Abstract
Tribbles homolog 2 (Trib2) is a pseudokinase that induces acute myelogenous leukemia (AML) in mice and is highly expressed in a subset of human AML. Trib2 has 3 distinct regions, a proline-rich N-terminus, a serine/threonine kinase homology domain, and a C-terminal constitutive photomorphogenesis 1 (COP1)–binding domain. We performed a structure-function analysis of Trib2 using in vitro and in vivo assays. The N-terminus was not required for Trib2-induced AML. Deletion or mutation of the COP1-binding site abrogated the ability of Trib2 to degrade CCAAT/enhancer-binding protein-α (C/EBP-α), block granulocytic differentiation, and to induce AML in vivo. Furthermore, COP1 knockdown inhibited the ability of Trib2 to degrade C/EBP-α, showing that it is important for mediating Trib2 activity. We also show that the Trib2 kinase domain is essential for its function. Trib2 contains variant catalytic loop sequences, compared with conventional kinases, that we show are necessary for Trib2 activity. The kinase domain mutants bind, but cannot efficiently degrade, C/EBP-α. Together, our data demonstrate that Trib2 can bind both COP1 and C/EBP-α, leading to degradation of C/EBP-α. Identification of the functional regions of Trib2 that are essential to its oncogenic role provides the basis for developing inhibitors that will block Trib functions in cancer.
Introduction
Tribbles (Trib) pseudokinases have recently emerged as important contributors to dysregulated signaling in malignant hematopoiesis. All 3 mammalian Trib homologs, Trib1, Trib2, and Trib3 (collectively termed “Trib1-3”), are associated with human malignancies.1-3 Trib1 and Trib2 function as oncogenes in acute myelogenous leukemia (AML) and cooperate with homeobox gene 9 (HoxA9) to accelerate the onset of AML in murine AML models.2,4 In particular, Trib2 is highly expressed in a specific subset of human AMLs that are associated with impaired CCAAT/enhancer-binding protein-alpha (C/EBP-α) function.2 These AMLs express both myeloid and T-cell markers, and some contain activating Notch1 mutations.5 As Trib2 is a direct transcriptional target of oncogenic Notch1, we hypothesize that the oncogenic Notch1 mutations are activating Trib2 expression in some of these tumors.5 These biphenotypic AMLs respond poorly to current therapies, suggesting that new therapeutic strategies are needed.6
Trib proteins interact with various signaling molecules and transcription factors, including activating transcription factor 4 (ATF4),7 p65,8 C-terminal interacting protein (CtIP),9 mitogen-activated protein kinase kinase (MAPKK),10 protein kinase B (AKT),11 and constitutive morphogenesis 1 (COP1).12 We recently demonstrated that Trib2 binds to and facilitates the degradation of C/EBP-α, and proposed that this process is a key feature of Trib2-induced AML.2 Trib2 (also termed c5Fw, TRB-2, GS3955, TRB2, and AW319517) has 3 clearly distinguishable regions, including an N-terminal portion, a central serine/threonine kinase–like domain (KD), and a C-terminal region that contains a COP1-binding site. The Trib2 N-terminus is the most divergent region of mammalian Trib proteins and is characterized by a high serine and proline content. The C-terminus contains a DQxVPx motif that is shared by Trib1-3 and binds the E3 ligase, COP1.12
Trib2 belongs to the pseudokinase family because it contains a central region that shares considerable homology with serine/threonine kinases, but has deviations in the catalytic loop that are likely to eliminate catalytic activity.13 Canonical kinases have an N-terminal lobe that binds adenosine triphosphate (ATP), a central hinge region, and a C-terminal lobe that binds substrates.14 The N-terminal lobe contains an invariant lysine residue critical for ATP binding that is conserved in Trib2, but the highly conserved sequence, HRDLKPEN, in the C-terminal subdomain, which is critical for phosphate transfer in active kinases, is altered to LRDLKLRK in Trib2 (the lysine at position 5 in this sequence is typical of serine/threonine kinases, whereas this position is an arginine residue in conventional tyrosine kinases). Also absent from Trib2 is the asparagine at position 8, which is predicted to stabilize the catalytic loop.14 The most striking difference between Trib2 and conventional kinases is the complete absence of a DFG motif in the C-terminal domain important for catalytic activity and chelation of Mg2+ ions.13 Consistent with these structural features, Trib proteins lack demonstrable serine/threonine kinase activity.15-17 Although Trib2 is a phosphoprotein, it is not autophos-phorylated, which is consistent with the idea that Trib2 lacks kinase activity.18
The Trib2 domains responsible for protein binding or functional/oncogenic activity are unknown. Here, we report the results of an in vivo structure/function study to dissect the role of Trib2 in the pathogenesis of AML and identify its functional regions. We show that the N-terminus of Trib2 is not essential for either hematopoietic cell differentiation or myeloid cell transformation. On the other hand, the integrity of the KD is required for both of these activities, and mutation of critical residues within the KD interferes with these activities. Binding of COP1 to the Trib2 C-terminus is essential for Trib-induced AML. In the absence of COP1 binding, C/EBP-α is no longer degraded, and leukemia does not occur. Together, our findings provide important new insights into structure-function relationships in Trib proteins and highlight the features of Trib2 to be targeted for the development of Trib2-directed inhibitors.
Methods
Constructs and retroviruses
A myc-tag was incorporated into the MigR1 retroviral vector (myc-tag, designated MM) by ligating a double-stranded oligo containing the myc tag into BglI/Xho1-digested MigR1. Trib2 deletion constructs were generated by ligating polymerase chain reaction (PCR)–amplified cDNA fragments from full-length MigR1-Trib22 into the MigR1-myc vector. Site-directed mutagenesis of the COP1-binding site at amino acids (aa) 326-331 and the catalytic domain (aa 173-180) was performed using the QuikChange kit (Stratagene) according to the manufacturer's instructions. HoxA9 was subcloned into the BglII/EcoR1-digested murine stem cell virus/internal ribosome entry site/nerve growth factor receptor (MSCV-IRES-NGFR) vector. The 2 shCOP1 constructs (clones V2MM_38006 and V2HS_260407; Open Biosystems) and scrambled shRNA (clone pSM2-NCshRNA; Open Biosystems) were subcloned into low-molecular-mass polypeptide (shRNA vector with green fluorescent protein [GFP]; gift from Scott Lowe, Cold Spring Harbor Laboratory).19 pcDNA-COP1 was kindly provided by M. Montminy (The Salk Institute, La Jolla, CA).
Immunoprecipitation and Western blot analysis
First, 1 mg of precleared lysates were incubated overnight with 20 μL of protein G beads coated with 5 μg anti-MYC 9E10 antibody. Whole cell lysates were prepared using modified radioimmune precipitation assay buffer (50mM Tris, pH 8.0, containing 0.5% Nonidet P-40 [NP-40], 0.25% sodium deoxycholate, 150mM NaCl, 1mM EDTA [ethylenediaminetetraacetic acid], 1mM phenylmethylsulfonyl fluoride [PMSF], 1mM sodium orthovanadate [Na3Vo4], 1mM NaF), supplemented with protease inhibitors (Complete EDTA-free; Roche). Antibodies used were anti–C/EBP-α (Sc-61; Santa Cruz Biotechnology), anti–C/EBP-β (Sc-7962; Santa Cruz Biotechnology), anti–phospho-AKT (4051; Cell Signaling Technology), anti-AKT (04-796; Millipore), anti-p44/42 MAPK (9102; Cell Signaling Technology), anti–phospho-p44/42 MAPK (9101; Cell Signaling Technology), anti-HA (hemagglutinin) (12CA5; Roche), anti-FLAG (M2; Sigma-Aldrich), anti-MYC (9E10), and anti–β-actin (A5316; Sigma-Aldrich).
