Abstract
The transcription factor network in Hodgkin lymphoma (HL) represents a unique composition of proteins found in no other hematopoietic cell. Among these factors, an aberrant expression of the T-cell transcription factor GATA3 is observed in B cell–derived Hodgkin and Reed/Sternberg (HRS) tumor cells. Herein, we elucidate the regulation and function of this factor in HL. We demonstrate binding of NFκB and Notch-1, 2 factors with deregulated activity in HL to GATA3 promoter elements. Interference with NFκB and Notch-1 activity led to decreased GATA3 expression, indicating a dependency of deregulated GATA3 expression on these transcription factors. Down-regulation of GATA3 in HL cell lines demonstrated its role in the regulation of IL-5, IL-13, STAT4, and other genes. A correlation between GATA3 and IL-13 expression was confirmed for HRS cells in HL tissues. Thus, GATA3 shapes the cytokine expression and signaling that is typical of HL. Conclusively, aberrant GATA3 expression in HRS cells is stimulated by the deregulated constitutive activity of NFκB and Notch-1, indicating a complex network of deregulated transcription factors in these cells. GATA3 activity significantly contributes to the typical cytokine secretion of and signaling in HRS cells, which presumably plays an essential role in HL pathogenesis.
Introduction
Hodgkin lymphoma (HL) is one of the most frequent malignant lymphomas in the Western world. Characteristic of classical HL (cHL) is the occurrence of a small number of the typical Hodgkin and Reed/Sternberg (HRS) tumor cells among a mixed cellular infiltrate that is essential for their survival. In this tumor microenvironment, CD4+ helper and regulatory T cells make up the largest populations of infiltrating hematopoietic cells.1 Although HRS cells have largely lost their B-cell phenotype,2 they derive in nearly all cases from germinal center B cells.3,4 HRS cells express a variety of markers that are not associated with B cells (eg, granzyme B, CCL17, STAT5).5-7 In a previous gene expression study of cHL cell lines, we observed that HRS cells also aberrantly express the T-cell transcription factor GATA3, which was not expressed by normal mature B cells or by any of several other types of B-cell lymphomas included in that analysis.8
GATA3 is an essential transcription factor for early T-cell development.9 During TH cell development, GATA3 primes cells for the TH2 phenotype by shutting down production of factors that promote TH1 development (eg, IFNγ),10 and by opening TH2 target loci by direct promoter activation (eg, IL-5, IL-13).11,12 Recently, GATA3 overexpression has been associated with the outgrowth of pancreatic cancer cells, whereas its expression in breast cancer seems to be a favorable prognostic factor.13,14
Similarly, Notch-1 induces TH2 development, albeit by relying on the presence of GATA3 for this function.15 Direct promoter transactivation of GATA3 by Notch-1 has been demonstrated in murine CD4+ T cells.16 Notch-1 is also expressed in HRS cells, and its activity in cHL leads to accelerated growth and reduced apoptosis, and contributes to the suppression of B-cell gene expression in HRS cells.17,18 Activation of Notch-1 in HRS cells appears to be mediated through its ligand Jagged-1, which is expressed on bystander cells in the tumor micromilieu of cHL.
Constitutive NFκB activity is another hallmark of HRS cells and plays a central role in the pathogenesis of HL.19,20 Multiple factors, including genetic lesions in genes encoding NFκB inhibitory factors, such as NFKBIA and TNFAIP3, contribute to the strong NFκB activity.1,21,22 NFκB activity was first linked to GATA3 expression in airway epithelial cells when p50 knockout mice were unable to express GATA323 and to generate TH2-cell generation.
Here, we investigate the mechanisms for aberrant GATA3 expression in HRS cells and its consequences for gene expression—in particular, for the cytokine-signaling network—in cHL. We applied functional assays to elucidate whether GATA3 is part of a transcription factor network driven by NFκB and Notch-1 activity in cHL.
Methods
Cell lines and primary tissue
Cell lines were purchased (Deutsche Sammlung von Mikroorganismen und Zellkulturen) and cultured in RPMI-1640 media supplemented with 10% fetal calf serum (Biochrom AG), 2mM glutamine (Invitrogen), and 1% penicillin G/streptomycin sulfate (Invitrogen). U-HO1 cells were cultured as described above but in Iscove Modified Dulbecco Media. Fresh frozen lymph nodes from patients diagnosed with cHL were acquired from the collection at the Senckenberg Institute of Pathology (University of Frankfurt, Germany). The Internal Review Board of the University of Frankfurt approved usage of biopsy material.
IL-13 suppression assays
For details, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Subcellular fractionation
For details, see supplemental Methods.
Gene expression arrays and statistical analysis
For details, see supplemental Methods.
Protein immunoblots
Immunoblots were performed by standard methods. For details, see supplemental Methods.
Lentivirus vector production
Lentiviral particles overexpressing IκBα superrepressor were generated as previously described.21 For further details and small hairpin RNA (shRNA) sequences, see supplemental Methods.
Semiquantitative RT-PCR
Reverse-transcription polymerase chain reaction (RT-PCR) was performed on total RNA prepared with the RNeasy Micro Kit (QIAGEN). After deoxyribonuclease I treatment, equal amounts of RNA were reverse transcribed using the Sensiscript RT Kit (QIAGEN) and random hexamers. PCR amplification was performed as previously described.24 A minimum number of cycles was performed to obtain a clear signal on agarose gel electrophoresis within the linear amplification phase. The following specific primer pairs were used: GATA3: 5′-ggcctcagccactcctacat-3′ and 5′-cactctttctcgtcctgccg-3′; GAPDH: 5′-cacagtccatgccatcac-3′ and 5′-caccaccctgttgctgta-3′; STAT4: 5′-atgtctcagtggaatcaagt-3′ and 5′-ctgtctttctgaaactgaag-3′; IL-13: 5′-cacccagaaccagaaggctc-3′ and 5′-ggcagaatccgctcagcatc-3′; IL-5: 5′-atgaggatgcttctgcattt-3′ and 5′-ggccgtcaatgtatttcttt-3′.
