Abstract
The Notch pathway is frequently activated in T-cell acute lymphoblastic leukemias (T-ALLs). Of the Notch receptors, Notch1 is a recurrent target of gain-of-function mutations and Notch3 is expressed in all T-ALLs, but it is currently unclear how these receptors contribute to T-cell transformation in vivo. We investigated the role of Notch1 and Notch3 in T-ALL progression by a genetic approach, in mice bearing a knockdown mutation in the Ikaros gene that spontaneously develop Notch-dependent T-ALL. While deletion of Notch3 has little effect, T cell–specific deletion of floxed Notch1 promoter/exon 1 sequences significantly accelerates leukemogenesis. Notch1-deleted tumors lack surface Notch1 but express γ-secretase–cleaved intracellular Notch1 proteins. In addition, these tumors accumulate high levels of truncated Notch1 transcripts that are caused by aberrant transcription from cryptic initiation sites in the 3′ part of the gene. Deletion of the floxed sequences directly reprograms the Notch1 locus to begin transcription from these 3′ promoters and is accompanied by an epigenetic reorganization of the Notch1 locus that is consistent with transcriptional activation. Further, spontaneous deletion of 5′ Notch1 sequences occurs in approximately 75% of Ikaros-deficient T-ALLs. These results reveal a novel mechanism for the oncogenic activation of the Notch1 gene after deletion of its main promoter.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a heterogeneous disease that is characterized by multiple subtypes. T-ALL affects both children and adults and results from the accumulation of blasts blocked at specific stages of T-cell differentiation. Treatment involves long-term and intensive combination chemotherapy, which are associated with severe side effects. Relapse is frequent in adult T-ALL with very unfavorable outcome. Less than 60% of patients survive longer than 5 years.1
T-ALL is associated with recurrent genetic and epigenetic abnormalities. Chief among them is the activation of the Notch pathway.2 The oncogenic effects of the Notch pathway are probably linked to its pleiotropic influence on T lymphocytes. Notch receptors are expressed on the surface of T cells, and upon binding to ligands expressed by neighboring cells, the heterodimeric receptor is cleaved by metalloproteases (at the S2 site) and γ-secretase (at the S3 site) to release an activated, intracellular form of Notch (ICN). Cleaved Notch then translocates to the nucleus where it binds to its downstream transcriptional mediator, recombination signal binding protein for immunoglobulin kappa J region (RBP-J), and coactivators to activate the transcription of Notch-dependent target genes.3-6
Of the 4 Notch receptors (Notch1-4), Notch1 appears to be the most important in human T-ALL. This was first demonstrated by the finding of a rare translocation in approximately 1% of T-ALL that fuses the sequences encoding the C-terminal portion of Notch1 downstream of the promoter of the β chain of the T-cell receptor (TCR), resulting in the expression of a constitutively active intracellular Notch1 protein that acts as a powerful oncogene.7 More recently, Aster and colleagues8 found that the Notch1 gene contains point mutations or deletions in approximately 70% of T-ALL cases, which are situated in the heterodimerization domain and the 3′ PEST domain. These mutations lead to increased cleavage of Notch1 as well as stabilization of the intracellular protein. Collectively, these studies suggest that Notch1 is the crucial receptor for oncogenic mutations. On the other hand, Notch3 mRNA expression is up-regulated in all T-ALL cases studied, also suggesting a role for this receptor in T-ALL development.9,10
A variety of murine T-ALL models have addressed the oncogenic role of Notch proteins carrying different mutations or deletions, usually by overexpressing them in retroviral systems or by transgenesis.11-15 These have provided valuable information about the potential of activated proteins to induce transformation, but they also come with caveats, as overexpression studies do not always resemble the physiologic disease. For example, overexpression of intracellular Notch1-3 are equally capable of inducing leukemia in the retroviral bone marrow transfer model,16 yet Notch2 does not appear to be affected in human T-ALL. Thus, the individual roles played by the specific Notch receptors in T-ALL progression remain poorly understood.
We recently described a spontaneous model of T-ALL in mice bearing a knockdown mutation of the Ikaros gene (IkL/L).17 These mice develop clonal T-cell lymphomas in the thymus with an early and reproducible time of onset, between 10-12 weeks of age, and they die at a median age of 18 weeks. Disease incidence is 100%. The tumor phenotype is predictable and shows an accumulation of blasts at the CD4+CD8+ (DP) and CD4−CD8+ (CD8 SP) stage of differentiation; αβ TCR/CD3 expression levels vary from low to intermediate. Interestingly, all tumors show high up-regulation of Notch target gene expression at the mRNA level. This is accompanied by the accumulation of intracellular Notch1 proteins that show point mutations or deletions in the PEST domain of the Notch1 gene. Tumor cell proliferation is strictly dependent on Notch signaling. IkL/L mice therefore develop a Notch-dependent T-ALL that is highly reminiscent of the human disease. Importantly, the Notch pathway genes were unmanipulated in this system.
Thus, the IkL/L mouse line is a relevant model for studying the role of Notch activation and the function of specific Notch receptors in the pathogenesis of T-ALL. Here, we deleted RBP-J, Notch3, and Notch1 in IkL/L mice and evaluated their genetic contribution to T-ALL initiation and progression. We show that Notch1 is the critical receptor in T-ALL and uncover a novel mechanism for the generation of oncogenic Notch1 proteins that involves deletion of its 5′ promoter.
Methods
Methods are available on the Blood Web site (see the Supplemental Materials link at the top of the online article). Research on mice at the IGBMC was approved by the Direction des Services Vétérinaires du Bas-Rhin. All microarray data are available on the Gene Expression Omnibus (National Center for Biotechnology Information) public database under accession number GSE23972.
Results
Genetic contribution of RBP-J, Notch3, and Notch1 to tumor progression in a spontaneous model of T-ALL
To determine the significance of Notch activation and to define the role of Notch receptors in T-ALL progression, we crossed IkarosL/L mice with animals carrying null mutations or floxed alleles for RBP-J, Notch3, and Notch1.18-20 To delete the floxed alleles, the CD4-Cre tg,21 which excises floxed sequences at the DN4 (CD4−CD8−CD44−CD25−) to DP stage of thymocyte differentiation, was chosen for 3 reasons: (1) because the constitutive null mutation is embryonic-lethal (eg, with RBP-J and Notch1), (2) because deletion at an earlier time point (eg, using lck-Cre) impairs T-cell differentiation and might introduce unnecessary side effects to our interpretation of the results,22 and (3) because the IkL/L tumors consistently express a αβ TCR and display a DP to CD8 SP cell-surface phenotype, suggesting that most tumor cells undergo some differentiation and will activate the CD4 promoter.
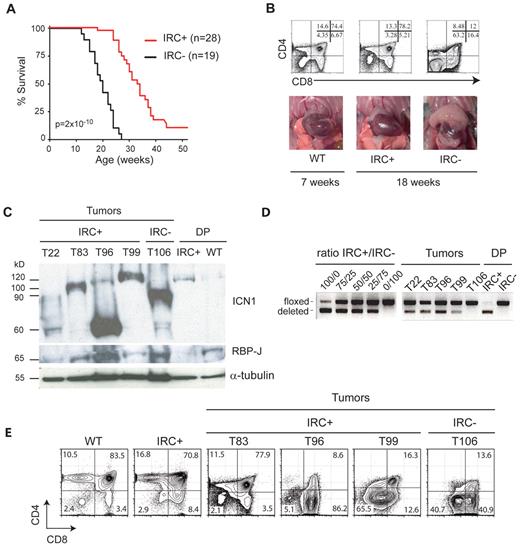
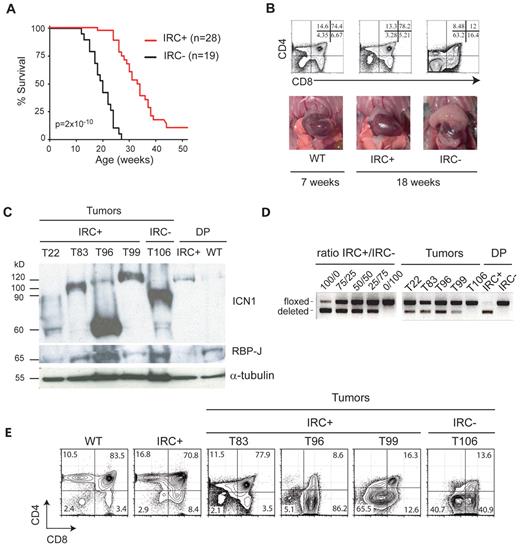
The effect of RBP-J inactivation was therefore studied in RBP-J–expressing IkL/LRBP-Jf/fCD4-Cre− (IRC−) and RBP-J–deleted IkL/LRBP-Jf/fCD4-Cre+ (IRC+) mice. In agreement with published results,23 CD4-Cre–mediated deletion of RBP-J in IkL/L mice did not affect T-cell differentiation and thymic cellularity (data not shown). In contrast, loss of RBP-J significantly pushed T-ALL–related death from a median of 18 weeks to 30 weeks (Figure 1A; P = 2 × 10−10). To determine whether this was due to a delay in tumor initiation or tumor growth, we analyzed the thymus of IRC+ and IRC− mice at 18 weeks of age. At this age, IRC− mice were either dead or showed pronounced thymic lymphomas with an abnormal CD4/CD8 phenotype (Figure 1B). In contrast, IRC+ mice were healthy and exhibited a normal thymic phenotype, similar to wild-type (WT). Nevertheless, IRC+ mice eventually died from T-ALL. IRC+ tumors expressed full-length RBP-J and intracellular, γ-secretase–cleaved Notch1 polypeptides (ICN1) of variable size (Figure 1C), suggesting a selection of PEST mutations in the Notch1 gene. These tumors showed low levels of deletion of the floxed sequences (Figure 1D), indicating that the CD4-Cre transgene had not been activated in most of the cells. Indeed, some IRC+ tumors were composed mostly of CD4−CD8− or CD4−CD8+ cells (see T99 and T96 in Figure 1E). Other tumors expressed CD4 but may have acquired resistance to the Cre-mediated deletion through another mechanism. Collectively, these results demonstrate that Notch activation is required for T-ALL initiation in IkL/L mice.