GST pull-down and binding assay
Tribbles constructs were amplified by PCR, ligated into pGEX 4T-1 (GE Healthcare), and expressed in BL21(DE3)pLysS cells (Stratagene). The cells were lysed by sonication in buffer A (25mM Tris 7.5, 200mM NaCl, 5mM dithiothreitol, and 5% glycerol). The soluble lysate was incubated with glutathione Sepharose beads (Amersham Biosciences) in the presence of protease inhibitors (Roche) for 3 hours at room temperature. The beads were washed with buffer B (buffer A with 1M NaCl). 35S-labeled C/EBP-α was in vitro translated (IVT) using the TnT Coupled Transcription/Translation kit (Promega), according to the manufacturer's instructions. Then, 10 μL of the IVT mix was incubated with 5 μL of glutathione S-transferase (GST)–Tribbles beads in buffer A, 1% bovine serum albumin (BSA), and 0.5% NP-40 for 1 hour at room temperature. The beads were washed 5 times with buffer B and resuspended in 20 μL sodium dodecyl sulfate (SDS) loading buffer. Next, 10 μL of each sample was loaded into a standard protein gel. The gel was stained with Coomassie Blue, then fixed in a solution of 40% water, 50% methyl alcohol (MeOH), and 10% acetic acid for 1 hour. The gel was incubated for 2-3 hours in a solution of 1M sodium salycilate in 10% glycerol, then dried and visualized by autoradiography.
32D differentiation assay
32D cells were maintained in Iscove modified Dulbecco medium containing 10% fetal bovine serum (FBS) and 10% WEHI-conditioned media. Cells were transduced with MigR1, FL-Trib2, or Trib2 mutants (GFP), and, 48 hours after transduction, 1 × 105 cells were plated in 5 ng/mL interleukin (IL)-3 or 25 ng/mL granulocyte colony-stimulating factor (G-CSF) and assessed for granulocytic differentiation by fluorescence-activated cell sorting analysis as previously described.2 For cell-cycle analysis, 1 × 105 sorted GFP+ 32D cells, transduced with MigR1, FL-Trib2, or Trib2 mutants, were plated in 5 ng/mL IL-3 or 25 ng/mL G-CSF 72 hours after sorting, fixed in ethanol, and stained with propidium iodide (P4170; Sigma-Aldrich) on days 2 and 5 of culture.
BMT
Bone marrow transplantation (BMT) experiments were performed as previously described previously.2 BM cells were collected from 6-10-week-old C57BL/6 mice 4 days after intravenous administration of 5-fluorouracil (5-FU; 5 mg) and retrovirally transduced ex vivo in the presence of IL-3, IL-6, and stem cell factor (SCF).2 Retroviral supernatants with equal titers were used to produce similar transduction efficiencies.20 Cells (0.2-1 × 106) were then injected intravenously into lethally irradiated (900 rads) B6 recipients. Chimeric mice were maintained on antibiotics for 2 weeks and assessed for engraftment 4-6 weeks after BMT, using GFP and NGFR as surrogate markers. Leukemic mice were euthanized after the development of severe cachexia and lethargy, signs that reliably predicted death from disease in several days time. Healthy controls were euthanized at the same time points. All experiments were performed in accordance with National Institutes of Health Guidelines for the Care and Use of Animals under an animal protocol approved by the University of Pennsylvania Animal Care and Use Committee.
Methylcellulose clonogenic assays
BM cells were collected from 6-week-old B6 mice 4 days after intravenous administration of 5-FU (5 mg) and retrovirally transduced (with MigR1, Trib2, dN, dC, KD, valine proline mutant (VPM), dNs, kinase domain mutant (KDM), and K177R) ex vivo in the presence of IL-3, IL-6, and SCF. Then, 25 000 unsorted cells were plated in triplicate in methylcellulose media (Methocult M3231; StemCell Technologies) supplemented with cytokines, including granulocyte-macrophage CSF (GM-CSF, 10 ng/mL; IL-3, 10 ng/mL; IL-6, 10 ng/mL; SCF, 20 ng/mL; PeproTech and BD Pharmingen). Next, 25 000 sorted GFP+ BM cells from MigR1, Trib2, dN, dC, and KD transplanted mice were plated in triplicate in methylcellulose media (Methocult M3434; StemCell Technologies). Colonies with > 50 cells were scored and assessed for 2 or 3 rounds of serial replating.
Flow cytometry
Cell suspensions were stained in phosphate-buffered saline (PBS)/2% FBS after blocking with nonspecific rat/mouse immunoglobulin G (IgG; Sigma-Aldrich). Cells were sorted on a MoFlo (Cytomation) cell sorter. Analytical flow cytometry was performed on a FACSCalibur (Becton Dickinson) and analyzed using FlowJo Version 8.8.6 software (TreeStar). Dead cells were excluded by a combination of forward and side scatter properties. The following antibodies were obtained from BD Pharmingen: phycoerythrin (PE)–anti–Gr-1 (RB6-8C5), biotin-anti-CD34 (RAM34), allophycocyanin (APC)–anti–c-Kit (2B8/CD117); and from Caltag: APC-anti–CD11b. Biotinylated anti-bodies were revealed with streptavidin–peridinin chlorophyll protein (BD Pharmingen). Biotinylated anti-NGFR antibody was produced from an established hybridoma line (HB-8737; ATCC).
Results
Trib2 mutant design and expression
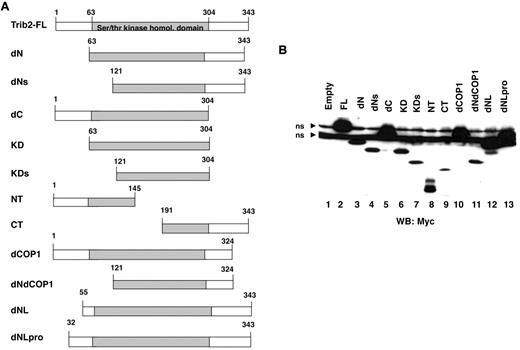
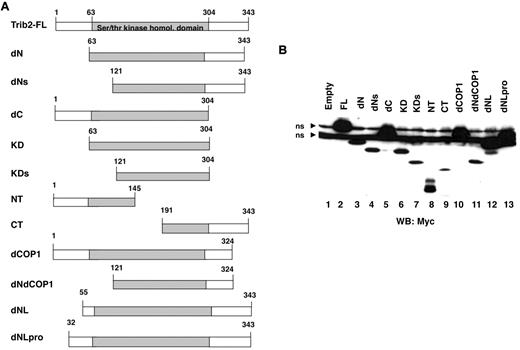
Trib2 has 3 domains, comprising distinct N- and C-termini and a central serine/threonine kinase homologous domain (aa 63-304). To map the Trib2 functional domains, we created a variety of mutants (Figure 1A). Several mutated Trib2 proteins remove all or part of the N-terminal region (dN, dNL, and dNLPro), whereas others remove all or part of the C-terminal region (dC, dCOP1). The dNdCOP1 mutant lacks both the N-terminal region and the C-terminal region beyond the COP1 site. Mutants were also made that express only the KD (KD) or portions of it (KDs). The NT and CT mutants express either the N- or C-terminal region and truncated KD sequences. Protein analysis of these mutants confirmed that proteins of the expected sizes were expressed (Figure 1B). Deletion of the N-terminus reduced protein expression levels (Figure 1B lane 3), and truncating Trib2 at aa 55 (dNL), instead of aa 63 (dN), restored the expression level of this mutant to full-length (FL) Trib2 expression levels (Figure 1B lane 12).