Immunohistochemistry
We performed immunohistochemical staining for GATA3 and IL-13 for 35 cases of cHL. Staining for GATA3 failed in 2 cases. Among the evaluable 33 cases, 26 (79%) were scored as positive for GATA3 expression in HRS cells. IL-13 staining was more difficult to apply and evaluate. One case was excluded from analysis because of inconsistent results. We concluded with a collection of 16 cases with evaluable staining for GATA3 and IL-13. For IL-13 evaluation, only a broad estimation of positive HRS cells was possible because of many IL-13+ non-HRS cells and considerable background staining of this secreted molecule. For GATA3 evaluation, in the first collection of 10 cases (including 4 consistently negative cases) a broad estimation of positivity was performed. When staining 6 additional cases, the frequency of GATA3 positivity was determined more precisely by counting positive and negative HRS cells (8-42 per case). Immunostaining for GATA3 and IL-13 were evaluated in a blinded fashion. The following antibodies were used for immunohistochemical staining on frozen tissue sections: anti-GATA3 (no. sc-268; Santa Cruz Biotechnology Inc) 1:50 dilution, anti–IL-13 (Ab16219; Abcam) 1:100 dilution. For antigen retrieval, sections were fixed with acetone for 10 minutes. Detection of antibody binding was performed with the REAL-AP system (REAL Detection System, Alkaline Phosphatase/RED, Rabbit/Mouse K5005; Dako) and the bridge goat-immunoglobulin biotin (Rabbit E0466; Dako) were used. Incubation with primary antibodies took place for 30 minutes at room temperature for both antibodies.
ELISA
A human IL-13 enzyme-linked immunosorbent assay (ELISA) kit was used according to manufacturer protocol (Thermo Fisher Scientific [Perbio]). In brief, cell culture supernatants of cells grown for 3, 5, or 7 days were separated from the cells at 4°C by centrifugation for 5 minutes at 300g. Triplicate measurements of each supernatant were performed.
MTT assay
After lentiviral transduction or treatment with IL-13–binding protein, cells were seeded at a density of 104 cells per well of a 96-well plate and grown for the indicated time points. Cell viability was assayed at indicated times in triplicate wells using the CellTiter96 AQueous One Solution Cell Proliferation Assay (Promega Corporation).
ChIP
To evaluate chromatin immunoprecipitation (ChIP), a ChIP-IT express kit (Active Motif) was used for HL cell lines according to manufacturer protocol. PCR conditions were established as described for semiquantitative RT-PCR. The following primer sequences were used to detect Notch-1 and NFκB binding sites in the GATA3 promoter according to GenBank entry NC_000010: −3762: 5′-cgctcccttcccccttcctt-3′; −3382: 5′-ggggatggagaagggcaacc-3′; −561: 5′-ttgctcagccagccccggct-3′; −386: 5′-gggagacctggatgagcccc-3′; −306: 5′-gcagaattgcagagtcgtcg-3′; +158: 5′-ggtgctgcccgttgagcacg-3′.
Cell sorting
Two to 10 days after transduction, virally infected cells were sorted using a DiVa Cell Sorter (Becton Dickinson). Besides the expression of shRNAs, the lentiviral particles used herein also expressed the green fluorescent protein (GFP) in a second expression cassette under the spleen focus-forming virus (SFFV) promoter. Therefore, equal numbers of living, that is, propidium iodide–negative, cells were sorted according to their GFP expression for further analysis.
Cell-cycle distribution flow cytometry
Cells were harvested at indicated time points, fixed in 70% ethanol, washed, stained with propidium iodide, and analyzed for their cell-cycle profile.
Quantitative real-time PCR
Quantitative TaqMan PCR (Invitrogen) was performed on complimentary DNA reverse transcribed from RNA of same numbers of GFP+-sorted cells. Real-time PCR was performed on an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) using TaqMan Gene Expression assays Hs00231122_m1 for GATA3, Hs01062014_m1 for Notch-1, Hs99999903_m1 for actin, Hs00154109_m1 for Birc3, Hs00164932_m1 for ICAM-1, Hs00236874_m1 for Lta (Invitrogen) and TaqMan Universal PCR master mix NoAmp Erase UNG (Invitrogen) according to manufacturer protocols. For quantitative PCR of ChIP samples, conditions as previously described were used with 4 custom designed assays generated by Applied Biosystems: assay −523NFκB (assay ID AIQJARO), assay −88NFκB (assay ID AIVI3F0), assay upstream Notch-1 (assay ID AI89I9B), assay downstream Notch-1 (assay ID AIAAYE4).
Mutational analysis of Notch-1
Details of the mutational analysis of Notch-1 are given in supplemental Methods.
Results
GATA3 is expressed in cHL cell lines and effectively down-regulated by lentiviral-mediated shRNA expression
Based on differential gene expression studies of cHL cell lines and various other B-cell lymphomas and normal B cells, we previously detected consistent and specific expression of multiple non-B cell–specific genes in HL cell lines.8 Among these up-regulated mRNAs, the T cell–specific transcription factor GATA3 was found with its expression validated on the protein level.8 Atayar and colleagues25 confirmed this finding in primary HL tissue, demonstrating that, in two-thirds of HL cases analyzed, at least 25% to more than 75% of HRS cells were positive for GATA3.
