouples DNA translocase activity from helicase activity. It will be of interest to determine whether FA carriers that harbor one A349P allele are characterized by hematopoietic stem cell/progenitor dysfunction or are predisposed to cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Johan de Winter (VU University Medical Center) for kindly providing the wild-type (VH10/hTert) and FA-J (EUFA30/hTert/SV40) fibroblasts and technical advice on cell transfection procedure. We thank Dr Sharon Cantor (University of Massachusetts Medical School) for FANCJ baculovirus construct. We thank Sarah Samira Subaran and Dr Fred E. Indig of the Confocal Imaging Facility, National Institute on Aging, National Institutes of Health for technical assistance with confocal microscopy. We thank Drs Senthil Perumal and Stephen Benkovic (Pennsylvania State University) for helpful advice on kinetic analysis of ATPase data.
This study was supported by the Intramural Research program of the National Institutes of Health, National Institute on Aging, and National Institute of Diabetes and Digestive and Kidney Diseases, the Fanconi Anemia Research Fund (R.M.B.), National Institutes of Health grant CA100839 (A.V.M.), and the Leukemia & Lymphoma Society Scholar Award 1054-09 (A.V.M.).
National Institutes of Health
Authorship
Contribution: Y.W. performed biochemical and genetic experiments; K.S.-y. and H.K. provided reagents; T.L. determined iron content; J.A.S. helped to purify proteins; J.S.D. and A.V.M. performed RAD51 displacement assays; A.N.S. performed some genetic assays; and Y.W. and R.M.B. designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.K. is Department of Molecular Oncology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Correspondence: Robert M. Brosh Jr, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, National Institutes of Health Biomedical Research Center, 251 Bayview Blvd, Baltimore, MD 21224; e-mail: broshr@mail.nih.gov.