Trib2 mutant design and expression. (A) Schematic of the Trib2 deletion mutants with the amino acid numbers indicated. (B) Expression of Trib2 mutant proteins. 293T cells were transfected with each myc-tagged mutant, and protein lysates were assessed for protein expression by Western blot and probed with the anti-myc antibody 9E10. ns indicates nonspecific binding by the antibody.
Trib2 mutant design and expression. (A) Schematic of the Trib2 deletion mutants with the amino acid numbers indicated. (B) Expression of Trib2 mutant proteins. 293T cells were transfected with each myc-tagged mutant, and protein lysates were assessed for protein expression by Western blot and probed with the anti-myc antibody 9E10. ns indicates nonspecific binding by the antibody.
Both the KD and C-terminal domains are required for Trib2 function
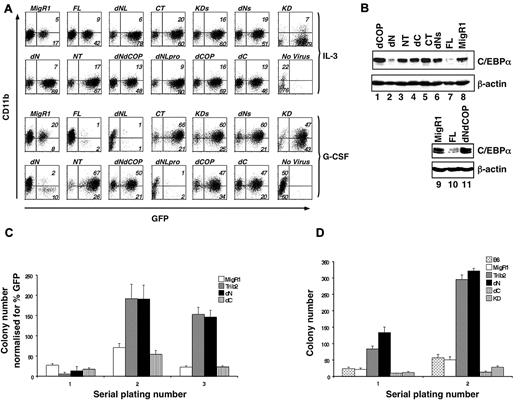
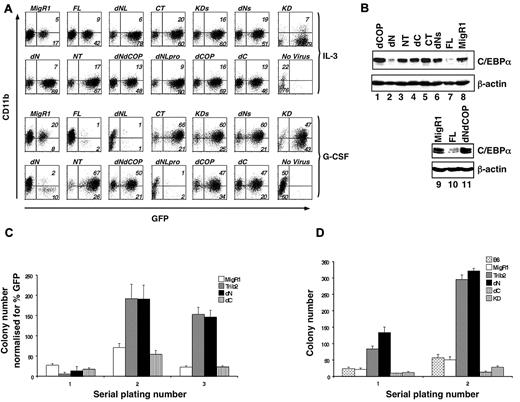
We previously showed that FL Trib2 blocks G-CSF–induced granulocytic differentiation and promotes degradation of C/EBP-α protein.2 To determine the regions of Trib2 required for these activities, we transduced the 32D myeloid cell line with retroviral supernatants expressing the various Trib2 mutant proteins. When these cells were treated with G-CSF to induce differentiation, as assessed by CD11b surface expression, FL Trib2-expressing cells failed to differentiate and demonstrated a markedly decreased percentage of GFP-expressing cells, compared with growth in IL-3 (Figure 2A).
The Trib2 N-terminus is dispensable for WT Trib2 activity. (A) Effect of Trib2 mutants on 32D cell differentiation. 32D cells were transduced with the retroviral vector (MigR1), WT Trib2 (FL), or Trib2 mutants and plated in equal numbers (1 × 105; day 0, 48 hours posttransduction) in IL-3 or G-CSF for 5 days. Transduced cells were identified by the expression of the GFP surrogate marker. CD11b expression was assessed after 5 days in culture. Data are representative of 3 independent experiments. (B) Effect of Trib2 mutants on C/EBP-α expression. Sorted GFP+ 32D cells transduced with the indicated retroviral constructs that had been cultured in IL-3 for 2 days were assessed for C/EBP-α protein expression by Western blot. β-actin is the protein loading control. (C) Serial replating ability of Trib2-transduced BM cells. 5-FU–treated BM cells transduced with the retroviral vector (MigR1) or Trib2 constructs were plated in equal number (25 000 unsorted cells in triplicate) in methylcellulose (M3231) containing IL-3, IL-6, SCF, and GM-CSF. The mean numbers of colonies normalized for GFP percentage (as a marker for transduced cells) ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments. (D) Serial replating ability of Trib2-transduced BM cells from chimeric mice. Then, 25 000 sorted GFP+ BM cells obtained from chimeric mice (MigR1, Trib2, dN, dC, and KD) 6 weeks after BMT and 25 000 control B6 total BM cells were plated in triplicate in methylcellulose (M3434, which contains SCF, IL-3, IL-6, and erythropoietin). The mean numbers of colonies ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments.
The Trib2 N-terminus is dispensable for WT Trib2 activity. (A) Effect of Trib2 mutants on 32D cell differentiation. 32D cells were transduced with the retroviral vector (MigR1), WT Trib2 (FL), or Trib2 mutants and plated in equal numbers (1 × 105; day 0, 48 hours posttransduction) in IL-3 or G-CSF for 5 days. Transduced cells were identified by the expression of the GFP surrogate marker. CD11b expression was assessed after 5 days in culture. Data are representative of 3 independent experiments. (B) Effect of Trib2 mutants on C/EBP-α expression. Sorted GFP+ 32D cells transduced with the indicated retroviral constructs that had been cultured in IL-3 for 2 days were assessed for C/EBP-α protein expression by Western blot. β-actin is the protein loading control. (C) Serial replating ability of Trib2-transduced BM cells. 5-FU–treated BM cells transduced with the retroviral vector (MigR1) or Trib2 constructs were plated in equal number (25 000 unsorted cells in triplicate) in methylcellulose (M3231) containing IL-3, IL-6, SCF, and GM-CSF. The mean numbers of colonies normalized for GFP percentage (as a marker for transduced cells) ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments. (D) Serial replating ability of Trib2-transduced BM cells from chimeric mice. Then, 25 000 sorted GFP+ BM cells obtained from chimeric mice (MigR1, Trib2, dN, dC, and KD) 6 weeks after BMT and 25 000 control B6 total BM cells were plated in triplicate in methylcellulose (M3434, which contains SCF, IL-3, IL-6, and erythropoietin). The mean numbers of colonies ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments.
When analyzed in the 32D differentiation assay, the 3 Trib2 variants with N-terminal deletions (dNLpro, dNL, and dN) functioned similarly to FL Trib2, and few GFP+ cells remained by day 5 in G-CSF (Figure 2A). In contrast, expression of mutants with Trib2 C-terminal deletions (dC and KD) abolished this activity as did a targeted deletion of the C-terminal 19 aa that includes the COP1-binding site (dCOP, aa 1-324; Figure 2A). The presence of the COP1 site was insufficient to maintain Trib2 function, as activity was lost in a construct that retained the COP1-binding site, but had part of the KD removed (dNs, aa 121-343; Figure 2A).