To obtain a more comprehensive picture of GATA3 expression, 7 HL cell lines were analyzed for protein abundance (Figure 1A). GATA3 expression was strongest in cHL cell lines U-HO1 and L-428, whereas a moderate expression was seen in KM-H2 and L-1236. No expression was detected in EBV+ L-591, the T cell–derived HDLM-2, and the lymphocyte-predominant HL cell line DEV. The surprising lack of GATA3 in the T cell–derived HDLM-2 has been described previously.8
GATA3 expression in classical Hodgkin lymphoma cell lines is effectively altered by small hairpin RNA (shRNA) knockdown. (A) Immunoblots of Hodgkin lymphoma cell lines L-428, L-591, L-1236, KM-H2, HDLM-2, U-HO1, and DEV. Same amount of protein (100 μg) were assayed for protein expression of GATA3. Jurkat T cells were used as positive controls. Actin was used as loading control to demonstrate equal loading. (B) Protein lysates of cell lines treated with lentiviral particles for 2 functional shRNAs (siGATA3 no. 1 and no. 2, lanes 3 and 4) and 1 control scrambled shRNA (siSCR, lane 2) for GATA3 were immunoblotted for GATA3 and actin expression. Lysates were prepared 3 days (L-1236), 4 days (U-HO1), or 5 days after infection (L-428, KM-H2). (C) Three days after infection, GFP+ cells were sorted. After RNA and complimentary DNA preparation, semiquantitative PCRs for GATA3 and GAPDH mRNAs were performed. In the No RT control, RNA of untreated cells was deposited into the complimentary DNA reaction without RT enzyme. A representative experiment of at least 3 independent repeated experiments is shown.
GATA3 expression in classical Hodgkin lymphoma cell lines is effectively altered by small hairpin RNA (shRNA) knockdown. (A) Immunoblots of Hodgkin lymphoma cell lines L-428, L-591, L-1236, KM-H2, HDLM-2, U-HO1, and DEV. Same amount of protein (100 μg) were assayed for protein expression of GATA3. Jurkat T cells were used as positive controls. Actin was used as loading control to demonstrate equal loading. (B) Protein lysates of cell lines treated with lentiviral particles for 2 functional shRNAs (siGATA3 no. 1 and no. 2, lanes 3 and 4) and 1 control scrambled shRNA (siSCR, lane 2) for GATA3 were immunoblotted for GATA3 and actin expression. Lysates were prepared 3 days (L-1236), 4 days (U-HO1), or 5 days after infection (L-428, KM-H2). (C) Three days after infection, GFP+ cells were sorted. After RNA and complimentary DNA preparation, semiquantitative PCRs for GATA3 and GAPDH mRNAs were performed. In the No RT control, RNA of untreated cells was deposited into the complimentary DNA reaction without RT enzyme. A representative experiment of at least 3 independent repeated experiments is shown.
To embark on the function of GATA3, we used shRNA to knockdown its expression in HL cell lines. Two different shRNAs directed against the GATA3 sequence were cloned into lentiviral vectors, which also encode GFP. Transduction of cHL cell lines expressing GATA3—namely U-HO1, L-428, KM-H2, and L-1236—with shRNA expressing viral particles resulted in transduction efficiencies of 30% in KM-H2, 50% in U-HO1 and L-428, and up to 80% in L-1236 3 days after infection (as measured by the fraction of GFP+ cells). Compared with a nonfunctional scrambled shRNA (SCR), the 4 transduced lines responded to both shRNAs directed against GATA3 with reduced GATA3 protein concentrations as early as 3 days (L-1236), 4 days (U-HO1), or 5 days after infection (L-428, KM-H2; Figure 1B). Reduced protein concentrations were heterogeneous. Both shRNAs induced a strong down-regulation of GATA3 protein to undetectable levels in KM-H2 and L-1236, whereas siGATA3 no. 1 was more potent than siGATA3 no. 2 in L-428 and U-HO1 cells. The inhibition of GATA3 by shRNA knockdown was also observed on mRNA levels in the 4 transduced cell lines, indicating that the knockdown is not limited to interference on the translational level but also extends to the transcriptional level (Figure 1C).
GATA3 inhibition leads to down-regulation of IL-5, IL-13, and STAT4
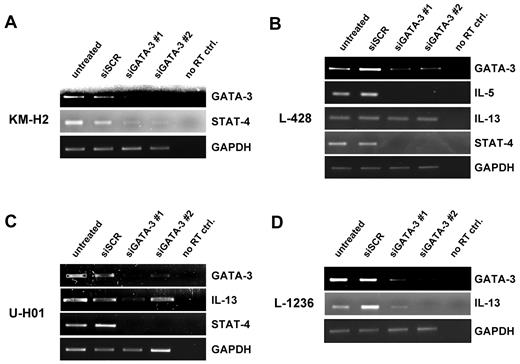
To test the consequences of GATA3 down-regulation on typical GATA3 target genes, we extended the semiquantitative RT-PCR analysis to 3 known GATA3 targets. Whereas the direct transactivation of IL-5 and IL-13 genes by GATA3 was first suggested in 1995,26,27 the suppressive effect of GATA3 expression on STAT4 has been described in developing TH1 and TH17 cells.28,29
In KM-H2 cells, loss of GATA3 concomitantly led to significantly diminished amounts of STAT4, as opposed to the inverse correlation described for T cells (Figure 2A). IL-5 could not be amplified from KM-H2 cells, as expected from our gene expression profiles, which showed a lack of IL-5 expression in this cell line.8 The expression of IL-13 in KM-H2 cells is somewhat controversial.30 In accord with data from Atayar and colleagues,25 we could not detect expression of IL-13 in KM-H2 cells.
