Abstract
The 5q− syndrome is the most distinct of all the myelodysplastic syndromes with a clear genotype/phenotype relationship. The significant progress made during recent years has been based on the determination of the commonly deleted region and the demonstration of haploinsufficiency for the ribosomal gene RPS14. The functional screening of all the genes in the commonly deleted region determined that RPS14 haploinsufficiency is the probable cause of the erythroid defect in the 5q− syndrome. A mouse model of the human 5q− syndrome has now been created by chromosomal engineering involving a large-scale deletion of the Cd74-Nid67 interval (containing RPS14). A variety of lines of evidence support the model of ribosomal deficiency causing p53 activation and defective erythropoiesis, including most notably the crossing of the “5q− mice” with p53-deficient mice, thereby ameliorating the erythroid progenitor defect. Emerging evidence supports the notion that the p53 activation observed in the mouse model may also apply to the human 5q− syndrome. Other mouse modeling data suggest that haploinsufficiency of the microRNA genes miR-145 and miR-146a may contribute to the thrombocytosis seen in the 5q− syndrome. Lenalidomide has become an established therapy for the 5q− syndrome, although its precise mode of action remains uncertain.
Introduction
The 5q− syndrome was first described by Van den Berghe et al in 1974.1 The well-known clinical features of the 5q− syndrome described in the first report are macrocytosis, anemia, a normal or high platelet count, and hypolobulated megakaryocytes in the bone marrow. Over the years, a female preponderance and a good prognosis with approximately 10% of patients transforming to acute myeloid leukemia (AML) have been well documented.2-5 The World Health Organization classification of myelodysplastic syndromes (MDSs) recognizes the 5q− syndrome as a distinct entity defined by a medullary blast count of less than 5% and the deletion of 5q [del(5q)] as the sole karyotypic abnormality.6 It should be noted that patients not correctly defined as 5q− syndrome (ie, with either additional karyotypic abnormalities or excess blasts) do not have a good prognosis.7 The majority of patients with the 5q− syndrome become transfusion dependent over time with the potential for iron loading.5,8
A recent paper by Patnaik et al2 determined that age, transfusion need at diagnosis, and dysgranulopoiesis were independent predictors of shortened survival in patients with the 5q− syndrome tightly defined according to World Health Organization criteria.
One of the features that first stimulated research on the 5q− syndrome was the localization of many genes relevant to hematopoiesis to the long arm of chromosome 5 (5q).4 The gene cluster at 5q31 includes interleukins 3, 4, 5, 9, 13, and 17β and granulocyte-monocyte colony stimulating factor.9 Several cytokine receptor genes are located on 5q, including colony-stimulating factor 1 receptor and platelet-derived growth factor-β.10
Bone marrow morphology
There is a good phenotype/genotype relationship in the 5q− syndrome such that it can often be predicted from the hematologic features and bone marrow morphology before the results of cytogenetic analysis are known.11 The study of Giagounidis et al8 presented a detailed analysis of the clinical and laboratory features of a group of MDS patients with the del(5q) including a detailed analysis of bone marrow morphology. Typically, the marrow from MDS patients is hypercellular, whereas this study showed that in 5q− syndrome, 20% of marrows were hypocellular, 40% were normocellular, and 40% hypercellular.8 Approximately half of all patients had hypoplastic erythropoiesis as defined by erythroid precursors of less than 20% in the bone marrow. Dysplasia in the red cell lineage was uncommon, with only 15% of patients having erythroid dysplasia in more than 10% of erythroid precursors.8 Dysplasia in the granulocytic lineage occurred in only 10% of all patients. All patients had mononuclear megakaryocytes with spherical nuclei.8 These observations now take on a new significance as the basis of phenotype comparisons with mouse models of the disease.