We next analyzed the ability of many of these mutants to degrade C/EBP-α, a critical functional feature of FL Trib2 (Figure 2B). Among the constructs tested, the only mutant that retained wild-type (WT) Trib2 activity and degraded C/EBP-α was the dN mutant (Figure 2B lane 2). This mutant retained both the entire KD and the COP1-binding site. Deletion of the 19 aa that include the COP1-binding site was sufficient to abrogate C/EBP-α degradation by Trib2 (Figure 2B lane 1). Thus, both the KD and C-terminal region containing the COP1-binding site were required for Trib2 to block differentiation and to degrade C/EBP-α.
FL Trib2 expression in primary BM cells allows cells to be serially replated in clonogenic methylcellulose assays.2 We assessed the ability of dN and dC mutants to serially replate primary BM cells (Figure 2C). Primary colony plates using unsorted BM did not reveal differences between control MigR1 and Trib2-transduced cells, presumably due to the 5-FU treatment and low transduction efficiencies (ie, colony number normalized to transduction efficiencies). However, in the selective environment of serial replating, FL Trib2 greatly increased the number of secondary and tertiary colonies. The dN mutant could similarly promote serial replating of the BM cells, whereas the dC mutant failed to function in this assay. Similarly, when GFP+ BM cells were sorted from mice 6 weeks after BMT with cells expressing either Trib2 FL, dN, dC, or KD, it was evident that dN retained Trib2 activity, whereas dC- and KD-expressing cells were unable to increase the number of colonies on primary plates and lost the ability to serially replate (Figure 2D). Together, these data demonstrate that the N-terminus of Trib2 is not required for differentiation inhibition, serial replating, or C/EBP-α degradation. Although the KD alone did not block 32D cell differentiation or promote serial replating, deletion into the KD (dNs) abrogated Trib2 function, indicating that an intact kinase domain is required. Thus, in these Trib2 functional assays, both an intact KD and COP1-binding site were necessary for activity.
Identification of aa in the COP1-binding site and kinase-like domain of Trib2 that are required for activity
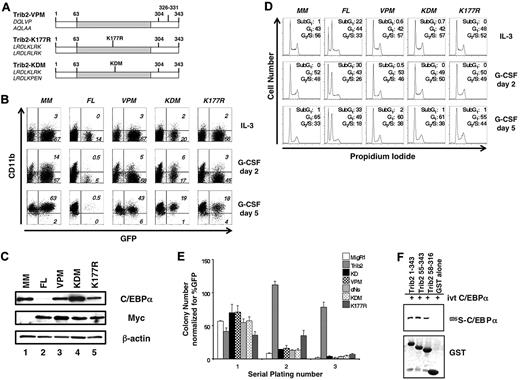
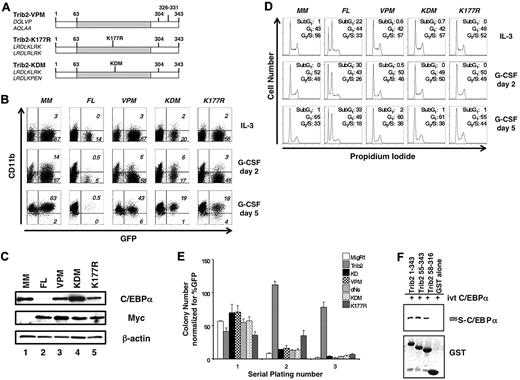
Next, we tested the importance of specific residues implicated in COP1 binding for various functional activities of Trib2 (Figure 3A), using a protein with DQLVP to AQLAA substitutions at residues 326-331 (designated VPM). These residues are required for the binding of COP1 to Trib312 ; thus, this mutant should reveal whether the effects of the C-terminal deletion (dCOP) were due to COP1 binding or resulted from other, as yet, unidentified functions encoded by the C-terminal 18 aa of the protein (aa 325-343). In addition, we mutated residues in the KD analogous to a key catalytic motif of enzymatically active kinases to further explore how the KD participates in Trib2 activity (Figure 3A). The sequence in the Trib2 KD, “LRDLKLRK,” is unlike the characteristic sequence of a typical ser/thr kinase catalytic loop, “HRDLKPEN.”13 The first K is the critical lysine residue that defines ser/thr kinases and is retained in Trib2. We mutated this lysine to arginine that specifies tyrosine kinases (K177R; Figure 3A).14 In addition, we changed the catalytic motif to resemble the canonical ser/thr motif (KDM mutant with the sequence LRDLKPEN; Figure 3A).
Both the Trib2 KD- and COP1-binding sites are required for Trib2 activity. (A) Schematic of Trib2 point mutants. Left, amino acid sequences show original sequence on top and mutated sequence on bottom. (B) Effect of Trib2 mutants on 32D cell differentiation. 32D cells transduced with the vector control (MigR1-myc; MM), WT Trib2 (FL) or the Trib2 mutants were plated in equal numbers (1 × 105; day 0, 48 hours posttransduction) in IL-3 or G-CSF. Transduced cells were identified by the expression of the GFP surrogate marker. CD11b expression was assessed after 2 and 5 days. Data are representative of 3 independent experiments. (C) Effect of Trib2 mutants on C/EBP-α expression. 32D cells were transduced with the indicated retroviral constructs, sorted for GFP 48 hours after transduction, and assessed for C/EBP-α, myc-tagged FL Trib2, and myc-tagged Trib2 mutant (VPM, KDM, and K177R) protein expression by Western blot, 3 days after sorting. MigR1-Myc is the empty vector (MM). β-actin is the protein loading control. (D) Effect of Trib2 mutants on 32D cell cycle. 32D cells were transduced with the vector control (MigR1-myc; MM), WT Trib2 (FL), or the Trib2 mutants, sorted for GFP 48 hours later, and 1 × 105 GFP+ cells were plated in 5 ng/mL IL-3 or 25 ng/mL G-CSF 3 days after sorting. Cell cycle was assessed by propidium iodide staining on days 2 and 5. (E) Effect of Trib2 mutants on serial replating by transduced BM. 5-FU–treated BM cells transduced with the MigR1 retroviral vector, WT Trib2, or the indicated Trib2 mutants were plated in equal number (25 000 unsorted cells in triplicate) in methylcellulose (M3231), containing IL-3, IL-6, SCF, and GM-CSF. Colonies with > 50 cells were scored and assessed over 3 rounds of serial replating. The mean numbers of colonies normalized for GFP percentage, (as a marker for transduced cells) ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments. (F) Interaction of Trib2 mutants and C/EBP-α. GST pull-down of Trib2 constructs incubated with IVT 35S C/EBP-α. Top panel: autoradiograph of 35S C/EBP-α. Bottom panel shows Coomassie gel of GST proteins.