RBP-J deletion delays T-ALL development. (A) Survival curves of IkL/LRBP-Jf/fCD4-Cre+ (IRC+) mice and IkL/LRBP-Jf/fCD4-Cre− (IRC−) mice. The statistical significance was calculated by log-rank test. Note that the 2 IRC+ mice killed at 52 weeks did not show signs of disease. (B) Thymocyte CD4/CD8 profiles as determined by flow cytometry (top) and photos of the thoracic cavity (bottom) from 18-week-old IRC+ and IRC− mice and a 7-week-old WT mouse. (C) Western blot of RBP-J and ICN1 (Val1744 antibody) expression in a panel of thymic tumors from IRC+ mice. Control samples are an IRC− tumor and sorted CD4+CD8+ (DP) cells from 4-week-old IRC+ and WT mice. α-Tubulin was used as a loading control. The variable sizes of the ICN1 proteins are likely due to C-terminal truncations. Note that the deletion of the RBP-J floxed sequences effectively leads to loss of RBP-J proteins in nontransformed IRC+ DP cells. (D) PCR analysis of the deletion of the floxed sequences in the samples shown in C. The left panel shows amplification of control samples consisting of mixes of DNA from IRC+ and IRC− thymocytes, at the indicated ratio. (E) CD4/CD8 profiles of samples shown in panel C, except for T22 where the FACS profile was not available.
RBP-J deletion delays T-ALL development. (A) Survival curves of IkL/LRBP-Jf/fCD4-Cre+ (IRC+) mice and IkL/LRBP-Jf/fCD4-Cre− (IRC−) mice. The statistical significance was calculated by log-rank test. Note that the 2 IRC+ mice killed at 52 weeks did not show signs of disease. (B) Thymocyte CD4/CD8 profiles as determined by flow cytometry (top) and photos of the thoracic cavity (bottom) from 18-week-old IRC+ and IRC− mice and a 7-week-old WT mouse. (C) Western blot of RBP-J and ICN1 (Val1744 antibody) expression in a panel of thymic tumors from IRC+ mice. Control samples are an IRC− tumor and sorted CD4+CD8+ (DP) cells from 4-week-old IRC+ and WT mice. α-Tubulin was used as a loading control. The variable sizes of the ICN1 proteins are likely due to C-terminal truncations. Note that the deletion of the RBP-J floxed sequences effectively leads to loss of RBP-J proteins in nontransformed IRC+ DP cells. (D) PCR analysis of the deletion of the floxed sequences in the samples shown in C. The left panel shows amplification of control samples consisting of mixes of DNA from IRC+ and IRC− thymocytes, at the indicated ratio. (E) CD4/CD8 profiles of samples shown in panel C, except for T22 where the FACS profile was not available.
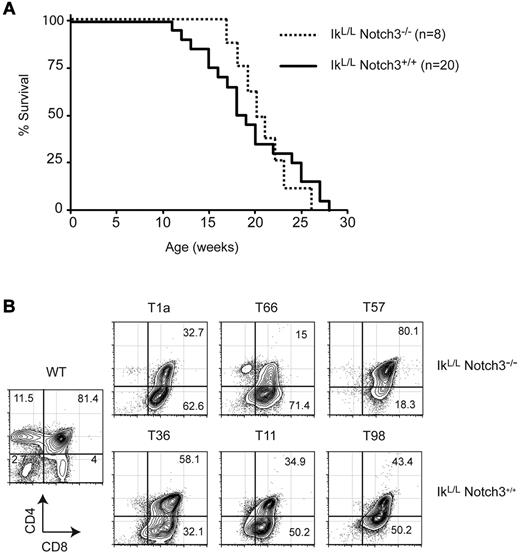
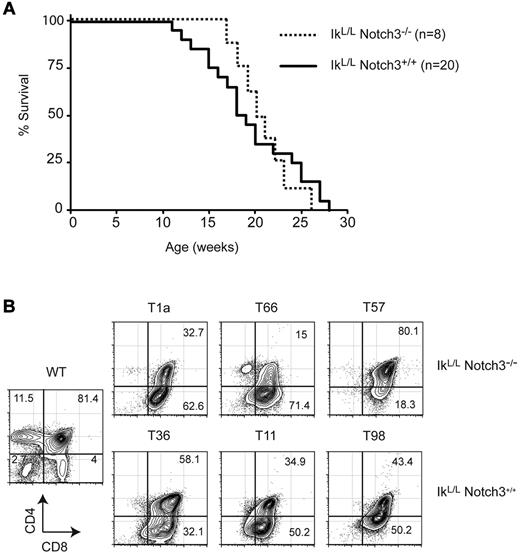
Notch3 is highly expressed in human T-ALL and in IkL/L tumors.9,10,17 We therefore evaluated the role of Notch3 in tumor progression by crossing IkL/L mice with Notch3-null animals. IkL/L mice died from T-ALL with similar kinetics and similar tumor profiles (DP/CD8 phenotype, low levels of αβ TCR) in the presence or absence of Notch3 (Figure 2 and not shown), indicating that Notch3 has minimal impact on T-ALL induced by Ikaros deficiency.
Notch3 is dispensible for leukemogenesis in IkL/L mice. (A) Survival curves of IkL/LNotch3+/+ and IkL/LNotch3−/− mice. (B) Representative CD4/CD8 profiles of thymic lymphomas from IkL/LNotch3+/+ and IkL/LNotch3−/− mice.
Notch3 is dispensible for leukemogenesis in IkL/L mice. (A) Survival curves of IkL/LNotch3+/+ and IkL/LNotch3−/− mice. (B) Representative CD4/CD8 profiles of thymic lymphomas from IkL/LNotch3+/+ and IkL/LNotch3−/− mice.