Cellular basis
The 5q− syndrome is a disorder of the human hematopoietic stem cell (HSC). In the study of Jaju et al,12 using combined immunophenotyping and fluorescent in situ hybridization, 1 of 3 cases of the 5q− syndrome was identified as having B-cell involvement. The work of Nilsson et al13 showed that in 9 patients, none had T-cell involvement but one had B-cell involvement. The data strongly supported the 5q− aberration being present in HSCs with a combined lympho-myeloid potential: in all patients a minimum of 94% of cells in the minor CD34+CD38− HSC compartment were 5q−, and in 3 of 5 patients 5q aberrations were found in a large fraction of purified CD34+CD19+ pro-B cells.13 These data plus more recent data on gene expression suggest that a lympho-myeloid HSC is the primary target of 5q deletions in MDS and that 5q deletions represent an early event in MDS pathogenesis.14
The cellular basis of the anemia and macrocytosis of the 5q− syndrome remains unclear. A recent study by Garderet et al15 enabled the analysis of cell proliferation and differentiation at a single cell level and determination of the enucleation capacity of erythroid precursors. The erythroid commitment of del(5q) clones was not altered, and their terminal differentiation capacity was preserved since they achieved final erythroid maturation (enucleation stage). The drop in red blood cell production was secondary to the decrease in the erythroid progenitor cell pool and to impaired proliferative capacity.15
There is also some evidence for an abnormal bone marrow stroma in the 5q− syndrome, and after treatment with lenalidomide there is a substantial improvement in the hematopoiesis-supporting potential of bone marrow stroma.16 The recent demonstration in the mouse that bone progenitor dysfunction induces myelodysplasia and secondary leukemia is of great interest.17 One of the paradoxes of all types of MDS is that the cells have a proliferative advantage in the bone marrow and yet grow poorly in vitro or in immunodeficient mice. This phenomenon remains unexplained and hampers research into MDS.
Genomic stability in the 5q− syndrome
The del(5q) is the sole karyotypic abnormality in the 5q− syndrome, and the relative clinical stability of this disorder may relate to the infrequency of additional abnormalities. In a study of patients with MDS and the del(5q), 3 genes known to be associated with disease progression in MDS were studied: FLT3, NRAS, and p53. No cases with the 5q− syndrome were found to harbor mutations of these genes, although p53 mutation was a feature of more advanced MDS cases with the del(5q).18 Similarly, Crescenzi et al19 described the deletion of several tumor suppressor genes in MDS patients with both the del(5q) and additional karyotypic changes. However, no additional deletions were observed in patients with the 5q− syndrome.19 Mutation of the JAK2 gene has been described in patients with the 5q− syndrome.20 A recent report described mutation of the JAK2 and MPL genes in 6% and 3%, respectively, of MDS patients with the 5q− syndrome; however, neither the phenotype nor prognosis was affected.2
A recent genome-wide single nucleotide polymorphism (SNP) analysis of 42 MDS patients with del(5q) determined whether additional genomic abnormalities occur. SNP array analysis can identify both copy number changes and regions of uniparental disomy (UPD). Copy number changes in addition to the del(5q) were found in approximately 50% of the 21 del(5q) MDS patients but in none of the 21 patients with the 5q− syndrome using the 50K Affymetrix SNP arrays. This study also showed that while small regions of UPD ( > 2 Mb) are common both in the 5q− syndrome and del(5q) MDS, large regions of UPD ( > 10 Mb) are found in del(5q) MDS but not in the 5q− syndrome.21
Cytogenetics and molecular mapping of the CDR
The del(5q) in the 5q− syndrome is considered to mark the location for a gene(s), the loss of which may affect important processes such as growth control and normal hematopoiesis.4 The 5q deletion is a large interstitial deletion, and most of the reported deletions fall into 1 of the following 3 types: del(5)(q13q31), del(5)(q13q33), and del(5)(q22q35). The cytogenetic characterization of 5q deletions present in the 5q− syndrome and other MDS and AML has not revealed any consistent differences in the breakpoints of the deletions between the different diseases.8