Both the Trib2 KD- and COP1-binding sites are required for Trib2 activity. (A) Schematic of Trib2 point mutants. Left, amino acid sequences show original sequence on top and mutated sequence on bottom. (B) Effect of Trib2 mutants on 32D cell differentiation. 32D cells transduced with the vector control (MigR1-myc; MM), WT Trib2 (FL) or the Trib2 mutants were plated in equal numbers (1 × 105; day 0, 48 hours posttransduction) in IL-3 or G-CSF. Transduced cells were identified by the expression of the GFP surrogate marker. CD11b expression was assessed after 2 and 5 days. Data are representative of 3 independent experiments. (C) Effect of Trib2 mutants on C/EBP-α expression. 32D cells were transduced with the indicated retroviral constructs, sorted for GFP 48 hours after transduction, and assessed for C/EBP-α, myc-tagged FL Trib2, and myc-tagged Trib2 mutant (VPM, KDM, and K177R) protein expression by Western blot, 3 days after sorting. MigR1-Myc is the empty vector (MM). β-actin is the protein loading control. (D) Effect of Trib2 mutants on 32D cell cycle. 32D cells were transduced with the vector control (MigR1-myc; MM), WT Trib2 (FL), or the Trib2 mutants, sorted for GFP 48 hours later, and 1 × 105 GFP+ cells were plated in 5 ng/mL IL-3 or 25 ng/mL G-CSF 3 days after sorting. Cell cycle was assessed by propidium iodide staining on days 2 and 5. (E) Effect of Trib2 mutants on serial replating by transduced BM. 5-FU–treated BM cells transduced with the MigR1 retroviral vector, WT Trib2, or the indicated Trib2 mutants were plated in equal number (25 000 unsorted cells in triplicate) in methylcellulose (M3231), containing IL-3, IL-6, SCF, and GM-CSF. Colonies with > 50 cells were scored and assessed over 3 rounds of serial replating. The mean numbers of colonies normalized for GFP percentage, (as a marker for transduced cells) ± SEM are shown. Data are representative of triplicate cultures from 2 independent experiments. (F) Interaction of Trib2 mutants and C/EBP-α. GST pull-down of Trib2 constructs incubated with IVT 35S C/EBP-α. Top panel: autoradiograph of 35S C/EBP-α. Bottom panel shows Coomassie gel of GST proteins.
We assessed the effect of these point mutants on granulocytic differentiation in the 32D assay. Both the VPM and KDM mutants failed to block granulocytic differentiation in the 32D assay (Figure 3B), indicating a critical role for the COP1-binding site and the KD in Trib2 activity. In contrast, K177R-transduced cells exhibited a decrease in the percentage of GFP+ cells between days 2 and 5 (Figure 3B). Neither the VPM, KDM, or K177R mutants decreased C/EBP-α expression to the extent of FL Trib2 (Figure 3C and data not shown).
As the percentage of GFP+ 32D cells expressing either FL Trib2 or the K177R point mutant decreased at day 5 of G-CSF treatment, we assessed the cell-cycle profile of 32D cells expressing our series of point mutants. 32D cells transduced with the empty vector, VPM, KDM, or K177R remained viable under both IL-3 and G-CSF culture conditions and exhibited a progressive increase in the fraction of cells in the G1 phase of the cell cycle throughout culture in G-CSF. In contrast, FL Trib2-expressing cells demonstrated an increase in cell death at the expense of cells in the G2/S-phase of the cell cycle, with this effect being more pronounced under differentiating conditions (Figure 3D). Although the K177R mutant did not promote apoptosis, the percentage of GFP+ cells decreased between days 2 and 5, suggesting that this mutant influenced the kinetics of cell growth.
To further explore the roles of the COP1-binding site and KD in transformation, we performed serial replating colony assays with our extended series of Trib2 mutants (KD, dNs, VPM, KDM, and K177R). 5-FU–treated BM cells were transduced with titer-matched retroviral supernatants, and unsorted populations were plated in methylcellulose. Although colony numbers were similar on the primary plates, only the FL Trib2-expressing cells demonstrated enhanced colony-forming ability on the secondary plating (Figure 3E). Unlike the other mutants, which showed a significant decrease in colony number by the secondary plating, the K177R-expressing colonies did not demonstrate a significant decline in cell number. However, by the tertiary replating, the K177R mutant cells (similar to the other Trib2 mutants) were unable to form colonies (a 2-log decrease, compared with FL Trib2). Together, these data demonstrate that the Trib catalytic core motif and COP1-binding site are required for WT Trib2 activity. Furthermore, the ability to efficiently degrade C/EBP-α correlated with the ability of Trib2 to block differentiation and serially replate.
We previously showed that Trib2 forms a complex with C/EBP-α in 293T cells.2 To determine whether this interaction would be direct, we incubated IVT 35S-labeled C/EBP-α with purified GST-Trib2 protein, GST-dNL (aa 55-343) or a mutant lacking the N-terminus and COP1-binding site (aa 58-316; Figure 3F). Both FL Trib2 (aa 1-343) and each of the mutants bound to IVT C/EBP-α (Figure 3F). Thus, the ability of Trib2 to bind C/EBP-α is not itself sufficient to promote C/EBP-α degradation.
Trib2 induces C/EBP-α degradation via COP1
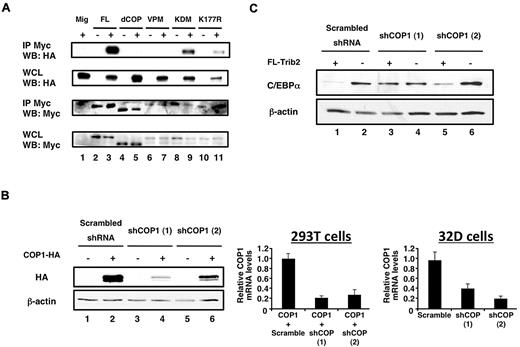
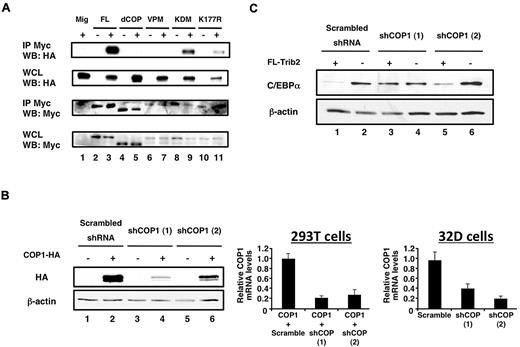
Our data suggest that an intact COP1-binding site is necessary for Trib2 to degrade C/EBP-α. C/EBP-α degradation correlated with the ability of COP1 to bind FL Trib2, as neither dCOP nor the VPM point-mutant–bound COP1 (Figure 4A lanes 3, 5, and 7). Both the KDM and K177R mutants bound COP1; however, it was not possible to quantify whether there would be a qualitative difference in COP1 binding for the K177R or KDM mutants, because the steady-state levels of these proteins are consistently lower under these assay conditions (Figure 4A lanes 3, 9, and 11).