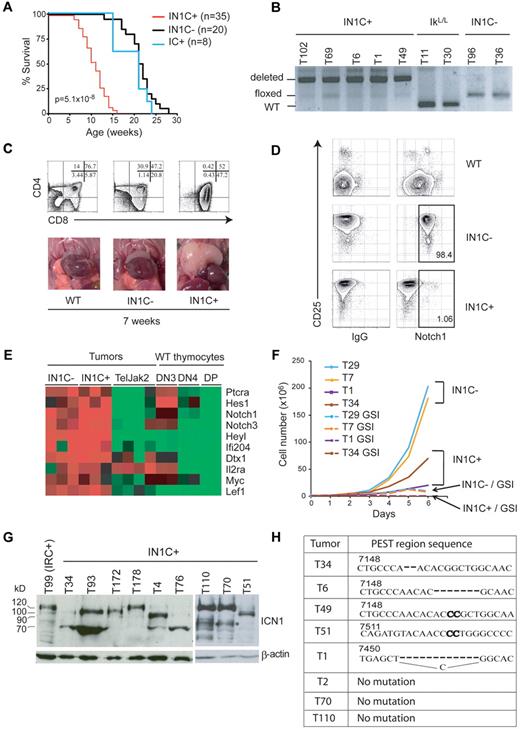
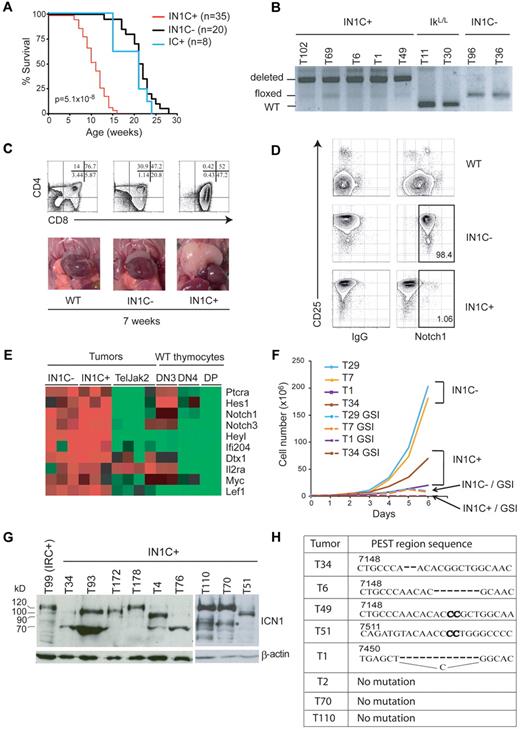
To investigate the role of Notch1, IkL/L mice were crossed with animals expressing floxed Notch1 alleles and CD4-Cre to generate IkL/LNotch1f/fCD4-Cre+ (IN1C+) and IkL/LNotch1f/fCD4-Cre− (IN1C−) mice. Unexpectedly, Notch1-deleted IN1C+ mice died significantly faster and developed T-ALL earlier than their IN1C− littermates (11 weeks for IN1C+ vs 20 weeks for IN1C−; Figure 3A,C). Furthermore, there was no detectable difference in tumor phenotype: both IN1C+ and IN1C− tumors exhibited similar CD4/CD8/CD3 profiles, showed biased TCR β chain rearrangement, and could transfer disease with similar kinetics in irradiated primary and secondary recipients (supplemental Figure 1 and not shown). In addition, the acceleration in tumor development was strictly dependent on the presence of both the floxed Notch1 alleles and the CD4-Cre tg, as IkL/LNotch1+/+CD4-Cre+ mice also died near 20 weeks of age (Figure 3A blue curve). These results suggest that deletion of the Notch1 floxed alleles is an oncogenic event.
Deletion of Notch1 accelerates T-ALL development. (A) Survival curves of IkL/LNotch1f/fCD4-Cre+ (IN1C+), IkL/LNotch1f/fCD4-Cre− (IN1C−), and IkL/LNotch1+/+CD4-Cre+ (IC+) mice. The P value corresponds to the statistical difference between the survival of IN1C+ and IN1C− mice by log-rank test. (B) PCR analysis of the deletion of the floxed sequences in a panel of IN1C+ tumors. (C) Thymocyte CD4/CD8 profiles (top) and photos of the thoracic cavity (bottom) from 7-week-old WT, IN1C−, and IN1C+ mice. (D) Surface Notch1 and CD25 expression of IN1C+ and IN1C− tumor cells and WT thymocytes. The immunoglobulin G isotype control is shown in the left panels. Similar results were observed in all IkL/L/IN1C− and IN1C+ mice analyzed (n > 10). (E) Transcriptome profiling of Notch target genes in 3 IN1C+ and 3 IN1C− tumors using Affymetrix 430 2.0 arrays. The data were normalized with those from leukemic T cells of Tel-Jak2 tg mice24 and from WT DN3, DN4, and DP thymocytes using the Robust Microarray Analysis algorithm. Red and green colors indicate high and low expression, respectively. (F) Proliferation of IN1C+ and IN1C− cell lines in the absence or presence of γ-secretase inhibitor over 6 days. Representative of 3 independent experiments. (G) Western blot of ΙCN1 expression in IN1C+ tumors using the Val1744 antibody. β-actin was used as a loading control. T99 is a IRC+ tumor that expresses ICN1 proteins of the normal 120 kDa size. The asterisk in the right panel points to likely degradation products. (H) PEST region sequences of IN1C+ tumors. The bold nucleotides correspond to insertions in the WT sequence. ICN1 proteins from samples T34, T6, and T49 are shown in supplemental Figure 1C; T51, T70 and T110 are shown in Figure 3G; T1 and T2 correspond to IN1C+ cell lines described in supplemental Figure 2. Numbering according to the Notch1 reference sequence NM_008714.
Deletion of Notch1 accelerates T-ALL development. (A) Survival curves of IkL/LNotch1f/fCD4-Cre+ (IN1C+), IkL/LNotch1f/fCD4-Cre− (IN1C−), and IkL/LNotch1+/+CD4-Cre+ (IC+) mice. The P value corresponds to the statistical difference between the survival of IN1C+ and IN1C− mice by log-rank test. (B) PCR analysis of the deletion of the floxed sequences in a panel of IN1C+ tumors. (C) Thymocyte CD4/CD8 profiles (top) and photos of the thoracic cavity (bottom) from 7-week-old WT, IN1C−, and IN1C+ mice. (D) Surface Notch1 and CD25 expression of IN1C+ and IN1C− tumor cells and WT thymocytes. The immunoglobulin G isotype control is shown in the left panels. Similar results were observed in all IkL/L/IN1C− and IN1C+ mice analyzed (n > 10). (E) Transcriptome profiling of Notch target genes in 3 IN1C+ and 3 IN1C− tumors using Affymetrix 430 2.0 arrays. The data were normalized with those from leukemic T cells of Tel-Jak2 tg mice24 and from WT DN3, DN4, and DP thymocytes using the Robust Microarray Analysis algorithm. Red and green colors indicate high and low expression, respectively. (F) Proliferation of IN1C+ and IN1C− cell lines in the absence or presence of γ-secretase inhibitor over 6 days. Representative of 3 independent experiments. (G) Western blot of ΙCN1 expression in IN1C+ tumors using the Val1744 antibody. β-actin was used as a loading control. T99 is a IRC+ tumor that expresses ICN1 proteins of the normal 120 kDa size. The asterisk in the right panel points to likely degradation products. (H) PEST region sequences of IN1C+ tumors. The bold nucleotides correspond to insertions in the WT sequence. ICN1 proteins from samples T34, T6, and T49 are shown in supplemental Figure 1C; T51, T70 and T110 are shown in Figure 3G; T1 and T2 correspond to IN1C+ cell lines described in supplemental Figure 2. Numbering according to the Notch1 reference sequence NM_008714.
Notch1 activation in Notch1-deleted tumors
Tumor development in IN1C+ mice could mean that the Notch1 alleles of the tumor cells had escaped deletion by the CD4-Cre tg. We evaluated the deletion efficiency of the floxed alleles in a panel of primary tumors from IkL/L, IN1C−, and IN1C+ mice by polymerase chain reaction (PCR) analysis (Figure 3B) and found that the IN1C+ tumors showed high levels of deletion, suggesting that most IN1C+ tumor cells had efficiently deleted both Notch1 alleles. We also evaluated the cell-surface expression of Notch1 on primary IN1C+ tumors by flow cytometry. In these experiments, IN1C− tumors uniformly expressed high levels of CD25, a putative Notch target gene, and surface Notch1 (Figure 3D). In contrast, IN1C+ tumors still expressed CD25 but not Notch1 (Figure 3D), indicating that Notch1 was no longer expressed on the surface of IN1C+ tumor cells.
That IN1C+ tumor cells expressed CD25 suggested that the Notch pathway was still active in these tumors. To test this possibility and to determine whether additional cooperative oncogenic pathways were activated in IN1C+ tumors, we analyzed the transcriptome profiles of 3 IN1C+ and 3 IN1C− tumors, and compared them with those of Tel-Jak2–induced T-cell tumors (that exhibit low levels of Notch target gene expression17 ), as well as with WT DN3 (CD4−CD8−CD44−CD25+), DN4, and DP thymocytes. Notch target genes were strongly up-regulated in both IN1C+ and IN1C− tumors (Figure 3E), suggesting Notch activation in both cases. In addition, IN1C+ tumors did not show a deregulation of genes associated with other known oncogenic pathways (data not shown). To determine whether IN1C+ tumors were dependent on Notch signaling for proliferation, cell lines generated from IN1C+ and IN1C− tumors were cultured in the presence of a γ-secretase inhibitor (GSI) (see supplemental Figure 2 for characterization of the cell lines). In all cases, GSI treatment arrested cell proliferation (Figure 3F). Together, these results indicate that the Notch pathway remains a dominant oncogenic pathway in Notch1-deleted T cells.