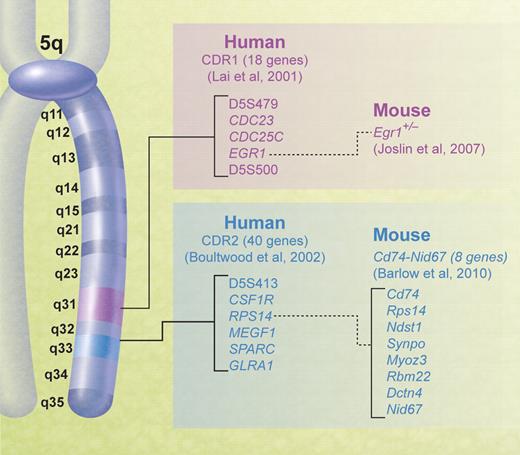
The large size of the typical chromosomal deletion in the 5q− syndrome has proved problematic in determining the molecular basis of the disease as the chromosomal region deleted contains hundreds of genes. The first step in the analysis of the molecular basis of diseases associated with large chromosomal deletions is the determination of a critical deleted region (CDR) since this is the region of deletion shared by all patients and localizes the gene(s) relevant for further study. Most 5q− syndrome patients share the large deletions referred to above, and so the majority of patients are not informative in terms of narrowing the CDR. Hence, a CDR is often based on exceptional patients with relatively small deletions. The 5q deletion breakpoints have been accurately determined in a large number of cases with the 5q− syndrome, but the 5q− syndrome CDR is largely based on 3 cases with small deletions (although visible by conventional cytogenetic analysis) of 5q.21-24 The phenotype of the 5q− syndrome is well established, and therefore the phenotype of these patients with small deletions could be confirmed as being typical of the syndrome.22 A strong phenotype/genotype relationship is generally not seen in other chromosome deletions in leukemia (including the association of 5q− and AML), and this introduces uncertainty to their analysis as to whether the determination of CDRs in exceptional cases can have a general applicability.
The CDR of the 5q− syndrome was established using molecular mapping and fluorescent in situ hybridization techniques by Boultwood and colleagues.23,24 The CDR was subsequently narrowed to the approximately 1.5-Mb interval at 5q32-q33 flanked by the DNA marker D5S413 and the GLRA1 gene (Figure 1).22 Genomic annotation of the CDR of the 5q− syndrome was performed, and several promising candidate genes mapping within the CDR were noted, including the tumor suppressor gene SPARC, RPS14, a component of the 40S ribosomal subunit, and several microRNA genes.22,25 All the 40 genes within the CDR were sequenced, and no mutations were found.25 The gene sequencing is critical to understanding the pathogenesis of the 5q− syndrome. For example, if Knudsen's 2-hit model applied to the 5q− syndrome, there would be a deletion of one gene on one chromosome (ie, the del5q) and a mutation of the remaining copy of the same gene. The lack of mutations in any of the genes mapping to the CDR suggests that haploinsufficiency (a dosage effect resulting from the loss of a single allele of a gene)26 is the basis of the 5q− syndrome. There has been growing recognition of haploinsufficiency as a cancer model27,28 during the past decade, and this is probably the correct model for the 5q− syndrome.
Chromosomal map of human 5q showing the positions of the CDRs in myeloid malignancies and corresponding mouse models.
Chromosomal map of human 5q showing the positions of the CDRs in myeloid malignancies and corresponding mouse models.
Expression analysis in hematopoietic stem cells from patients with the 5q− syndrome
The transcriptome of the CD34+ cells of patients with the 5q− syndrome has been determined using gene expression profiling.25 As expected, there are multiple genes down-regulated in the 5q− syndrome that map to the long arm of chromosome 5, consistent with a gene dosage effect.25 The CD34+ cells of 5q− syndrome patients have a distinct gene expression profile, suggesting a common underlying pathophysiologic basis to the disease.25,29
The majority of genes mapping to the CDR of the 5q− syndrome at 5q32-q33 show a reduction in expression levels consistent with the deletion of one allele in patients with the 5q− syndrome.25 Candidate genes identified as showing haploinsufficiency in the HSCs of patients with the 5q− syndrome by Boultwood and colleagues25 include the tumor suppressor gene SPARC and RPS14. Moreover, 2 genes mapping to the CDR, RBM22, and CSNK1A1 showed a reduction in gene expression of > 50% in some patients with the 5q− syndrome, consistent with the down-regulation of the remaining allele.25 The RBM22 gene, encoding a highly conserved RNA-binding protein, is the most significantly down-regulated gene mapping to the CDR of the 5q− syndrome. RBM22 has been shown to play a role in the regulation of gene splicing and apoptosis.30