GATA3 downstream targets IL-5, IL-13, and STAT4 are influenced by GATA3 down-regulation. Semiquantitative RT-PCR on sorted GFP+ cells as in Figure 1C on KM-H2 (A), L-428 (B), U-HO1 (C), and L-1236 (D) cells. Messenger RNA of IL-5, IL-13, and STAT4 were amplified. A representative experiment of at least 3 independent repeated experiments is shown for each cell line.
GATA3 downstream targets IL-5, IL-13, and STAT4 are influenced by GATA3 down-regulation. Semiquantitative RT-PCR on sorted GFP+ cells as in Figure 1C on KM-H2 (A), L-428 (B), U-HO1 (C), and L-1236 (D) cells. Messenger RNA of IL-5, IL-13, and STAT4 were amplified. A representative experiment of at least 3 independent repeated experiments is shown for each cell line.
The L-428 cell line expresses all 3 downstream target genes. Although IL-5 and STAT4 expression was diminished to undetectable levels on down-regulation of GATA3 (Figure 2B), IL-13 expression remained unaltered.
In U-HO1 cells, IL-13 was strongly down-regulated after reduction of GATA3 with 1 of the 2 GATA3-specific shRNAs (Figure 2C). In addition, STAT4 expression was no longer detectable in these cells—even when GATA3 was incompletely knocked down (Figure 2C lane 4). IL-5 could not be detected in U-HO1 cells by RT-PCR.
The L-1236 cell line lacks IL-5 expression.8 IL-13 expression in these cells is robustly decreased when GATA3 expression is depleted. Although we expected to find STAT4 expressed in L-1236 cells,8 we were unable to amplify its mRNA even when extended PCR conditions were applied. Taken together, GATA3 activity directly influences various cytokines and activators in a number of different cHL cell lines.
To obtain a more comprehensive overview of GATA3 regulated genes in cHL cell lines, we performed a genechip analysis comparing gene expression in GATA3-specific shRNA-transduced L-1236 and U-HO1 cells compared to samples transduced with vectors encoding scrambled shRNAs. According to a quantitative TaqMan pretest assay, the down-regulation of GATA3 in this experiment was moderate but statistically significant, with down-regulation by a factor of 3.5- to 7.2-fold in L-1236 cells and 5.5- to 7.5-fold in U-HO1 cells (data not shown). According to genechip analysis, GATA3 transcription levels were down-regulated to 55% for UHO-1 cells and 69% for L-1236. Regarding the 3 GATA3 targets (IL-5, IL-13, and STAT4) analyzed in detail by specific quantitative RT-PCR (see Figure 2), the 3 genes were not detected above background levels in L-1236 cells. Therefore, genechips were not informative for regulation. Signals for IL-13 and STAT4 were detected at low levels in UHO-1 cells, but no significant regulation was observed, although this result was consistently seen in multiple earlier specific RT-PCR experiments. This finding may be explained through the moderate down-regulation of GATA3 observed in this experiment. In U-HO1 cells, however, IL-5 was the most down-regulated gene (2.05-fold). Thus, we can add IL-5 to the genes that are regulated by GATA3 in UHO-1 cells.
The gene expression profiles of all siGATA3 and siSCR-treated samples for U-HO1 and L-1236 cells were analyzed for genes with up- or down-regulated expression levels of more than 1.5-fold and false discovery rates of less than 0.25 (supplemental Table 1). In this analysis, the siGATA3-treated samples of U-HO1 and L-1236 were together compared with control samples, to focus on genes consistently regulated by GATA3 (directly or indirectly) in both lines. Ten genes were down-regulated and 18 genes were up-regulated. It is noteworthy that known functions of the 28 consistent GATA3 targets include apoptosis regulation, regulation of transcription, cell-cycle control, and others, indicating that GATA3 not only regulates components of cytokine signaling but is involved in the regulation of multiple cellular processes in HRS cells.
GATA3 abundance correlates with IL-13 expression in HL
Because the connection between IL-13 and GATA3 was inconsistent in the cHL cell lines (observed in 2 of 4 cell lines), we wanted to clarify whether a correlation between these 2 factors exists in HL, especially because a prosurvival activity has been attributed to IL-13 expression in HL.20,30,31 Therefore, we performed immunohistochemical staining for both proteins in 35 cases that represent the histologic subtypes nodular sclerosis or mixed cellularity of cHL. Of these, 16 cases with evaluable staining for both GATA3 and IL-13 were included in the final collection (see supplemental Methods, supplemental Table 2). All cases were at least 60% positive in HRS cells for IL-13. Generally, the cases with enhanced positivity of 80%-90% also showed elevated levels of GATA3 positivity of at least 70%. We used the Pearson product moment correlation to determine whether there was a statistically significant correlation between nuclear GATA3 positivity and IL-13 expression in our collection (Figure 3). Under the hypothesis of a T distribution, the correlation is significant with a P value of .0025. The correlation of 0.619 indicates that there is a linear correlation between GATA3 and IL-13 expression.
GATA3 and IL-13 expression correlate in primary classical Hodgkin lymphoma tissue. Sixteen primary cases of classical Hodgkin lymphoma were analyzed for their correlation of GATA3 and IL-13 after immunohistochemical staining. Samples 1, 5, 6, 8, 9, 11-13, and 15 are of the nodular sclerosis subtype. Samples 2-4, 7, 10, 14, and 16 are of the mixed cellularity subtype. The Pearson product moment correlation resulted in a P value of .0025. The T-distribution hypothesis test resulted in a correlation coefficient of 0.619 (cor = 1 for a perfect linear correlation). In this analysis, 3 cases with only cytoplasmic staining for GATA3 were considered “negative,” because only nuclear GATA3 can bind to the IL-13 promoter.