Finally, we asked whether activated Notch1 proteins were still expressed in IN1C+ tumors, using an antibody specific for γ-secretase–cleaved Notch1 (Val1744). Strikingly, all IN1C+ tumors expressed ICN1 (Figure 3G). These proteins varied in size, were shorter than the expected 120 kDa in 60%-70% of the cases, and were similar to the truncated ICN1 proteins in IkL/L tumors,17 suggesting the presence of PEST domain mutations at the DNA level. This was confirmed by sequencing the Notch1 transcripts in 8 IN1C+ tumors, which revealed heterozygous frame-shift mutations in the 3′ Notch1 sequence that would result in the production of truncated Notch1 proteins lacking the PEST domain in 5 cases (Figure 3H). Thus, the Notch1 gene is still a target of oncogenic mutations despite the deletion of floxed sequences and the absence of surface Notch1 in IN1C+ T cells.
Deletion of the Notch1 promoter/exon 1 is oncogenic in Ikaros-deficient T cells
In the course of this study, we came across intriguing results from Aster and colleagues, who discovered a panel of murine T-cell lines that did not express surface Notch1, were sensitive to GSI for growth, and exhibited 5′ deletions in the Notch1 gene (see companion paper by Ashworth et al25 ). Their results, as well as those from Tsuji et al,26 suggested that Notch1 could be transcribed from cryptic intragenic promoters if the conserved 5′ promoter is lost and that the resulting proteins may be oncogenic. We therefore revisited the Notch1 floxed mutation. In this mutation, the loxP sites were placed around a 3.5-kb sequence encompassing the conserved promoter and exon 1, which encodes the leader peptide responsable for surface Notch1 expression.20 Previous analyses have shown that Cre-mediated deletion of the floxed sequence leads to the loss of Notch1 proteins in thymocytes,21 indicating that the floxed mutation is null.
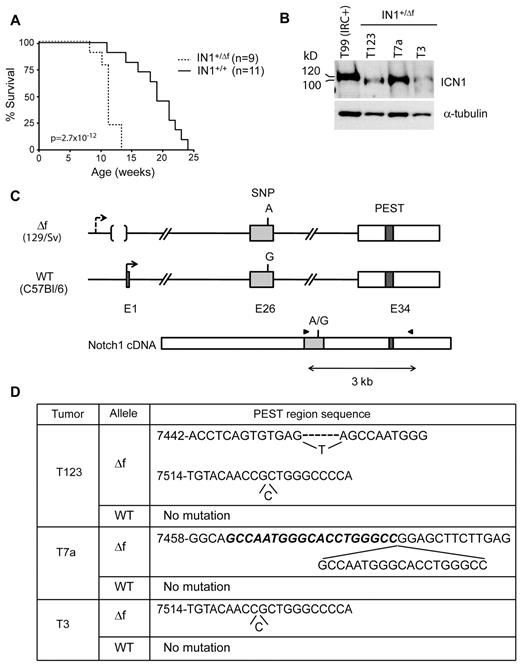
To determine whether deletion of the Notch1 floxed sequences leads to a null allele or an oncogenic allele on an Ikaros-deficient background, we generated IkL/L mice heterozygote for the floxed allele. Floxed Notch1 mice were bred with cytomegalovirus-Cre tg mice to delete the floxed sequences in the germline (the germline-deleted allele was designated N1Δf). Notch1+/ Δf mice were then crossed with IkL/L animals to obtain IkL/LNotch1+/ Δf (IN1+/ Δf) mice. Strikingly, IN1+/ Δf mice died from T-ALL at a median age of 11 weeks, while IkL/LNotch1+/+ littermates died near 20 weeks (Figure 4A). Thus, deletion of the Notch1 promoter/exon 1 on a single allele significantly accelerates tumorigenesis.
Oncogenic effect of deleting floxed Notch1 sequences from a single allele. (A) Survival curves of IkL/LNotch1+/Δf (IN1+/Δf) and IkL/LNotch1+/+ (IN1+/+) mice. The statistical significance was calculated by log-rank test. (B) Western blot of ICN1 expression in IN1+/Δf tumors. The control sample is the IRC+ T99 tumor shown in Figure 1C, which expresses ICN1 proteins of normal size. α-Tubulin was used as a loading control. (C) Strategy to identify the allele harboring the PEST domain mutation. A single nucleotide polymorphism in exon 26 (rs27201809; Mouse Genome Informatics database) distinguishes the WT and deleted alleles, which are derived from the C57Bl/6 and 129/Sv strains, respectively. RT-PCR amplification and sequencing of exon 26 and the PEST region in exon 34 identifies the allele carrying the mutation. (D) Association of PEST region mutations with the N1Δf allele in the 3 tumors shown in B. T3 had a single nt insertion. T7a had a duplication of 19 nt (in bold and italic). T123 had 2 separate mutations, both in the N1Δf allele: a deletion of 6 nt, which were replaced by a single T, and a single nt insertion. Numbering according to the Notch1 reference sequence NM_008714.
Oncogenic effect of deleting floxed Notch1 sequences from a single allele. (A) Survival curves of IkL/LNotch1+/Δf (IN1+/Δf) and IkL/LNotch1+/+ (IN1+/+) mice. The statistical significance was calculated by log-rank test. (B) Western blot of ICN1 expression in IN1+/Δf tumors. The control sample is the IRC+ T99 tumor shown in Figure 1C, which expresses ICN1 proteins of normal size. α-Tubulin was used as a loading control. (C) Strategy to identify the allele harboring the PEST domain mutation. A single nucleotide polymorphism in exon 26 (rs27201809; Mouse Genome Informatics database) distinguishes the WT and deleted alleles, which are derived from the C57Bl/6 and 129/Sv strains, respectively. RT-PCR amplification and sequencing of exon 26 and the PEST region in exon 34 identifies the allele carrying the mutation. (D) Association of PEST region mutations with the N1Δf allele in the 3 tumors shown in B. T3 had a single nt insertion. T7a had a duplication of 19 nt (in bold and italic). T123 had 2 separate mutations, both in the N1Δf allele: a deletion of 6 nt, which were replaced by a single T, and a single nt insertion. Numbering according to the Notch1 reference sequence NM_008714.
Like IN1C+ tumors, IN1+/ Δf tumors expressed truncated ICN1 proteins (Figure 4B), suggesting a selection of oncogenic PEST mutations. To determine whether the PEST mutations occurred on the WT allele, or the deleted one, we exploited the observation that these alleles differed by a single nucleotide polymorphism at position 5179 in exon 26 (numbering according to the Notch1 reference sequence NM_008714). The deleted allele, derived from the 129/Sv strain (the parental strain of the GS-1 embryonic stem cell line used to engineer the mutation), had an “A” at this position, while the WT allele (derived from the C57Bl/6 strain; see “Methods”) had a “G” (Figure 4C). We amplified cDNA fragments spanning exons 26-34 (that comprises both the single nucleotide polymorphism and the PEST domain sequences) from 3 IN1+/ Δf tumors, subcloned, and sequenced the amplification products. All 3 tumors showed PEST mutations in the 129/Sv allele, while the C57Bl/6 allele showed no mutations (Figure 4D). These data suggest that the PEST mutations are strongly selected for on the N1Δf allele compared with the WT allele. Thus, our results indicate that loss of the Notch1 promoter/exon 1 is oncogenic on an Ikaros-deficient background and that the deleted allele is sensitive to subsequent PEST mutations.
Truncated transcripts encoding constitutively active Notch1 proteins are expressed in IN1C+ tumors
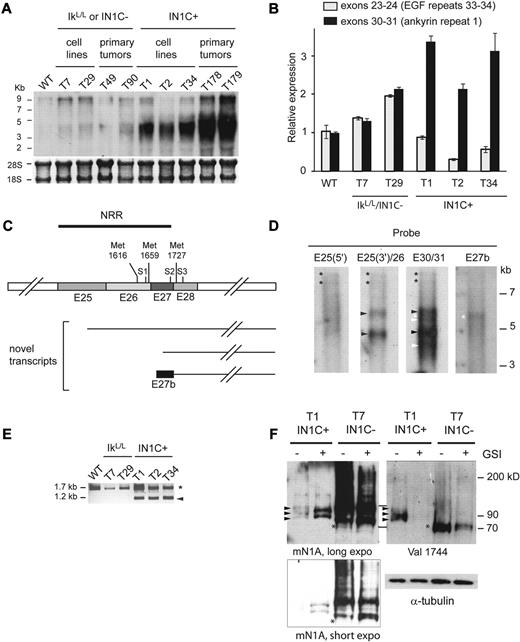
To investigate how oncogenic Notch1 proteins are generated after deletion of the main promoter, we compared the Notch1 transcripts in IN1C+ tumors with those in IN1C− or IkL/L tumors and WT thymocytes by Northern blot using a probe corresponding to exon 34. As expected, this probe revealed full-length approximately 9-kb transcripts in WT thymocytes, IkL/L, and IN1C− tumor cells (Figure 5A). In contrast, the majority of the transcripts in the IN1C+ tumors were between 4-6 kb. Some 9-kb transcripts were still detected in IN1C+ tumors; these may have initiated from the cryptic 5′ 1a and 1b promoters upstream of the main promoter,26 with subsequent splicing to 5′ exons, as detected by reverse-transcription (RT)–PCR (supplemental Figure 3). We further analyzed the relative levels of Notch1 transcripts containing 5′ and/or 3′ sequences in a ratiometric RT-quantitative PCR assay, using primers to amplify exons 23-24 (which encodes part of the extracellular domain) or exons 30-31 (which encodes part of the cytoplasmic domain). These assays showed that the ratio of 5′ to 3′ transcripts were similar in the IkL/L or IN1C− cell lines (Figure 5B). In contrast, there was a clear bias toward the 3′ transcripts in the IN1C+ cell lines. Thus, deletion of the proximal promoter results in the transcription of truncated 3′ Notch1 transcripts in IN1C+ cells.