Several genes including SPAG6, WIG-1, and BMI-1 are commonly up-regulated in the CD34+ cells of patients with the 5q− syndrome.25 Interestingly, WIG-1, a p53-induced gene, encodes a growth inhibitory protein,31 BMI-1 is necessary for maintenance of adult self-renewing HSC,32 and SPAG6 is markedly overexpressed in pediatric AML.33
Several significantly deregulated canonical gene pathways in the CD34+ cells of patients with the 5q− syndrome have been identified including Wnt/β-catenin signaling, protein ubiquitination, aminoacyl-tRNA biosynthesis, cell cycle:G1/S checkpoint, actin cytoskeleton signaling pathways,25 and most recently the p53 pathway.34
Candidate genes and disease mechanisms
RPS14
The RSP14 gene is a strong candidate gene for the 5q− syndrome based on evidence from several sources.35,36 The small subunit protein rpS14, the yeast homologue of the bacterial S11 protein, directly binds helix 28 of 18S rRNA and is essential for the assembly of 40S ribosomal subunits.37,38 The yeast ribosomal protein rpS14 is necessary for the endonucleolytic cleavage that removes 200 nucleotides from the 3′ end of 20S pre-rRNA to generate mature 18S rRNA and functional 40S ribosomal subunits.39 Upon depletion of rpS14, ribosomal proteins and rRNA destined for 40S subunits are rapidly degraded, whereas 60S subunits assemble at normal rates.38 The specific function of RPS14 in humans remains largely unknown; however, it is most probable that RPS14 is essential for the assembly of 40S ribosomal subunits.
Haploinsufficiency for RPS14 in the CD34+ cells of patients with the 5q− syndrome and the analogy with Diamond–Blackfan anemia (DBA) was noted some years ago.25,35 DBA is a disorder caused by haploinsufficiency for the related ribosomal gene RPS19 (also required for the maturation of 40S ribosomal subunits).40 DBA is a broad developmental disease characterized by anemia, bone marrow erythroblastopenia, and an increased malignancy.41 Mutations in RPS19 are found in approximately 25% of patients with DBA and lead to haploinsufficiency of RPS19.40 RPS19 protein and mRNA analysis confirm that expression from the normal RPS19 allele is not sufficient to compensate for the defective allele.40 Deficiency of RPS19 has been shown to block proliferation of immature erythroid progenitor cells.42 Interestingly, DBA has been associated with mutations now in 7 ribosomal protein genes, RPS19, RPS24, RPS17, RPL35A, RPL5, RPL11, and RPS7, in approximately 43% of patients.43
The anemia in DBA and the 5q− syndrome is a result of a failure of erythropoiesis, and both disorders show haploinsufficiency for ribosomal proteins RPS19 and RPS14, respectively, required for the maturation of 40S ribosomal subunits. The pivotal report from Ebert et al35 in 2008 identified RPS14 as a 5q− syndrome gene by an RNA interference screen of each gene within the CDR. RNA interference allows the functional characterization of deletions in leukemia, modeling hemizygous or homozygous inactivation depending on the efficacy of knockdown. The development of short hairpin RNA (shRNA) lentiviral expression vectors enables the use of RNA interference in both quiescent and proliferating progenitor cells. Three to 5 unique, lentivirally expressed shRNAs targeting each of the 40 genes in the region were introduced into normal CD34+ human bone marrow hematopoietic cells, and the effects of each shRNA on hematopoietic differentiation were determined. The knockdown of RPS14 recapitulated the phenotype of the 5q− syndrome: a block in erythroid differentiation (leading to erythroid cell apoptosis) with relative preservation of megakaryocyte differentiation as measured by fluorescence-activated cell sorter analysis.35 Forced expression of an RPS14 cDNA in primary bone marrow cells from patients with the 5q− syndrome rescued the phenotype.35 Thus, haploinsufficiency of RPS14 is the probable cause of the erythroid defect that characterizes the 5q− syndrome. RPS14 haploinsufficiency caused a block in the processing of preribosomal RNA and in the formation of the 40S ribosomal subunit.35 This ribosomal processing defect is highly analogous to the functional defect seen in DBA.