GATA3 and IL-13 expression correlate in primary classical Hodgkin lymphoma tissue. Sixteen primary cases of classical Hodgkin lymphoma were analyzed for their correlation of GATA3 and IL-13 after immunohistochemical staining. Samples 1, 5, 6, 8, 9, 11-13, and 15 are of the nodular sclerosis subtype. Samples 2-4, 7, 10, 14, and 16 are of the mixed cellularity subtype. The Pearson product moment correlation resulted in a P value of .0025. The T-distribution hypothesis test resulted in a correlation coefficient of 0.619 (cor = 1 for a perfect linear correlation). In this analysis, 3 cases with only cytoplasmic staining for GATA3 were considered “negative,” because only nuclear GATA3 can bind to the IL-13 promoter.
For unknown reasons, 3 cases showed only cytoplasmic localization of GATA3. In addition, 3 other cases demonstrated cytoplasmic and nuclear positivity (supplemental Table 2). Therefore, we analyzed the GATA3-expressing cell lines for subcellular localization of this factor (supplemental Figure 1A). GATA3 was exclusively found in the nucleus of all 4 HL cell lines and not seen in the cytoplasm, which is also true when GATA3 was knocked down by shRNAs in L-1236 cells (supplemental Figure 1B).
To further evaluate the association between GATA3 and IL-13 under cell culture conditions, we transduced HL cell lines with shRNAs for GATA3 and monitored IL-13 levels in the cell culture medium by ELISA (Figure 4A). Protein lysates from cells corresponding to the cell culture medium samples used for ELISA were assayed for GATA3 protein expression in immunoblots (Figure 4B). GATA3 proteins significantly decreased as early as 3 days after infection whereas secreted IL-13 in L-1236 cells started to diminish at approximately 5 days after infection. At 7 days after infection, IL-13 levels were reduced from 325 pg/mL to approximately 200 pg/mL for both shRNA constructs and, hence, to approximately 60% of the cytokine level in the cell culture medium of scrambled shRNA transduced cells. As expected, in accordance with the results of the semiquantitative RT-PCR (Figure 2B), L-428 did not show reduced levels of secreted IL-13 as a consequence of GATA3 depletion (data not shown). Moreover, the amount of IL-13 in KM-H2 culture medium was below detection (7 pg/mL) in the ELISA (data not shown) and consequently verifies the results of our semiquantitative RT-PCR (Figure 2A), where IL-13 could not be amplified due to lack of IL-13 mRNA. In conclusion, IL-13 and GATA3 correlate in vitro and in vivo in HL, indicating that GATA3 contributes to IL-13 expression in HRS cells, although HL cell lines show a heterogeneous picture.
GATA3 regulates IL-13 expression in classical Hodgkin lymphoma cell lines. (A) Cell culture supernatant of siSCR, siGATA3 no. 1, and siGATA3 no. 2 transduced L-1236 cells 3, 5, and 7 days after infection were analyzed for IL-13 concentration by enzyme-linked immunosorbent assay in triplicate. Statistical significance was determined using a 2-tailed t test. (B) Protein lysates from cells corresponding to cell culture supernatants in (A) were prepared and 100 μg of protein were loaded onto SDS-PAGE. GATA3 abundance was detected with antibody sc-268. Actin served as loading control. One representative experiment of 3 independent experiments is shown.
GATA3 regulates IL-13 expression in classical Hodgkin lymphoma cell lines. (A) Cell culture supernatant of siSCR, siGATA3 no. 1, and siGATA3 no. 2 transduced L-1236 cells 3, 5, and 7 days after infection were analyzed for IL-13 concentration by enzyme-linked immunosorbent assay in triplicate. Statistical significance was determined using a 2-tailed t test. (B) Protein lysates from cells corresponding to cell culture supernatants in (A) were prepared and 100 μg of protein were loaded onto SDS-PAGE. GATA3 abundance was detected with antibody sc-268. Actin served as loading control. One representative experiment of 3 independent experiments is shown.
GATA3 expression has no prosurvival effect in HL
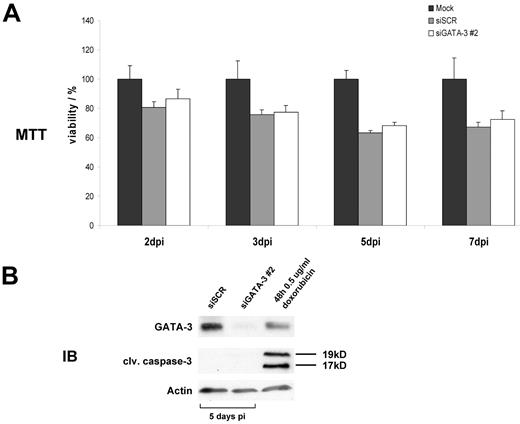
Because we established that IL-13 expression is dependent on GATA3 activity in HL and because IL-13 cytokine expression is often described as a prosurvival factor for HRS cells,30,32,33 we embarked on determining whether GATA3 likewise has a prosurvival and antiapoptotic function in HRS cells. On transduction of L-1236 cells with shRNAs targeting GATA3, the amount of metabolically active and, thus, healthy dividing cells was measured in an MTT assay (Figure 5A). Although a decrease of viable cells after GATA3 knockdown was observed 3, 5, and 7 days after infection (compared with mock-infected cells set as 100% viable cells) the effect was not significantly different from cells treated with nonfunctional scrambled control shRNA. The effects in MTT assays measured for L-428 and KM-H2 cells were similar to the results in L-1236 cells (data not shown). To further validate these findings, we performed immunoblots on transduced cells, analyzing those cells for cleaved caspase-3 as a marker for apoptotic cells. L-1236 cells did not show any signs of apoptosis up to 1 week after lentiviral transduction (Figure 5B) although GATA3 was fully depleted. Results for L-428 and KM-H2 were in accord with findings for L-1236 (data not shown). Therefore, GATA3 does not influence the survival of HL cells in our models. However, depletion of IL-13 activity, as previously described,34 reduced the viability of L-1236 cells (supplemental Figure 2A) whereas none of the other 3 cell lines reacted on the IL-13 binding protein. The reduced amount of metabolically active cells is caused by apoptosis as a result of an increased number of cells with sub-G1 content (supplemental Figure 2B). Therefore, GATA3 may not be the only factor involved in IL-13 regulation for HL and additional interference with other unknown upstream IL-13 regulators would be needed to produce similar effects as those seen with IL-13 binding protein.