The Trib2 COP1-binding site and COP1 are required for C/EBP-α degradation. (A) Interaction of Trib2 and COP1. 293T cells were transfected with COP1-HA (lane 1), myc-tagged Trib2-FL (lane 2), COP1-HA and myc-tagged Trib2-FL (lane 3), myc-tagged dCOP (lane 4), COP1-HA and myc-tagged dCOP (lane 5), myc-tagged VPM (lane 6), COP1-HA and myc-tagged VPM (lane 7), myc-tagged KDM (lane 8), COP1-HA and myc-tagged KDM (lane 9), myc-tagged K177R (lane 10), or COP1-HA and myc-tagged K177R (lane 11). Proteins were immunoprecipitated using the Myc 9E10 antibody, and Western blotting was performed with HA (top panels) and Myc (bottom panels) antibodies on immunoprecipitates (IP), and on whole cell lysates (WCLs). (B) COP1 knockdown decreases COP1 expression. Left panel, 293T cells were transfected with scrambled shRNA, or 1 of 2 different shCOP1 plasmids alone or together with COP1-HA. Western blotting was performed with HA antibody to detect COP1 expression. β-actin served as the loading control. Middle panel, 293T cells were cotransfected with COP1-HA and either the scrambled shRNA or each of the shCOP1 plasmids and analyzed by real-time RT-PCR for COP1 expression. Error bars denote SEM of each sample measured in triplicate. Right panel, sorted GFP+ 32D cells transduced with either shCOP1 or scrambled shRNA-expressing retrovirus were analyzed for endogenous COP1 knockdown by real-time RT-PCR. Error bars denote SEM of each sample measured in triplicate. (C) COP1 knockdown impairs the effects of Trib2 on C/EBP-α expression. Sorted GFP+ tNGFR+ 32D cells transduced with the indicated retroviral constructs were assessed for C/EBP-α protein expression by Western blot. The shRNA constructs were expressed from the low-molecular-mass polypeptide retroviral vector, which expresses GFP as a surrogate marker, whereas Trib2 was expressed from a version of MigR1 that expresses truncated NGFR as a surrogate marker.37 The cells that were not transduced with FL-Trib2 were transduced with the empty NGFR vector (lanes 2, 4, and 6). β-Actin was the protein loading control.
The Trib2 COP1-binding site and COP1 are required for C/EBP-α degradation. (A) Interaction of Trib2 and COP1. 293T cells were transfected with COP1-HA (lane 1), myc-tagged Trib2-FL (lane 2), COP1-HA and myc-tagged Trib2-FL (lane 3), myc-tagged dCOP (lane 4), COP1-HA and myc-tagged dCOP (lane 5), myc-tagged VPM (lane 6), COP1-HA and myc-tagged VPM (lane 7), myc-tagged KDM (lane 8), COP1-HA and myc-tagged KDM (lane 9), myc-tagged K177R (lane 10), or COP1-HA and myc-tagged K177R (lane 11). Proteins were immunoprecipitated using the Myc 9E10 antibody, and Western blotting was performed with HA (top panels) and Myc (bottom panels) antibodies on immunoprecipitates (IP), and on whole cell lysates (WCLs). (B) COP1 knockdown decreases COP1 expression. Left panel, 293T cells were transfected with scrambled shRNA, or 1 of 2 different shCOP1 plasmids alone or together with COP1-HA. Western blotting was performed with HA antibody to detect COP1 expression. β-actin served as the loading control. Middle panel, 293T cells were cotransfected with COP1-HA and either the scrambled shRNA or each of the shCOP1 plasmids and analyzed by real-time RT-PCR for COP1 expression. Error bars denote SEM of each sample measured in triplicate. Right panel, sorted GFP+ 32D cells transduced with either shCOP1 or scrambled shRNA-expressing retrovirus were analyzed for endogenous COP1 knockdown by real-time RT-PCR. Error bars denote SEM of each sample measured in triplicate. (C) COP1 knockdown impairs the effects of Trib2 on C/EBP-α expression. Sorted GFP+ tNGFR+ 32D cells transduced with the indicated retroviral constructs were assessed for C/EBP-α protein expression by Western blot. The shRNA constructs were expressed from the low-molecular-mass polypeptide retroviral vector, which expresses GFP as a surrogate marker, whereas Trib2 was expressed from a version of MigR1 that expresses truncated NGFR as a surrogate marker.37 The cells that were not transduced with FL-Trib2 were transduced with the empty NGFR vector (lanes 2, 4, and 6). β-Actin was the protein loading control.
To determine whether COP1 would be necessary to degrade C/EBP-α, we performed COP1 knockdown experiments. Our 2 different retroviral shCOP1 constructs decreased the expression of exogenously expressed COP1 protein and mRNA in 293T cells and endogenous COP1 mRNA in 32D cells (Figure 4B). When Trib2 and shCOP1 were coexpressed in 32D cells, C/EBP-α degradation by Trib2 was impaired, in comparison to cells expressing a scrambled shRNA (Figure 4C lanes 1, 3, and 5). shCOP1 (1) was a more potent inhibitor of Trib2-induced C/EBP-α degradation than shCOP1 (2), consistent with the effect on exogenous COP1 protein in transfected 293T cells. Although shCOP1 (2) decreased COP1 mRNA to a slightly greater extent than shCOP1 (1) in transduced 32D cells, it is possible that posttranscriptional activities of this shRNA caused it to have a milder effect on endogenous COP1 protein in 32D cells. Together, these data demonstrate that COP1 is an important mediator of Trib2-dependent C/EBP-α degradation.
Trib2 leukemogenic activity is retained in the dN mutant, but lost in the absence of the COP1-binding site
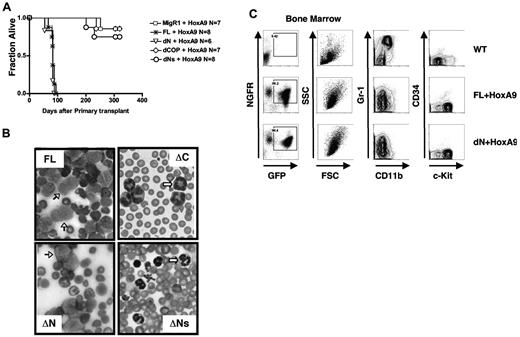
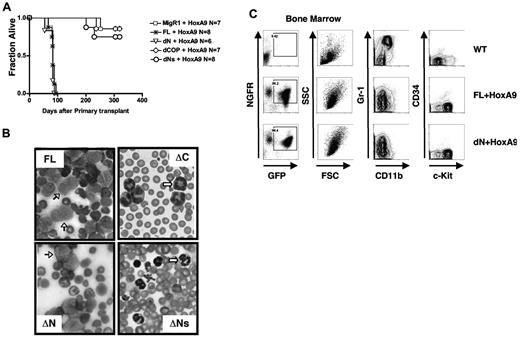
Trib2 is an oncogene that causes AML in the murine BMT model, with a latency of ∼ 175 days.2,21 HoxA9 also induces AML in the murine BMT model, with a latency of ∼ 170 days; however, the penetrance of acute leukemia can be incomplete.22 Disease latency is greatly decreased when HoxA9 and Trib2 are coexpressed (83 ± 8 days).21 To assay the leukemogenic potential of the Trib mutants in a context that would strongly select for leukemia, we performed the in vivo BMT leukemia assay by coexpressing the Trib2 mutants and HoxA9. In order to readily identify transduced cells, Trib2 was introduced into murine hematopoietic stem cells on a retroviral vector that expressed GFP as a surrogate marker and HoxA9 was coexpressed with tNGFR as a surrogate marker. Mice transplanted with cells expressing dN developed AML, with similar latencies to FL Trib2 (79 days ± 12 days) with 100% penetrance (Figure 5A). Mice transplanted with cells expressing dCOP or dNs were unable to accelerate and/or increase the penetrance of HoxA9-induced AML. All mice that developed leukemia had AML with phenotypes similar to those previously described,2,21 consisting of splenomegaly, high white blood cell counts with circulating blasts, hypercellular BM with excess blasts, lymphadenopathy, and hepatic infiltration (Figure 5B and data not shown). The majority of BM cells from FL/HoxA9 and dN/HoxA9 leukemic mice were GFP+tNGFR+, indicating expression of both HoxA9 and Trib2 (Figure 5C left panels). The immunophenotypes of the FL and dN-Trib2-induced AML were similar (Gr-1low/intermediateCD11blow/intermediateCD34neg/lowc-Kithigh; Figure 5C middle and right panels).