Truncated Notch1 transcripts and proteins in IN1C+ tumors. (A) Northern blot of total RNA (10 μg) from primary tumors and cell lines of the indicated genotypes using a probe for exon 34 (top). Methylene blue staining of 18S and 28S ribosomal RNA was used as a loading control (bottom). (B) RT-quantitative PCR of Notch1 transcripts, using primers to amplify exons 23-24 (encoding the extracellular EGF repeats 33/34) and exons 30-31 (encoding the first intracellular ankyrin repeat). Results were normalized to hypoxanthine-guanine phosphoribosyltransferase levels and to those of WT thymocytes, for which the exon 23-24 mRNA level was arbitrarily fixed at 1. Data represent the mean of 2 experiments. (C) Scheme summarizing the results of the 5′-RACE experiments performed on the T1 and T34 IN1C+ cell lines. The organization of the Notch1 transcript in the region of interest is shown, with the position of the S1, S2, and S3 cleavage sites, and the putative methionines that could be used for translation initiation. cDNAs identified by 5′-RACE are shown (see supplemental Figure 5 for sequence details). (D) Northern blot of Notch1 transcripts in poly(A)+ RNA (1.5 μg each) from the T1 cell line, hybridized with probes from the indicated exons/introns. E25(5′) corresponds to nt 4279-4741; E25(3′)/26 corresponds to nt 4758-5246; E30/31 corresponds to nt 5715-6179 (reference sequence NM_008714). E27b corresponds to a 628 nt region from intron 27 that includes the 113 nt sequence of exon 27b (see supplemental Figure 5). Autoradiograms for E25(3′)/26 and E30/31 were exposed for 18 hours; those for E25(5′) and E27b were exposed for 44 hours. Asterisks indicate transcripts initiating from 5′ promoters; black arrowheads indicate transcripts initiating from exon 25; white arrowheads indicate transcripts initiating downstream of exon 26 (likely in exon 27); white asterisks indicate transcripts containing exon 27b. (E) RT-PCR of exon 27b–containing transcripts in the indicated samples. cDNA was amplified using a forward primer located within exon 27b and a reverse primer from exon 31. The arrowhead indicates the correctly spliced transcripts; the asterisk indicates likely splicing intermediates that had not excised intron 27. (F) Western blot of Notch1 expression in total cell extracts from the T1 IN1C+ and T7 IN1C− cell lines, cultured in the presence or absence of GSI for 3 days. The membrane was first analyzed with the Val1744 antibody, and then with the mN1A Ab. α-tubulin was used as a loading control. Long (10 minutes) and short (30 seconds) exposures are shown for the mN1A Ab. The lines between the top 2 panels indicate the positions of the molecular weight markers used to align the blots. Asterisks indicate the γ-secretase–cleaved proteins from the T7 cell line; arrowheads indicate the γ-secretase–cleaved proteins from the T1 cell line. Note that the ICN1 proteins in the T7 line do not completely disappear after GSI treatment, probably due to the increased stability of the truncated proteins in this cell line. All data are representative of > 2 independent experiments.
Truncated Notch1 transcripts and proteins in IN1C+ tumors. (A) Northern blot of total RNA (10 μg) from primary tumors and cell lines of the indicated genotypes using a probe for exon 34 (top). Methylene blue staining of 18S and 28S ribosomal RNA was used as a loading control (bottom). (B) RT-quantitative PCR of Notch1 transcripts, using primers to amplify exons 23-24 (encoding the extracellular EGF repeats 33/34) and exons 30-31 (encoding the first intracellular ankyrin repeat). Results were normalized to hypoxanthine-guanine phosphoribosyltransferase levels and to those of WT thymocytes, for which the exon 23-24 mRNA level was arbitrarily fixed at 1. Data represent the mean of 2 experiments. (C) Scheme summarizing the results of the 5′-RACE experiments performed on the T1 and T34 IN1C+ cell lines. The organization of the Notch1 transcript in the region of interest is shown, with the position of the S1, S2, and S3 cleavage sites, and the putative methionines that could be used for translation initiation. cDNAs identified by 5′-RACE are shown (see supplemental Figure 5 for sequence details). (D) Northern blot of Notch1 transcripts in poly(A)+ RNA (1.5 μg each) from the T1 cell line, hybridized with probes from the indicated exons/introns. E25(5′) corresponds to nt 4279-4741; E25(3′)/26 corresponds to nt 4758-5246; E30/31 corresponds to nt 5715-6179 (reference sequence NM_008714). E27b corresponds to a 628 nt region from intron 27 that includes the 113 nt sequence of exon 27b (see supplemental Figure 5). Autoradiograms for E25(3′)/26 and E30/31 were exposed for 18 hours; those for E25(5′) and E27b were exposed for 44 hours. Asterisks indicate transcripts initiating from 5′ promoters; black arrowheads indicate transcripts initiating from exon 25; white arrowheads indicate transcripts initiating downstream of exon 26 (likely in exon 27); white asterisks indicate transcripts containing exon 27b. (E) RT-PCR of exon 27b–containing transcripts in the indicated samples. cDNA was amplified using a forward primer located within exon 27b and a reverse primer from exon 31. The arrowhead indicates the correctly spliced transcripts; the asterisk indicates likely splicing intermediates that had not excised intron 27. (F) Western blot of Notch1 expression in total cell extracts from the T1 IN1C+ and T7 IN1C− cell lines, cultured in the presence or absence of GSI for 3 days. The membrane was first analyzed with the Val1744 antibody, and then with the mN1A Ab. α-tubulin was used as a loading control. Long (10 minutes) and short (30 seconds) exposures are shown for the mN1A Ab. The lines between the top 2 panels indicate the positions of the molecular weight markers used to align the blots. Asterisks indicate the γ-secretase–cleaved proteins from the T7 cell line; arrowheads indicate the γ-secretase–cleaved proteins from the T1 cell line. Note that the ICN1 proteins in the T7 line do not completely disappear after GSI treatment, probably due to the increased stability of the truncated proteins in this cell line. All data are representative of > 2 independent experiments.
To map the 5′ ends of the truncated transcripts, we performed 5′-RACE on IN1C+ tumor cells, using an assay that specifically detects full-length capped mRNA. A sequence in exon 31 was used as the 3′ anchor. Distinct 5′-RACE products were amplified from 2 IN1C+ cell lines ranging from 700-1200 bp (supplemental Figure 4 and data not shown). Sequencing of the subclones revealed a variety of 5′ ends that started from the end of exon 25 all the way to exon 29 (Figure 5C and see supplemental Figure 5 for sequence information). Several clones began in intron 27 and contained a 113 bp segment of this intron (hereafter named exon 27b) spliced to exon 28 and downstream exons (Figure 5C and supplemental Figure 5). To determine whether these sequences corresponded to the truncated transcripts identified in the Northern blot, we performed further Northern blots on poly(A)+ RNA from a IN1C+ cell line and used a series of probes covering exons 25-31 (Figure 5D). No transcripts were detected with a probe from the 5′ end of exon 25. Two major transcripts (approximately 4.5 and 6 kb) were detected with a probe from the 3′ end of exon 25 and exon 26 (black arrowheads). Three additional transcripts (approximately 4, 5.5, and 5.7 kb) were detected with a probe from exons 30-31 (white arrowhead and asterisk). Finally, a weak transcript was detected with an exon 27b probe (white asterisk). These results suggest that IN1C+ tumors predominantly express transcripts that start in exons 25-27, with a minor transcript starting in exon 27b. Interestingly, transcripts of different sizes were detected with 2 of the probes, suggesting that they also use different polyadenylation signals (see companion paper by Ashworth et al25 ).
The exon 27b–containing transcript was intriguing in that it includes intronic sequences not found in the normal full-length Notch1 mRNA. RT-PCR analyses readily amplified this product from all IN1C+ cell lines tested but not from lines with intact Notch1 5′ sequences (Figure 5E arrowhead). In addition, screening of > 40 RNA samples from a wide range of normal tissues, as well as from embryos and embryonic stem cell lines, did not reveal the presence of this transcript (data not shown). These results suggest that the exon 27b–containing transcripts may be specific to leukemic cells with 5′ promoter deletions.