GATA3 has no antiapoptotic or prosurvival effect in HL. (A) MTT assay on transduced L-1236 cells 2, 3, 5, and 7 days postinfection (dpi). Results for mock infected cells were set as 100% viable cells. Values presented are the mean of triplicate measurements. (B) Protein lysates of L-1236 cells after viral small hairpin RNA transfer were assayed for GATA3, cleaved caspase-3, and actin abundance 5 days after infection. Treatment with doxorubicin for 48 hours served as a positive control for apoptosis induction. One representative assay of 3 independent experiments is given.
GATA3 has no antiapoptotic or prosurvival effect in HL. (A) MTT assay on transduced L-1236 cells 2, 3, 5, and 7 days postinfection (dpi). Results for mock infected cells were set as 100% viable cells. Values presented are the mean of triplicate measurements. (B) Protein lysates of L-1236 cells after viral small hairpin RNA transfer were assayed for GATA3, cleaved caspase-3, and actin abundance 5 days after infection. Treatment with doxorubicin for 48 hours served as a positive control for apoptosis induction. One representative assay of 3 independent experiments is given.
GATA3 regulatory elements are occupied by Notch-1 and NFκB proteins
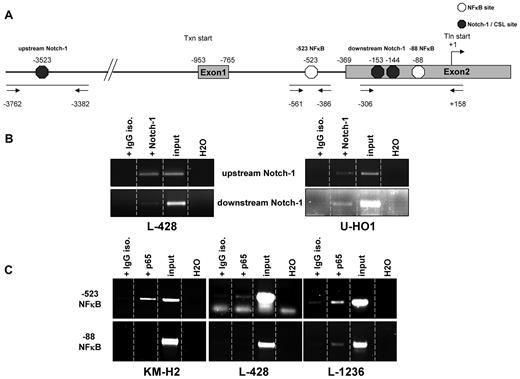
Although we could assign a functional role for GATA3 in cHL, the mechanisms by which its expression is regulated was hitherto enigmatic. Therefore, we analyzed putative regulatory elements surrounding the GATA3 promoter. Because of the reported linkage of GATA3 expression with NFκB activity in murine airway epithelial cells and direct GATA3 promoter activation by Notch-1 in murine T cells,16,23 we focused on potential binding sites for these 2 factors in the human GATA3 gene. We used the known binding sequences GGGRNNYYCC for NFκB35 and TGGGAA for Notch-1/CSL36 and identified several putative binding sites within or near the GATA3 promoter according to the GenBank entry NC_000010. Three Notch-1 binding sites at −3523, −153, and −144, relative to the translational start site set as +1, were identified. Two NFκB sites were identified, one at −88 and one within intron 1 at position −523 (Figure 6A). We performed ChIP assays to clarify whether any of these sites are occupied by Notch-1 and/or NFκB. After antibody treatment and consecutive PCRs, we observed that Notch-1 robustly occupied the more 5′ binding site termed “upstream Notch-1” as well as the more 3′ located site “downstream Notch-1” in L-428 and U-HO1 cells (Figure 6B). However, the PCR band intensity generated after precipitation with Notch-1 antibody was weak for the downstream binding site in L-428 cells.
GATA3 promoter and regulatory element sequences are occupied by NFκB subunit p65 and Notch-1 in vivo. (A) Schematic draft of the GATA3 gene. Exons 1 and 2 are depicted. The transcriptional start site (Txn) is at −953. Translational start site (Tln) is located in exon 2 at position +1. Exon 2 starts at −369. NFκB sites are indicated by white hexagons whereas black hexagons represent Notch-1/CSL sites. The 3 PCR amplicons are depicted by bars underlining putative binding sites. Arrows represent primers used for PCR. (B) Results of a representative chromatin ChIP PCR after antibody treatment of samples in the indicated lanes for putative Notch-1 binding sites. (C) As in (B) PCRs for NFκB ChIP. Each ChIP was repeated twice.
GATA3 promoter and regulatory element sequences are occupied by NFκB subunit p65 and Notch-1 in vivo. (A) Schematic draft of the GATA3 gene. Exons 1 and 2 are depicted. The transcriptional start site (Txn) is at −953. Translational start site (Tln) is located in exon 2 at position +1. Exon 2 starts at −369. NFκB sites are indicated by white hexagons whereas black hexagons represent Notch-1/CSL sites. The 3 PCR amplicons are depicted by bars underlining putative binding sites. Arrows represent primers used for PCR. (B) Results of a representative chromatin ChIP PCR after antibody treatment of samples in the indicated lanes for putative Notch-1 binding sites. (C) As in (B) PCRs for NFκB ChIP. Each ChIP was repeated twice.