The COP1-binding site and intact KD are necessary for the leukemogenic activity of Trib2. (A) Kaplan-Meier survival curve of mice receiving Trib2-FL, dN, dCOP, dNs, or MigR1 cotransduced with HoxA9 BM. The median survival of FL and dN mice was 83 and 79 days, respectively. (B) Wright-Giemsa–stained peripheral blood smears from mice receiving full-length Trib2 (FL, top left) and the ΔC (top right), ΔNs (bottom right), and ΔN mice (bottom left). Several leukemic blasts are indicated by the black arrows, and several metamyelocytes are shown by the white arrows. (C) Flow cytometric analysis of BM cells from C57BL/6 mice receiving HoxA9 and/or Trib2 transduced cells. Left panels, analysis of GFP (Trib2 mutants) and HoxA9-NGFR expression (percentages given) in cells obtained from BM of leukemic FL+HoxA9 and dN+HoxA9 mice, compared with C57BL/6 control mice. Flow cytometric analysis of Gr-1 and CD11b expression (middle panels) and CD34 and c-Kit expression (right panels) in the GFP+NGFR+ fractions. Representative plots are shown.
The COP1-binding site and intact KD are necessary for the leukemogenic activity of Trib2. (A) Kaplan-Meier survival curve of mice receiving Trib2-FL, dN, dCOP, dNs, or MigR1 cotransduced with HoxA9 BM. The median survival of FL and dN mice was 83 and 79 days, respectively. (B) Wright-Giemsa–stained peripheral blood smears from mice receiving full-length Trib2 (FL, top left) and the ΔC (top right), ΔNs (bottom right), and ΔN mice (bottom left). Several leukemic blasts are indicated by the black arrows, and several metamyelocytes are shown by the white arrows. (C) Flow cytometric analysis of BM cells from C57BL/6 mice receiving HoxA9 and/or Trib2 transduced cells. Left panels, analysis of GFP (Trib2 mutants) and HoxA9-NGFR expression (percentages given) in cells obtained from BM of leukemic FL+HoxA9 and dN+HoxA9 mice, compared with C57BL/6 control mice. Flow cytometric analysis of Gr-1 and CD11b expression (middle panels) and CD34 and c-Kit expression (right panels) in the GFP+NGFR+ fractions. Representative plots are shown.
Similar to previously published data, we did not observe 100% penetrance in the MigR1+HoxA9 control mice cohort (1 of 7 mice developed AML), which likely reflects the level of HoxA9 expression in the recipient mice.22,23 Similar to MigR1, only 1 of 8 dNs and 2 of 7 dCOP chimeric mice developed AML over the 1-year observation period. The AML latency in these cohorts was nearly twice as long as that in the FL Trib2/HoxA9 or the dN Trib2/HoxA9 mice; however, the general characteristics of the leukemias were similar to the HoxA9 AML (data not shown; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).21 All transductions were performed using an identical lot of HoxA9 retroviral supernatant and titer-matched Trib2 retroviral supernatants into donor cells derived from pooled BM cells from 5-FU–treated mice. In addition, GFP+tNGFR+ cells were readily identified in the peripheral blood of all recipient mice at 6-8 weeks after BMT. Moreover, both the percentage and number of circulating GFP+tNGFR+ cells were similar when either FL Trib2 or Trib2 mutants were used (supplemental Figure 2). Thus, the differences in disease penetrance and latency are likely to reflect differences in the oncogenic potential of the Trib mutants.
Discussion
In this report, we show that the Trib2 KD and COP1-binding region are required for inhibition of myeloid differentiation and induction of transformation. In contrast, the Trib2 N-terminus is not essential for these activities. Trib2 forms complexes with both COP1 and C/EBP-α, leading to C/EBP-α degradation. In the absence of an intact KD or COP1-binding site, Trib2 failed to block granulocytic differentiation, did not promote serial replating in methylcellulose clonogenic assays, and did not cause AML.
All of the Trib2 gain-of-function activities correlate with efficient C/EBP-α degradation, suggesting that this is an excellent marker to assess Trib2 oncogenic activity. Loss of C/EBP-α function is associated with myeloid transformation in a variety of murine models and human leukemias.24-26 Furthermore, we previously showed that Trib2-induced cell death can be prevented by exogenous C/EBP-α expression,2 and that the expression of a C/EBP-α DNA-binding mutant with dominant-negative activity in 32D cells results in a similar increase in cell death under differentiating conditions.27 Together, these data provide strong support for the hypothesis that C/EBP-α degradation is an important function of Trib2 oncogenicity.
While we have identified an important role for Trib2 in C/EBP-α degradation, which correlates with myeloid transfor-mation, apoptosis, and inhibition of cell proliferation, other functions of Trib2 may be involved. Previous studies identified roles for Trib proteins in MAPK activation,4,10 AKT function,11,28 and C/EBP-β activity.28,29 We investigated these particular activities in Trib2-transduced 32D cells. Despite reports that Trib proteins regulate MAPK signaling, we observed no differences in phopho-p44/42 MAPK activity in FL Trib2 expressing 32D cells (supplemental Figure 4A), a result that is similar to our published data showing that neither Trib1, Trib2, nor Trib3 differentially influences MAPK activation in hematopoietic cells.30 In contrast, Trib2 expression decreased the amount of C/EBP-β and phospho-AKT protein (supplemental Figure 4B-C). Thus, it is possible that our observed phenotypes are due to inputs from both C/EBP-α and C/EBP-β, and reinforces the findings that Tribbles proteins modulate multiple C/EBP family members. Although the mechanism by which Trib2 modulates AKT activity is unknown, previous work suggests that AKT is an important downstream mediator of IL-331 and G-CSF32 survival signals in 32D cells. Thus, the ability of FL Trib2 to impair AKT signaling in 32D cells may explain the observed increase in cell death and decrease in proliferation in FL Trib2-expressing cells, as reduced AKT signaling would make these cells refractory to survival factor stimulation. In addition to these pathways, multiple other pathways have been associated with Trib proteins; these include FOXO,33 nuclear factor-κB,8 and Mcl1.18 These pathways are known regulators of cell survival and proliferation and may also contribute to the observed phenotypes. The extent to which each of these activities influences Trib2 transformation would certainly be of future interest.