We next evaluated the Notch1 proteins synthesized from the truncated transcripts. Three methionines (aa 1616, 1659, 1727) are present between exons 26-28 (Figure 5C). Proteins initiating from these methionines would be predicted to lack the extracellular domain and the negative regulatory region but retain the S3 cleavage site, thereby creating a protein susceptible to γ-secretase cleavage in the absence of ligand. To test this hypothesis, we analyzed the Notch1 proteins from T1 IN1C+ and T7 IN1C− cells, treated or not with GSI, by Western blot (Figure 5F). Two anti-Notch1 antibodies were sequentially used: the Val1744 antibody that specifically detects γ-secretase–cleaved Notch1 and the mN1A antibody that detects the cdc10/ankyrin repeat domain of all Notch1 proteins before and after cleavage but which appears to detect γ-secretase–cleaved Notch1 with lower intensity in our hands. As expected, the untreated IN1C− sample contained mainly uncleaved Notch1 proteins (left panel, the larger size bands above the asterisk; see lower panel for a short exposure of the same membrane) and some γ-secretase–cleaved proteins (right panel, asterisk). Interestingly, less proteins were detected by the mN1A antibody from IN1C+ cells compared with IN1C− samples (left panel), suggesting that the truncated Notch1 transcripts are not efficiently translated. Nevertheless, cleaved Notch1 proteins were readily detected by the Val1744 antibody in the untreated IN1C+ sample (right panel, arrowheads), and similar size proteins were detected by both antibodies (left and right panels, arrowheads), suggesting that most of the truncated Notch1 proteins are rapidly cleaved. Proteins of slightly larger size were detected by the mN1A antibody in GSI-treated IN1C+ cells, indicating that these proteins contain the γ-secretase cleavage site. Together, our analyses at the RNA and protein levels indicate that IN1C+ tumors express novel transcripts that encode proteins constitutively activated by γ-secretase cleavage.
Loss of the Notch1 promoter induces transcription initiation in the 3′ region of the Notch1 locus and chromatin remodeling
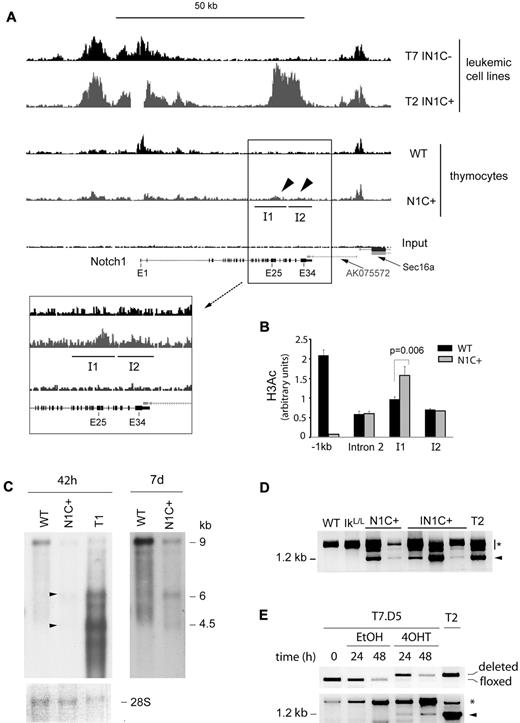
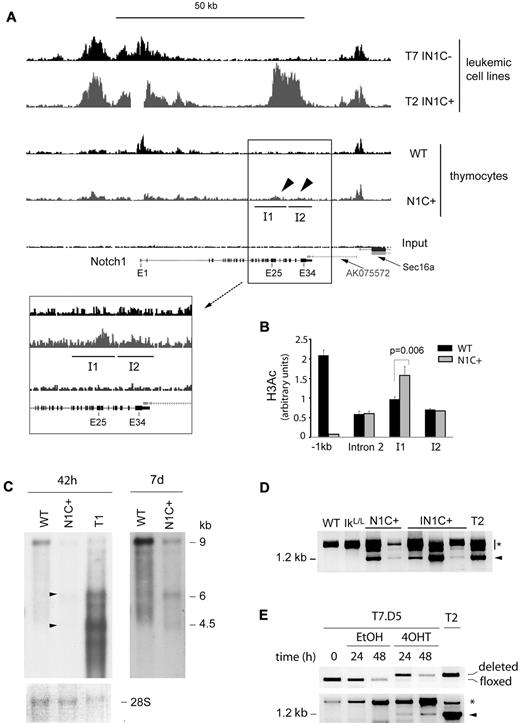
Our results suggest that transcription of the Notch1 gene is reorganized in Ikaros-deficient cells after deletion of the 5′ promoter. To map these changes, we evaluated the distribution of acetylated histone H3 (H3ac), a chromatin mark associated with active promoters, in the Notch1 locus of IN1C+ versus IN1C− tumor cells, by chromatin immunoprecipitation-sequence analysis (Figure 6A top 2 histograms). In IN1C− cells, where the main promoter is intact, we detected a broad approximately 15-kb region of H3 acetylation in the 5′ end of the Notch1 gene. In IN1C+ cells, this region of H3 acetylation was still present, indicating that the deleted floxed sequences are not required to target active epigenetic marks to the proximal Notch1 region. Strikingly, IN1C+ cells showed a novel domain of high H3 acetylation over a 10-kb region in the 3′ portion of the gene between exons 25-34. These results indicate a major reorganization of the epigenetic landscape in the Notch1 gene of IN1C+ tumor cells after deletion of the 5′ promoter, where de novo marks of transcriptional activation accumulate in the 3′ part of the gene.
Transcriptional reprogramming of the Notch1 locus after deletion of proximal promoter sequences. (A) Chromatin immunoprecipitation-sequence analysis of histone H3 acetylation in the Notch1 locus of T7 IN1C− and the T2 IN1C+ cells and WT and Notch1f/fCD4-Cre+ (N1C+) thymocytes. Top 4 histograms have a vertical scale of 150. The enlarged histograms in the inset have a vertical scale of 50. The regions identified as I1 and I2 correspond to islands of enriched tag density in the IN1C+ thymocytes, which were predicted by the “statistical model for identification of chip-enriched regions” algorithm in the N1C+, but not in the WT or input samples (see also Table 1).27 The gap in the T2 IN1C+ sample corresponds to the location of the floxed deletion. (B) Real-time PCR measurement of H3 acetylation in the WT and N1C+ samples shown in panel A at several positions along the Notch1 locus. Amplicons in intron 2 and islands I1 and I2 were located, respectively, at 29.3, 36.5, and 41.7 kb downstream of the transcription start site. (C) Northern blot of Notch1 transcripts in WT and N1C+ thymocytes and in the T1 IN1C+ cell line. An amount of 2 μg of poly(A)+ RNA was loaded for each sample, and the blots were hybridized with the E30/31 probe (see Figure 6D). The left panel shows a 42-hour exposure; the right panel shows a 7-day exposure of the WT and N1C+ lanes. Arrowheads indicate transcripts likely to have initiated from 3′ promoters in the N1C+ sample. A photo of the methylene blue staining of the membrane is shown as a loading control in the bottom panel. (D) RT-PCR of exon 27b–containing transcripts in nontransformed thymocytes from 3- to 4-week-old mice with the indicated genotypes. See Figure 5E for details. Samples were defined as nontransformed according to their CD4/CD8 profile and normal CD25 expression. (E) RT-PCR of exon 27b–containing transcripts in the T7.D5 Cre-ERT2+ clone cultured in the presence of 4OHT or vehicle for the indicated times (bottom). Cells were also analyzed for the deletion of floxed Notch1 sequences by PCR (top). T2 corresponds to a IN1C+ leukemic cell line. Similar results were obtained in 4 independent experiments. In panels D and E, the arrowhead indicates the specific product from a correctly spliced transcript; the asterisk indicates products that may correspond to splicing intermediates of transcripts initiated at upstream locations.