The ChIP for NFκB binding showed that the −523 site within intron 1 was occupied by the p65 activator subunit of NFκB in all 3 cell lines (Figure 6C). In L-428 and L-1236, binding was less pronounced than in KM-H2 cells. In addition, precipitation with the IgG isotype negative control antibody resulted in faint bands in these 2 lines, indicating some background signal. The −88 NFκB binding site was only weakly occupied by p65 in L-1236 cells compared with negative control samples. The other 2 cell lines lacked p65 binding. To validate these findings quantitatively, we performed additional ChIP experiments with L-428 and U-HO1 cells and evaluated the transcription factor binding using TaqMan PCR. These studies nicely validated for both cell lines the binding of Notch-1 and NFκB to their respective binding sites (supplemental Figure 3). Therefore, we demonstrated binding of both transcription factors at various promoter and/or regulatory elements in the GATA3 gene in HL cell lines, indicating that GATA3 expression might be regulated by these transcription factors.
Notch-1 and NFκB activity induce GATA3 expression in HL
To elucidate the effect of Notch-1 and NFκB binding for GATA3 expression, we interfered with the activity of Notch-1 by shRNA knockdown and diminished NFκB activity by expression of an IκBα superrepressor. Successful reduction of NFκB activity was validated by strong down-regulation (at least 4-fold) of the 3 typical NFκB target genes ICAM-1, Birc3, and Lta in 3 of the 4 transduced cell lines, and at least a moderate down-regulation (1.5- to 4-fold) in KM-H2 cells (Fig 7A). As a consequence of this decreased NFκB activity, GATA3 expression was consistently reduced in all 4 cell lines. In L-428 and L-1236 cells, GATA3 mRNA was down-regulated by a factor of approximately 1.5-fold whereas GATA3 expression was reduced by 2.5- to 3-fold in KM-H2 and U-HO1 cell lines. Therefore, NFκB activity regulates GATA3 expression in cHL cell lines.
NFκB activity and Notch-1 expression regulate GATA3 expression. (A) TaqMan PCR of 1 typical result of L-428, L-1236, KM-H2, and U-HO1 cells 5 days after viral transduction with IκBα superrepressor, GFP+ cell sorting, and RNA/complimentary DNA preparation. The 3 typical NKκB target Birc3, ICAM-1, and Lta were used as controls for NFκB activity. mRNA expression for all 4 analyzed genes is depicted as fold change down-regulation after normalization to actin housekeeping controls. (B) Immunoblot for Notch-1 expression of HDLM-2, KM-H2, L-428, L-1236, and U-HO1 cells. Jurkat cells served as positive controls. 100 μg protein were loaded. Actin was used as a loading control. (C) TaqMan RT-PCR as in (A) on KM-H2, L-428, and U-HO1 cells 5 days after delivery of small hairpin RNA against Notch-1. (D) Immunoblot for Notch-1 and GATA3 expression in L-428 and U-HO1 cells 5 days after transduction of indicated small hairpin RNA sequences. Actin served as loading control. Each lentiviral transduction was performed 3 times.
NFκB activity and Notch-1 expression regulate GATA3 expression. (A) TaqMan PCR of 1 typical result of L-428, L-1236, KM-H2, and U-HO1 cells 5 days after viral transduction with IκBα superrepressor, GFP+ cell sorting, and RNA/complimentary DNA preparation. The 3 typical NKκB target Birc3, ICAM-1, and Lta were used as controls for NFκB activity. mRNA expression for all 4 analyzed genes is depicted as fold change down-regulation after normalization to actin housekeeping controls. (B) Immunoblot for Notch-1 expression of HDLM-2, KM-H2, L-428, L-1236, and U-HO1 cells. Jurkat cells served as positive controls. 100 μg protein were loaded. Actin was used as a loading control. (C) TaqMan RT-PCR as in (A) on KM-H2, L-428, and U-HO1 cells 5 days after delivery of small hairpin RNA against Notch-1. (D) Immunoblot for Notch-1 and GATA3 expression in L-428 and U-HO1 cells 5 days after transduction of indicated small hairpin RNA sequences. Actin served as loading control. Each lentiviral transduction was performed 3 times.
To modulate Notch-1 activity, 2 different shRNA sequences were delivered via lentiviral transduction into cHL cells expressing Notch-1. To verify whether our cHL cell lines expressed sufficient levels of Notch-1 as previously published,37 we performed immunoblots on Jurkat T cells and on various cHL cell lines (Figure 7B). HDLM-2, KM-H2, L-428, and U-HO1 cells expressed Notch-1 protein even stronger than Jurkat leukemia cells. L-1236 cells had a weak expression similar to that of Jurkat cells. Hence, 3 cell lines, namely KM-H2, L-428, and U-HO1 expressed Notch-1 and GATA3 at sufficient levels and could thus be used for shRNA assays. Notch-1 expression could be efficiently down-regulated at least 4-fold in all 3 cell lines with siN-1 no. 1 shRNA as determined by quantitative TaqMan mRNA target gene measurement (Figure 7C). siN-1 no. 2 shRNA led to 2.5- to 3-fold down-regulation of Notch-1 in U-HO1 cells. In all 3 cell lines, we observed that, as a consequence of altered Notch-1 expression, GATA3 decreased 2.5- to 3-fold when cells were transduced with siN-1 no. 1 shRNA against Notch-1. ShRNA siN-1 no. 2 had similar effects on GATA3 expression when used in U-HO1 cells (Figure 7C). Moreover, in immunoblots, we detected that, after Notch-1 protein level reduction, GATA3 protein expression was drastically diminished in L-428 and U-HO1 cell lines when siNotch-1 no. 1 or siNotch-1 no. 2 was used (Figure 7D). In conclusion, besides the influence of NFκB activity on GATA3 expression, we demonstrated that the T cell transcription factor Notch-1 also modulates expression of GATA3 in HL-derived cell lines.