The 32D studies suggest that Trib2 exerts strong proapoptotic and antiproliferative functions on the cells in differentiating conditions; however, Trib2 induced significant apoptosis even in IL-3 conditions. These findings differ from the effect of expressing a dominant-negative C/EBP-α mutant in 32D cells, which blocked differentiation and promoted death in G-CSF conditions, but did not induce significant apoptosis in IL-3 conditions.27 Although these data suggest that the effect of Trib2 in 32D cells is not solely dependent on C/EBP-α, the extent to which these other Trib-induced functions influence myeloid transformation remains to be determined.
Although we cannot rule out that other activities of Trib2 are required for myeloid transformation, degradation of C/EBP-α appears to be particularly important. Indeed, in a separate study assessing Trib1 and Trib3 family members, we found that oncogenic activity correlated with C/EBP-α degradation, as Trib1, but not Trib3, efficiently degraded C/EBP-α and only the former induced AML in the murine BMT model.30 These data support both a correlative and causal relationship between Trib2 and C/EBP-α degradation in myeloid transformation.
We previously found that Trib2 and HoxA9 synergize to accelerate the onset of AML in the murine BMT model.21 We used the HoxA9 cooperativity model to assay the oncogenic activity of the Trib2 mutants, as it provides more sensitivity to identify weak leukemogenic activity than the induction of de novo leukemia. When coexpressed with HoxA9, both Trib2 FL and dN increased AML penetrance to 100% and decreased disease latency by half. In contrast, neither dCOP nor dNs enhanced the leukemogenic activity of HoxA9. The failure of the dCOP and dNs mutants to function in any of our multiple assays of Trib2 differentiation and transformation further highlights the importance of the Trib2 KD and COP1-binding site. Although data from our group and others show that Trib2 can induce apoptosis in cell lines,34 Trib2 clearly functions as an oncogene in myeloid transformation, as demonstrated in both our colony and BMT assays, by inducing gain-of-function activities, such as C/EBP-α degradation. This illustrates a limitation of cell-line assays, where the effects of Tribbles on survival and differentiation may obscure its transforming effects.
Although the Trib KD does not function as a conventional kinase,15-18 our results reveal an essential role for this domain in Trib2 oncogenic functions. Our data also suggest that the importance of specific residues in the KD may vary, depending on context. In Drosophila, for example, mutation of the lysine residue in the KD catalytic loop (K266R) behaved similarly to WT dTrib to degrade string and induce a premature pause in the embryonic cell cycle.16 In a separate study in myeloid BAF3 and TF-1 cells, mutation of the analagous lysine at aa 177 to either alanine (K177A) or arginine (K177R) had no effect on Trib apoptotic functions.18 In contrast, mutation of the catalytic loop (K177R and KDM) in our experiments interfered with Trib2 activity. However, the K177R mutant did exhibit weak activity in C/EBP-α degradation, inhibition of AKT activation, 32D differentiation, and the BM colony assays. It is possible that the assays used in our studies are more sensitive to mild defects in function than other assays. Alternatively, the importance of these particular residues may vary with the nature of the Trib degradation target. Although some of the details of Trib function remain to be elucidated, our data clearly show that the functions of the Trib2 KD are essential for myeloid transformation.
Trib2 is classified as a pseudokinase; however, these functions are poorly understood. It is possible that Trib2 is a kinase whose substrates have yet to be identified. Alternatively, Trib2 may function as a decoy kinase that binds substrates and prevents other kinases from binding and phosphorylating their targets. A third possibility is that Trib2 functions as an adapter that binds multiple proteins and brings them together. Although our data do not rule out Trib2 functioning as a kinase or decoy molecule, we favor a model in which Trib2 functions as an adapter that binds both the COP1 E3 ligase and the substrate C/EBP-α resulting in C/EBP-α degradation. In this model, Trib2 would not only bind both the enzyme and the substrate, but would also provide the proper configuration for degradation to occur. Nevertheless, it remains to be determined whether C/EBP-α binding by Trib2 is essential for its degradation.
COP1 is an E3 ubiquitin ligase that functions alone or in a protein complex to promote substrate-specific ubiquitination and degradation. Several mammalian COP1 substrates have been described. COP1 E3 ligase activity is sufficient to degrade p53,35 whereas COP1 inhibits c-Jun independently of degradation.36 Relevant to our studies, binding of COP1 to Trib3 stimulated lipolysis in adipose tissue by degrading acetyl-coenzyme A carboxylase (ACC), an important regulator of fatty acid synthesis.12 Thus, both Trib2 and Trib3 promote protein degradation. It appears that all 3 mammalian Tribs bind COP1 as the binding site is conserved; however, it is not known if they share the same target substrates for delivery to COP1.
Our studies also show that the COP1 binding is important for Trib2-mediated C/EBP-α degradation. Not only must the binding site on Trib2 be intact, but our knockdown experiments suggest that COP1 is important for C/EBP-α degradation in the cell types tested. These data also demonstrate that COP1 binding is crucial for Trib2-mediated transformation, where it is likely to function as an E3 ligase, and suggest that blocking the Trib2:COP1 interaction would inhibit Trib2-transforming functions.
To summarize, we have provided a detailed analysis of the Trib2 oncogene and demonstrated how the various domains and key residues within those domains contribute to leukemic transformation. Our data show that Trib2 forms complexes with both the substrate, C/EBP-α, and enzyme, COP1, suggesting that it may function as an adaptor for protein degradation. Both the KD- and COP1-binding sites are required for Trib2-induced myeloid differentiation and transformation, whereas the N-terminus is nonessential for these activities. As Trib2 oncogenic activity requires binding both C/EBP-α and COP1, disrupting either of these interactions with molecules that block protein:protein interactions may be therapeutically beneficial.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank members of the Pear Lab for their important contributions to these studies. We are also grateful to the UPenn Stemmler and BRB Animal Facilities, UPenn Abramson Cancer Center Flow Cytometry, and AFCRI cores.
This work was supported by grants from the Science Foundation Ireland (to K.K.; PIYRA) and National Institutes of Health (NIH CA093615; to W.S.P.) and pilot project funds from the Abramson Cancer Center at the University of Pennsylvania NCI Cancer Center Support Grant CA-016520 (to W.S.P.). Individual support was provided by a Leukemia & Lymphoma Society Special Fellow Award (to K.K.), NIH T32 (T32CA009140; to P.H.D.), and an NIH-funded Penn Prep Award (R25GM071745) to M.E.V.
National Institutes of Health
Authorship
Contribution: K.K., W.B., P.H.D., M.E.V., O.S., and S.U. performed experiments; K.K., W.B., and S.U. made the figures; and K.K., W.B., S.C.B., and W.S.P. designed research, analyzed data, and wrote the mansucript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K.K. is Department of Biochemistry, BioSciences Institute, University College Cork, Cork, Ireland.
Correspondence: Warren S. Pear, Department of Pathology and Laboratory Medicine, Abramson Family Cancer Research Institute, University of Pennsylvania, Philadelphia, PA 19104; e-mail: wpear@mail.med.upenn.edu; or Karen Keeshan, RM1.08 Biosciences Institute, Department of Biochemistry, University College Cork, Cork, Ireland; e-mail: k.keeshan@ucc.ie.
References
Author notes
K.K. and W.B. contributed equally to this study.