Transcriptional reprogramming of the Notch1 locus after deletion of proximal promoter sequences. (A) Chromatin immunoprecipitation-sequence analysis of histone H3 acetylation in the Notch1 locus of T7 IN1C− and the T2 IN1C+ cells and WT and Notch1f/fCD4-Cre+ (N1C+) thymocytes. Top 4 histograms have a vertical scale of 150. The enlarged histograms in the inset have a vertical scale of 50. The regions identified as I1 and I2 correspond to islands of enriched tag density in the IN1C+ thymocytes, which were predicted by the “statistical model for identification of chip-enriched regions” algorithm in the N1C+, but not in the WT or input samples (see also Table 1).27 The gap in the T2 IN1C+ sample corresponds to the location of the floxed deletion. (B) Real-time PCR measurement of H3 acetylation in the WT and N1C+ samples shown in panel A at several positions along the Notch1 locus. Amplicons in intron 2 and islands I1 and I2 were located, respectively, at 29.3, 36.5, and 41.7 kb downstream of the transcription start site. (C) Northern blot of Notch1 transcripts in WT and N1C+ thymocytes and in the T1 IN1C+ cell line. An amount of 2 μg of poly(A)+ RNA was loaded for each sample, and the blots were hybridized with the E30/31 probe (see Figure 6D). The left panel shows a 42-hour exposure; the right panel shows a 7-day exposure of the WT and N1C+ lanes. Arrowheads indicate transcripts likely to have initiated from 3′ promoters in the N1C+ sample. A photo of the methylene blue staining of the membrane is shown as a loading control in the bottom panel. (D) RT-PCR of exon 27b–containing transcripts in nontransformed thymocytes from 3- to 4-week-old mice with the indicated genotypes. See Figure 5E for details. Samples were defined as nontransformed according to their CD4/CD8 profile and normal CD25 expression. (E) RT-PCR of exon 27b–containing transcripts in the T7.D5 Cre-ERT2+ clone cultured in the presence of 4OHT or vehicle for the indicated times (bottom). Cells were also analyzed for the deletion of floxed Notch1 sequences by PCR (top). T2 corresponds to a IN1C+ leukemic cell line. Similar results were obtained in 4 independent experiments. In panels D and E, the arrowhead indicates the specific product from a correctly spliced transcript; the asterisk indicates products that may correspond to splicing intermediates of transcripts initiated at upstream locations.
We then asked whether H3 acetylation was specific to tumor cells, and whether Ikaros deficiency was required for the transcriptional reorganization. To address these questions, we analyzed the H3ac distribution in the Notch1 locus between WT and Ikaros-expressing Ik+/+Notch1f/fCD4-Cre+ (N1C+) thymocytes (Figure 6A bottom 2 histograms). N1C+ cells showed low but distinct enrichment of H3ac between exons 25-34 compared with WT (Figure 6A, see arrowheads and inset). Using the “statistical model for identification of chip-enriched regions” algorithm,27 which predicts islands of enriched tag frequency, we identified 2 islands of H3ac enrichment in the 3′ region of the Notch1 gene in N1C+ thymocytes that were not present in WT cells (labeled I1 and I2 in Figure 6A; see Table 1 for relevant parameters). Although H3 acetylation in these regions was reduced compared with IN1C+ leukemic cells, the enrichment over WT or input was significant (P < 10−10; Table 1) and was confirmed by real-time PCR for island I1 (Figure 6B; enrichment was not confirmed for I2, which probably reflects the smaller increase of H3 acetylation at this island). Thus, deletion of the 5′ promoter appears to initiate chromatin reorganization in the Notch1 locus and promote the acquisition of activation marks in the 3′ region of the gene. Importantly, this occurs in the presence of WT levels of Ikaros and in the absence of transformation.
Chromatin reorganization in the 3′ region of the Notch1 gene in N1C+ thymocytes was accompanied by low levels of transcription from that region. Transcripts of 4.5 and 6 kb, similar to the major transcripts in IN1C+ tumor cells, were detected by Northern blot in N1C+ cells (Figure 6C). In addition, exon 27b–containing transcripts were detected by RT-PCR in N1C+ and nontransformed IN1C+ cells but not in WT or IkL/L thymocytes (Figure 6D). As N1C+ animals do not develop T-ALL, these results suggest that transcription from 3′ cryptic promoters is initiated in nontransformed T cells. Transcription from 3′ promoters appears to be T-cell specific, as analysis of Notch1-expressing tissues from N1+/Δf mice (with a heterozygous germline deletion of the floxed sequences) revealed exon 27b–containing transcripts only in the thymus (supplemental Figure 6).
To determine whether deletion of the 5′ promoter could directly induce transcription initiation from 3′ sites, we established an inducible system to delete the floxed Notch1 sequences in vitro. The IN1C− cell line, T7, in which the Notch1 promoter/exon 1 sequences are floxed but not deleted, was transduced with a retroviral vector encoding green fluorescent protein and Cre-ERT2, a tamoxifen-inducible fusion protein between the Cre recombinase and the ligand binding domain of the estrogen receptor. Green fluorescent protein–positive cells were cloned and expanded. The T7.D5 clone, which did not show detectable levels of deletion before tamoxifen (4-hydroxytamoxifen [4OHT]) treatment but showed efficient deletion after 24 hours in 4OHT-supplemented medium (> 90% deletion; Figure 6E), was chosen for further study. T7.D5 cells were cultured in the presence of 4OHT, or vehicle, for 24 or 48 hours. We analyzed exon 27b–containing transcripts by RT-PCR, the ratio of 5′ to 3′ Notch1 transcripts by RT-quantitative PCR, and H3 acetylation by chromatin immunoprecipitation. Although the 5′ to 3′ transcript ratio and the H3 acetylation levels did not vary significantly in 4OHT-treated cells (not shown), exon 27b–containing transcripts were detected as early as 24 hours after 4OHT treatment. Thus, deletion of the 5′ promoter directly reprograms the Notch1 locus to begin transcription from 3′ promoters.
Spontaneous deletions of 5′ genomic sequences in Ikaros-deficient primary tumors
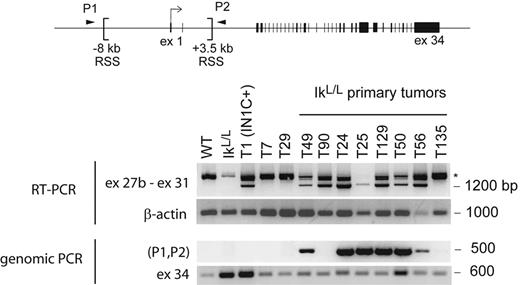
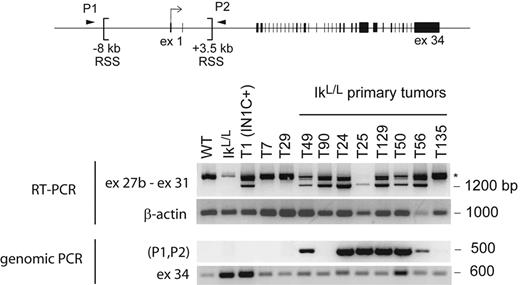
Intriguingly, our analysis of primary IkL/L or IN1C− tumors revealed low levels of approximately 4-kb transcripts (Figure 5A). To determine whether transcription from 3′ sites was common in Ikaros-deficient T-ALL, we analyzed 8 primary IkL/L or IN1C− tumors using the exon 27b–specific RT-PCR assay. Strikingly, exon 27b–containing transcripts were detected in 7 of the tumors (Figure 7). As RAG-mediated deletion of the sequences between the 2 cryptic recombination signal sequences at −8191 and +3575 bp of the transcription initiation site is always associated with transcription from 3′ sites (see companion paper by Ashworth et al25 ), we tested if similar deletions occurred in the endogenous Notch1 locus of IkL/L tumors by PCR (P1 and P2 in Figure 7). An approximately 500-bp fragment was amplified from 6 of 7 tumors that expressed the exon 27b–containing transcripts but not from premalignant IkL/L thymocytes, the T135 primary T-ALL, or the T29 and T7 cell lines (Figure 7). These results indicate that 5′ deletions and 3′ Notch1 transcripts occur spontaneously in approximately 75% of Ikaros-deficient T-ALL.
Presence of 3′ Notch1 transcripts and 5′ genomic deletions in primary IkL/L tumors. The indicated RNA or DNA samples were analyzed for the presence of exon 27b–containing transcripts by RT-PCR (top) and genomic deletion of sequences between 2 recombination signal sequences (RSS) present in the 5′ region of the Notch1 locus by PCR (bottom). RAG-mediated deletion of the sequences between the RSS sites moves the sequences of the PCR primers P1 and P2 closer together, allowing amplification of a 500-bp fragment. The IkL/L sample was extracted from premalignant thymocytes of a 7-week-old mouse; T7 and T29 are cell lines derived from IN1C− and IkL/L tumors, respectively; T49 and T90 are the primary tumors from Figure 5A. β-Actin and exon 34 of Notch1 served as control RT-PCR and PCR reactions, respectively. *Products that may correspond to splicing intermediates of transcripts initiated upstream.