Moreover, similarities in the defective gene expression patterns observed in the CD34+ cells of patients with DBA and patients with the 5q− syndrome were recently reported by Pellagatti et al,29 including the down-regulation of multiple ribosomal genes (eg, RPL28) and genes involved in translation initiation (eg, EEF1D) and the up-regulation of several pro-apoptotic genes (eg, BAX). Using hierarchical clustering, patients with the 5q− syndrome could be separated both from other patients with refractory anemia and healthy controls solely on the basis of the deregulated expression of ribosomal- and translation-related genes, further suggesting that the 5q− syndrome represents a disorder of aberrant ribosome biogenesis.29 The down-regulation of multiple ribosomal genes in MDS patients with the del(5q) has recently been confirmed.44
Several other bone marrow failure syndromes are associated with defects in factors associated with ribosome synthesis including Shwachman-Diamond syndrome,45 dyskeratosis congenita, and cartilage hair hypoplasia.46 The demonstration of RPS14 as a 5q− syndrome gene adds to the body of evidence suggesting that defective ribosomal biogenesis may have a more general relevance in leukemogenesis.47,48
p53-dependent disease mechanism
As a key regulator of cell growth and cell death, the level of p53 is precisely regulated in cells. P53 is maintained at a low level during normal cell growth and is activated in response to various cellular stresses. Impaired ribosomal biogenesis, such as that resulting from haploinsufficiency of certain ribosomal proteins, for example RPS19 and RPS6 (and most probably RPS14), can cause nucleolar stress.49,50 In response to this stress, several ribosomal proteins including RPL5 and RPL11 bind to MDM2, the key regulator of p53,51,52 and block MDM2-mediated p53 ubiquitination and degradation.53,54 The increased half-life, from minutes to hours, quickly leads to higher levels of p53. Activated p53 then promotes the transcription of its many target genes, resulting in p53-dependent cell cycle arrest or apoptosis.55,56 In this way, the ribosomal protein-MDM2-p53 signaling pathway provides a molecular switch monitoring the integrity of ribosomal biogenesis.57
There is mounting evidence that up-regulation of the p53 pathway is a common response to deficiency of ribosomal proteins in various diseases.47 Indeed, the study of animal models of human disorders of defective ribosome biogenesis, including DBA and Treacher Collins syndrome, consistently show that ribosomal stress leads to activation of the p53 pathway and that this mechanism underlies the phenotypes characteristic of these disorders.50,58,59
The recent report of a “5q− mouse” is a major step forward in understanding the pathophysiologic basis of the 5q− syndrome. Barlow et al36 generated this mouse model using large-scale chromosomal engineering. Haploinsufficiency of the Cd74-Nid67 interval caused macrocytic anemia, prominent erythroid dysplasia, and monolobulated megakaryocytes in the bone marrow. The Cd74-Nid67 interval on mouse chromosome 18 is syntenic with a region within the CDR of the human 5q− syndrome22 and contains 8 known genes, 2 of which have been excluded (Ndst1 and Cd74), leaving Rps14, Synpo, Myoz3, Dctn-4, Rbm22, and Nid67 as candidates.36 It is most probable that Rps14 is the major gene in relation to the phenotype, taking into account the other lines of evidence from the in vitro screening35 and global gene expression analysis in the 5q− syndrome,29 although other genes within this region or in the CDR may also play a role.36,60
The “5q− mouse” has a defective bone marrow–progenitor development, the appearance of bone marrow cells expressing high amounts of the tumor suppressor p53 and increased bone marrow cell apoptosis. To investigate these findings further, the “5q− mouse” was crossed with p53-deficient mice. Significantly, this rescued the progenitor cell defect, restoring hematopoietic stem cell bone marrow populations.36 These data suggest that a p53-dependent mechanism underlies the pathophysiology of the 5q− syndrome.