Discussion
HRS cells are B cell–derived in nearly all cases, although these cells have largely lost their B-lineage–specific gene expression program and regularly express a variety of non–B cell markers,2 including GATA3,8,25 which, in the hematopoietic system, is specifically expressed in T cells and among mature B cell lymphomas found only in HRS cells of cHL.8 We studied the consequences of aberrant GATA3 expression in HRS cells and mechanisms for its deregulated expression. Three known GATA3 target genes (IL-5, IL-13, and STAT4) were analyzed specifically for altered expression after GATA3 depletion. However, we could not analyze each of these genes in all 3 cell lines because HL lines showed a surprisingly heterogeneous expression pattern of IL-5, IL-13, and STAT4. It is important, however, that with one exception (IL-13 in L-428), down-regulation of GATA3 led a consistent down-regulation of the 3 genes in all instances where they were initially expressed. The finding that STAT4 is positively regulated by GATA3 expression in HL was unexpected and in contrast to previous findings in TH1 and TH17 cells.28,29 However, we consistently observed a positive regulation of STAT4 by GATA3 in 3 cell lines, indicating that its regulation in HRS cells seems to be inverse to the situation in normal T cells. The lack of responsiveness of L-428 regarding IL-13 transcription levels upon GATA3 down-regulation indicates that, in this cell line, IL-13 expression is regulated by a set of transcription factors that overcome the need for GATA3. The consequences of GATA3 expression for IL-13 expression was further analyzed at the protein level, showing that GATA3 down-regulation in L-1236 cells caused a reduced secretion of IL-13 into the culture medium.
The connection between IL-13 and GATA3 was further studied on the protein level in vivo. Although most of the cases expressed both GATA3 and IL-13, we could nevertheless identify a statistically significant correlation between the fraction of GATA3+ and IL-13+ HRS cells in the HL cases. This result indicated that, also, in vivo, GATA3 is involved in the regulation of IL-13 expression and is an important, albeit not the only, positive modulator of its expression level.
Previously, a prosurvival effect was appointed to IL-13 in HRS cells,30,32,33 which we could confirm in L-1236 cells (supplemental Figure 2). Therefore, we tested the effect of GATA3 knockdown on survival and apoptosis induction in HL cell lines. However, neither by measuring cell viability in an MTT assay, nor by immunoblot analysis for cleaved caspase-3, could we detect clear signs of apoptosis in any of the 3 HL lines analyzed. Thus, the partial down-regulation of IL-13 by down-regulation of GATA3 is not strong enough to translate into a direct apoptotic effect for HL cells. Main functions of GATA3 in HRS cells may relate to the modulation of the microenvironment by influencing cytokine-signaling pathways, which cannot be tested in in vitro HL cell lines in the absence of the typical HL micromilieu.
Genechip analysis revealed that GATA3 regulates multiple further genes in HRS cells. Therefore, the effects of GATA3 in HRS cells are not only limited to cytokine expression and signaling but extend to numerous other cellular functions.
Experiments designed to reveal the mechanisms for aberrant expression of GATA3 in HRS cells showed that the NFκB and Notch-1 transcription factors reside at binding sites in the vicinity of the transcriptional start site of GATA3. The NFκB site within intron 1 is robustly bound by p65-activator subunits of NFκB in 4 cell lines, and the −88NFκB site in exon 2 was bound by NFκB in 2 of the cell lines. The Notch-1 sites in the promoter of GATA3 at −3523 and the sites in exon 2 are occupied by Notch-1 proteins in each of the cell lines tested (Figure 6 and supplemental Figure 3). Inhibition of NFκB activity or down-regulation of Notch-1 expression showed that this binding is functionally relevant, as inhibiting these transcription factors caused a marked down-regulation of GATA3. Thus, the deregulated, constitutive activity of NFκB and Notch-1 in HRS cells contributes to the aberrant expression of GATA3. Activating Notch-1 mutations in HL cell lines as a cause for its constitutive activity can be ruled out because a mutational analysis of 5 HL cell lines did not reveal any activating mutations (data not shown). A picture begins to emerge in which multiple transcription factors influence each other, together causing the “reprogramming” of HRS cells with a hyperactivated phenotype of these cells that is rather unique among lymphoid malignancies. It remains to be determined which initial factor causes the deregulation of numerous signaling pathways and transcription factors, and whether there are master regulators that stabilize the typical HRS cell gene-expression pattern.
Therefore, it can be seen that GATA3 influences multiple cytokines and activators (IL-5, IL-13, and STAT4) in cHL. By contributing to the regulation of these and additional factors, GATA3 influences gene expression in HRS cells as well as their interaction with other cells in the microenvironment, thus shaping tumor physiology. Furthermore, it appears that constitutively active transcription factors NFκB and Notch-1 contribute to deregulated GATA3 expression, a process that activates the expression of the STAT4 transcription factor, pointing to a multicomponent, interrelated, and deregulated transcription factor network in HRS cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Andrea Wind and Katharina Waldhelm for excellent technical assistance. We also thank Klaus Lennartz for support during fluorescence-activated cell sorting, and Gwen Lorenz and Roland Schmitz for help with Notch-1 mutation analysis. Finally, we are indebted to Ralf Lieberz and Sylvia Hartmann for their expertise and performance of immunohistochemical staining.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (KU 1315/5-2).
Authorship
Contribution: J.S. designed and performed research, analyzed data and wrote the manuscript; C.D. performed statistical analysis; M.-L.H. provided biopsy material and supervised and evaluated immunohistochemical staining; and R.K. designed and supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ralf Küppers, Institute of Cell Biology (Cancer Research), University of Duisburg-Essen, Medical School, Virchowstr 173, D-45122 Essen, Germany; e-mail: ralf.kueppers@uk-essen.de.