Presence of 3′ Notch1 transcripts and 5′ genomic deletions in primary IkL/L tumors. The indicated RNA or DNA samples were analyzed for the presence of exon 27b–containing transcripts by RT-PCR (top) and genomic deletion of sequences between 2 recombination signal sequences (RSS) present in the 5′ region of the Notch1 locus by PCR (bottom). RAG-mediated deletion of the sequences between the RSS sites moves the sequences of the PCR primers P1 and P2 closer together, allowing amplification of a 500-bp fragment. The IkL/L sample was extracted from premalignant thymocytes of a 7-week-old mouse; T7 and T29 are cell lines derived from IN1C− and IkL/L tumors, respectively; T49 and T90 are the primary tumors from Figure 5A. β-Actin and exon 34 of Notch1 served as control RT-PCR and PCR reactions, respectively. *Products that may correspond to splicing intermediates of transcripts initiated upstream.
Discussion
Ikaros-deficient IkL/L mice represent an important model to study the natural evolution of Notch-dependent T-ALL, as they develop a disease highly reminiscent of the human condition. Although induced Notch-dependent models have been described, based on the overexpression of activated Notch1 or Notch3, their capacity to recapitulate the progressive activation of this pathway is limited. Few spontaneous models have been reported or studied extensively. In this study, we demonstrate by a genetic approach that activation of the Notch pathway is key to tumor initiation in IkL/L mice. Our results show that Notch activation is dependent on RBP-J, thereby implicating the transcription of Notch target genes in the leukemic process.
Our results also reveal that novel Notch1 transcripts are transcribed upon deletion of the 5′ main promoter to generate truncated proteins lacking the extracellular domain. Importantly, the truncated Notch proteins are predicted to lack the negative regulatory region but retain the S3 γ-secretase cleavage site, suggesting that they are membrane bound and γ-secretase sensitive. Indeed, IN1C+ tumor cells require γ-secretase activity for proliferation. The oncogenic effect of the promoter deletion is therefore similar to that of mutations targeting the negative regulatory region (which decreases ligand dependency) or chromosomal translocations where the breakpoint occurs in intron 24.7 All lead to the synthesis of constitutively active, γ-secretase–sensitive Notch1 proteins.
Surprisingly, Notch3 is not required for tumorigenesis in IkL/L mice. The role of this receptor in T-ALL is controversial, and it is currently unclear how Notch3 contributes in leukemogenesis. Our results suggest that Notch3 is dispensable for the initiation and progression of IkL/L tumors. Notch3 is expressed in all cases of human T-ALL and has been shown to modulate the splicing of Ikaros transcripts to favor those encoding dominant-negative Ikaros isoforms.9,28 However, we were unable to detect dominant-negative Ikaros isoforms in a group of human T-ALL samples, suggesting that Notch3 may be not sufficiently active in these leukemias to influence Ikaros splicing.29
We demonstrate that deletion of the main Notch1 promoter is an oncogenic event that accelerates T-ALL. Our data also indicate that 5′ deletions of the Notch1 promoter occur spontaneously and are common in IkL/L leukemias as well as in other murine T-ALL models (companion paper by Ashworth et al,25 Tsuji et al26 ). Promoter deletion reprograms the epigenetic landscape around the Notch1 locus and leads to the accumulation of active chromatin marks in the 3′ part of the gene. Importantly, this occurs in nontransformed thymocytes. Thus, transcriptional reprogramming predates leukemia initiation. Although deletion of the Notch1 promoter strongly promotes tumorigenesis, it is not sufficient for tumor initiation, as N1C+ mice or animals carrying one deleted allele in the germline do not develop T-ALL. Other events, such as PEST mutations, are likely to synergize with the promoter deletion to increase Notch1 activation. It is also likely that Ikaros deficiency facilitates Notch1 transcription, as Ikaros silences Notch target gene transcription at the molecular level.17,30,31 Thus, strong activation of Notch1 appears to require multiple hits.
How 3′ cryptic promoters are activated remains to be elucidated. In yeast, aberrant transcription from cryptic initiation sites within the coding region has been linked to defects in reforming the nucleosome structure during transcription elongation. Moreover, histone deacetylation during transcription appears to be important for correct nucleosome positioning and prevention of cryptic initiation.32-37 Our results show that aberrant transcription from the Notch1 coding region is accompanied by a significant increase in H3 acetylation along the 3′ part of the gene, suggesting a loss of deacetylation dynamics in the absence of the 5′ promoter. Interestingly, Ikaros binds to a region of exon 25 in the Notch1 gene of WT thymocytes (supplemental Figure 8). Ikaros has been shown to associate with Sin3 and the NuRD histone deacetylase complex and specifically with histone deacetylase complex 1 and 2.38,39 Thus, Ikaros may function to recruit deacetylase complexes to the Notch1 coding region during transcription to prevent intragenic initiation.
Why does deletion of the Notch1 promoter stimulate 3′ transcription if initiation from cryptic sites is linked to ongoing transcription? Transcription may still occur from the alternative 5′ promoters 1a or 1b, both of which are active in IN1C+ tumor cells (supplemental Figure 3). Deletion of the main promoter may also change the topology of the Notch1 locus and facilitate the nucleation of new transcription initiation complexes at the 3′ sites. It will be important to determine whether the complexes normally involved in Notch1 transcription are required for 3′ promoter activation. Indeed, the −5-kb and +1.4-kb ICN1 binding sites are not deleted in IN1C+ cells.40 Thus, stimulation by RBP-J/ICN1 complexes at these sites could establish a forward feeding mechanism that leads to high levels of 3′ transcription in IN1C+ cells. The deletion of the 5′ ICN1 binding sites in some IkL/L primary tumors might explain why 3′ transcription is lower in these tumors (Figure 5A). Other events, as yet undefined, may also contribute. Indeed, chromatin acetylation and 3′ transcriptional activity are higher in IN1C+ leukemic cells than in premalignant IN1C+ thymocytes or in nontransformed N1C+ cells. Further, 4OHT-induced deletion of the 5′ Notch1 promoter triggers the expression of exon 27b–containing transcripts but not robust chromatin reorganization. Identification of the mechanism behind the strong activation of the 3′ Notch1 promoter will be important for understanding T-ALL progression.
In conclusion, the Notch pathway appears to be subject to multiple checkpoints in developing T cells. In the WT situation, Notch is activated after ligand interaction, cleaved Notch proteins are rapidly degraded by the proteasome, and transcriptional repressors such as Ikaros can silence Notch target gene transcription, even in the presence of Notch signals (Kleinmann et al30 and our unpublished data). In the scenario described here, the Notch pathway is subject to multiple hits. First, when Ikaros function is diminished, Notch target genes are less efficiently silenced. In addition, when the 5′ promoter is deleted, truncated proteins are generated that do not require ligand interaction for cleavage, resulting in constitutive Notch activation. Finally, somatic mutations in the PEST domain prevent the rapid degradation of these oncogenic proteins in the cytoplasm. The end result is that the Notch pathway becomes the driver of the transformation process during T-ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank A. Gurney for the surface anti-Notch1 antibody; W. Pear for the MigR1 vector; G. Nolan for the Eco-Phoenix cells; A. Krust, N. Ghyselinck, and E. Mohier for RNAs from normal mouse tissues; D. Metzger and A. Joutel for plasmids and/or mice; the IGBMC transcriptome and sequencing platform; M. Seif, C. Tomasetto, and E. Daguenet for help; P. Marchal for technical assistance; C. Ebel for cell sorting; D. Dembélé for help with the microarray and chromatin immunoprecipitation-sequence data analyses; and S. Falcone and M. Gendron for animal husbandry.
This work was supported by grants from the Institut National du Cancer (INCa, S.C. and P.K.), La Ligue Contre le Cancer (S.C.; équipe labellisée La Ligue 2007, 2010), the Agence Nationale de la Recherche (S.C.), the Association pour la Recherche sur le Cancer (S.C.), the Fondation de France (P.K.), and institute funding from Inserm, CNRS, and l'Université de Strasbourg. R.J. received a predoctoral fellowship from La Ligue Régionale Contre le Cancer; J.M. and A.-S.G.L.L. received predoctoral fellowships from the Ministère de la Recherche et de la Technologie (MRT) and La Ligue Nationale Contre le Cancer (A.-S.G.L.L.). A.M.-G. received a predoctoral fellowship from the Conacyt Association of Mexico. A.O. received postdoctoral fellowships from INCa and the Fondation pour la Recherche Médicale.
Authorship
Contribution: R.J., J.M., A.M.-G., A.O., S.C., and P.K. designed and performed experiments, analyzed data, and wrote the paper; B.J. and S.L.G. performed high-throughput sequencing; A.-S.G.L.L., J.G., T.A., and J.C.A. shared data and protocols; T.H., T.G., and F.R. provided key reagents; and J.C.A. contributed to scientific discussions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Susan Chan and Philippe Kastner, IGBMC, 1 rue Laurent Fries, 67404 Illkirch cedex, France; e-mail: scpk@igbmc.fr.
References
Author notes
R.J., J.M., and A.M.-G. contributed equally to this study.