Importantly, activation of p53 and up-regulation of the p53 pathway has recently been reported in the human 5q− syndrome.34 This most probably results from haploinsufficiency of the ribosomal gene RPS14. Ten genes in the p53 pathway are significantly deregulated in CD34+ cells from 5q− syndrome patients: FAS, CD82, WIG1, CASP3, SESN3, TNFRSF10B, MDM4, BAX, DDB2, and BID. All these genes were expressed at higher levels in 5q− syndrome compared with healthy controls, with the exception of MDM4 (a negative regulator of p53), which was expressed at lower levels in 5q− syndrome patients.34 Moreover, many of the most significantly up-regulated known genes in 5q− syndrome are p53 targets, including WIG-1, RPS27L, PHLDA3, BCL11B, FDXR, FAS, and BAX.25,29
An immunohistochemical analysis of p53 protein expression in bone marrow trephine sections from patients with 5q− syndrome showed moderate to strong p53 expression in all 3 cases of 5q− syndrome studied while only rare p53-positive cells were observed in normal bone marrow. By double immunostaining, it was shown that clusters of p53-positive cells with the morphologic appearance of erythroblasts were positive for glycophorin in all 3 patients with the 5q− syndrome.34
Activation of p53 as a result of ribosomal stress in the 5q− syndrome is likely to play an important role in the defective erythropoiesis and increased apoptosis observed in this disorder.34,36 Thus, p53 activation represents an important potential therapeutic target in the 5q− syndrome, and several p53 inhibitors are available including pifithrin.61 However, this will be a therapeutic option in humans only if this intervention does not abrogate the critical tumor suppressor function of p53.
In human disorders involving haploinsufficiency of 40S ribosomal proteins, it has been suggested that RPL11 up-regulation (or signaling components involved in this process) could be clinically targeted without interfering with other pathways involved in p53 induction. This proposal is based on the observation that disruption of biogenesis of 40S ribosomes leads to p53 induction mediated by rpL11.53,54 The increased free RPL11 generated after ribosomal stress is due to the cell selectively increasing the translation of mRNAs with a polypyrimidine tract at their 5′-transcriptional start site (5′-TOP mRNAs), including that encoding RPL11.54 Thus, if specific agents that target the signaling components involved in RPL11 up-regulation and/or that regulate 5′-TOP mRNA translation could be developed, there is the potential to alleviate the block in ribosomal biogenesis without involving other pathways involved in p53 induction.54 This is a key step in developing specific therapies for diseases associated with reduced ribosomal protein expression. Interestingly, rapamycin has been shown to selectively attenuate the translation of 5′-TOP mRNA, and an inhibitory effect of rapamycin on the translation of rpL11 mRNA has been demonstrated.54,62
miR-145 and miR-146a as mediators of the 5q− syndrome
Interestingly, work has been published recently on the identification of the microRNA (miRNAs) genes miR-145 and miR-146a as mediators of the 5q− syndrome phenotype.60 The miR-145 gene maps within the CDR of the 5q− syndrome,25 and the miR-146a gene maps adjacent to the distal boundary of the CDR and would also be deleted in virtually all patients with the human 5q− syndrome. Therefore, miR-145 and miR-146a are candidate genes on this basis.25,60 The report by Starczynowski and colleagues60 described the down-regulation of miR-145 and miR-146a in the CD34+ cells of 3 patients with the 5q− syndrome and identified TIRAP and TRAF6 as respective targets of these miRNAs. Knockdown of miR-145 and miR-146a together or enforced expression of TRAF6 in mouse HSCs resulted in thrombocytosis, mild neutropenia, and megakaryocytic dysplasia.60 Therefore, it was suggested that the thrombocytosis observed in some patients with the 5q− syndrome may be the result of deficiency of miR-145 and miR-146a.60 Although clear evidence for haploinsufficiency for RPS14 has been widely reported,25,35 normal expression levels of miR-145 were described in most 5q− syndrome patient CD34+ cell samples in a recent study.25 It has been shown that p53 enhances the posttranscriptional maturation of several miRNAs with growth suppressive function, including miR-145, in response to DNA damage.63 Another study has found that p53 transcriptionally induces the expression of miR-145 by interacting with a potential p53 response element in the miR-145 promoter.64 These findings offer a possible explanation why reduced levels of miR-145 have not been observed in the CD34+ cells of all 5q− patients (ie, while 1 allele of miR-145 is lost as a result of the del(5q), p53 activation may cause an increased expression of the remaining allele). Further work is needed to clarify these issues. A detailed study of the hematology of mice with knockouts for these miRNA genes would be very informative.65
Other candidate genes
Many candidate genes have been proposed for the 5q− syndrome and/or MDS/AML with a del(5q). Interestingly, the first candidate gene proposed was a gene for hemoglobin by Van den Berghe et al1 in their original publication. Subsequently, the presence of hematopoietic growth factor genes and receptors localized to 5q gave rise to other candidate genes being proposed such as GM-CSF.66 More recent candidate genes include IRF1,67 CTNNA1,68 SPARC,25,69,70 RBM22,25 EGR1,71 APC,72 and NPM1.73 Table 1 draws attention to the localization of candidate genes in particular CDRs and the phenotypes of mice with knockouts of these genes.
Cooperating events in the development of the 5q− syndrome
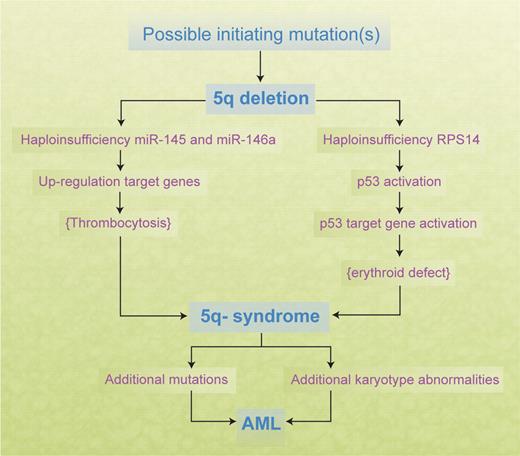
Mounting data strongly suggest that the RPS14 gene is an important gene in relation to the erythroid differentiation defect of the 5q− syndrome,35,36 and there is evidence suggesting that loss of the miRNA genes miR-145 and miR146a may play a role in the abnormalities of the megakaryocyte lineage observed.60 Thus, several cooperating genetic events may be necessary in the development of the 5q− syndrome (Figure 2). However, whether RPS14 or other identified candidate genes are causal genes in relation to producing a clonal hematopoietic disorder, and whether haploinsufficiency of an additional gene or genes is involved, remain important questions. Haploinsufficiency of other genes localized within the CDR22 such as the tumor suppressor gene SPARC could play a role in establishing clonal dominance. Clearly, further mouse knockout models might prove very informative in relation to all these questions.
Cooperating genetic events in the development of the 5q− syndrome and progression to AML.
Cooperating genetic events in the development of the 5q− syndrome and progression to AML.
Relationship of 5q− syndrome to other leukemias
The relationship of the 5q− syndrome to other leukemias possessing the del(5q) is clearly of importance as other types of MDS such as secondary MDS with del(5q) or MDS with additional karyotypic abnormalities to del(5q) have a poor prognosis in striking contrast to the good prognosis of the 5q− syndrome.4,5
The 1-1.5-Mb CDR at 5q31 identified in AML and the more advanced forms of MDS by Lai and coworkers74 is distinct from the CDR of the 5q− syndrome. This more proximal CDR is essentially defined by small numbers of patients with atypical 5q deletions, although a large group of patients with the del(5q) was studied in total.74,75 The AML CDR at 5q31 is flanked by D5S479 and D5S500 and contains 18 genes including the candidate gene EGR1 (Figure 1).74 No inactivating mutations have been described in any candidate genes mapping to this interval.76 However, it has been shown that Egr1-deficient mice treated with a DNA alkylating agent to induce secondary cooperating mutations developed immature T-cell lymphomas or a myeloproliferative disorder at increased rates and with shorter latencies than that of wild-type littermates.71 Low or absent expression of CTNNA1, a putative tumor suppressor gene also mapping to 5q31, has been described in a proportion of MDS/AML patients with a del(5q), and it has been suggested that markedly reduced expression levels of CTNNA1 may play an important role in MDS/AML with the del(5q).68 However, bone marrow cells from patients with the 5q− syndrome show expression levels consistent with the loss of one CTNNA1 allele only.14,77
The determination of CDRs in deletions associated with leukemia has been very informative, but the emphasis is now clearly moving toward the characterization of the role of specific genes in the 5q− syndrome which have been shown by mouse models or functional studies to be of biologic significance. In addition, other genes located on 5q but outside the CDRs have been published as having possible importance in these leukemias; these include NPM173 and APC.72 Interestingly, we have observed low levels of NPM1 expression in advanced MDS with the del(5q) but not in the 5q− syndrome. Whether low levels of NPM1 gene expression play a role in disease progression remains to be determined.
Although several genes have been postulated to have a role in these more severe forms of MDS or AML with a del(5q), the genes deleted in the majority of these cases are completely overlapping with those deleted in the 5q− syndrome since there is no difference in the location or magnitude of the 5q deletion.4,5 Of course, it is highly likely that the deletion of several genes along chromosome 5q may have various phenotypic effects. It may well be that the critical differences between these leukemias lie with mutations and other genomic aberrations on chromosomal regions outside 5q. Notably, the del(5q) in AML, particularly secondary AML, invariably occurs together with other karyotypic abnormalities and frequently as part of a complex karyotype. Interestingly, mutations with loss of function of p53 are significantly associated with the del(5q) in therapy-related MDS and therapy-related AML after previous treatment with alkylating agents and are associated with genetic instability.78 Moreover, an association between p53 mutation and advanced primary MDS with the del(5q) has been made.18 Intriguingly, a 5q− syndrome patient with a small clone of p53 mutated cells in the bone marrow, identified using a sensitive sequencing approach at time of diagnosis, was recently described. This subclone expanded during treatment with lenalidomide, with subsequent leukemic transformation.79
Lenalidomide
Until recent years, most patients with the 5q− syndrome were treated with best supportive care only. However, the thalidomide analog lenalidomide has been shown to have dramatic therapeutic effects in patients with the 5q− syndrome and other patients with MDS and a del(5q).80-83 An early study by List et al84 showed that lenalidomide has hematologic activity in patients with low-risk MDS who have no response to erythropoietin. The response rate was higher among patients with a clonal interstitial deletion involving chromosome 5q31.1 (83% compared with 57% among those with a normal karyotype and 12% among those with other karyotype abnormalities).
A further study showed that lenalidomide can reduce transfusion requirements and reverse cytologic and cytogenetic abnormalities in patients who have MDS with the 5q31 deletions.85 The patients in this study had a chromosome 5q31 deletion that was either isolated or accompanied by additional cytogenetic abnormalities, a disease of low- or intermediate-1 risk according to the International Prognostic Scoring System, and transfusion-dependent anemia. Interestingly, the response rate to lenalidomide was not significantly influenced by age, sex, French-American-British type, International Prognostic Scoring System, or cytogenetic pattern. A multivariate analysis showed that the hematologic response to lenalidomide was adversely affected by thrombocytopenia and a high requirement for transfusion. Among the 85 patients who could be evaluated, 62 had cytogenetic improvement, and 38 of the 62 had a complete cytogenetic remission.85
The US Food and Drug Administration approved the use of lenalidomide “for the treatment of patients with transfusion-dependent anemia due to low- or intermediate 1-risk myelodysplastic syndromes associated with a deletion 5q cytogenetic abnormality with or without additional cytogenetic abnormalities.” In contrast, the European Medicines Agency has refused marketing authorization for lenalidomide intended for the treatment of anemia due to MDS. The European Medicines Agency observed that “the high incidence of transformation to AML observed in the single-arm trial (MDS-003) is of concern since it could indicate an increased risk associated with lenalidomide. Given the noncomparative nature of the data, there is insufficient evidence to establish that the benefits outweigh the risks.”86
The study of Gohring et al87 shows that patients with del(5q) who fail to achieve sustained erythroid or cytogenetic remission after treatment with lenalidomide have an increased risk for clonal evolution and AML progression. Three and 5 years after study entry, the cumulative incidence of AML for patients with a cytogenetic response was 10% and 21%, respectively, and for patients without cytogenetic response, it was 46% and 60%. Progression to AML occurred in 7 of the 19 (37%) patients with MDS with isolated del(5q) and a normal blast count. This seems high compared with historical data regarding the frequency of transformation of the 5q− syndrome to AML and to the more recent data of Patnaik et al.2 It is clear that large, randomized, placebo-controlled studies are needed to answer the questions relating to the safety of lenalidomide in MDS. It would be valuable to predict which patients respond to lenalidomide, and an important first step in this direction was the establishment of an erythroid differentiation signature which predicts response to lenalidomide in MDS.88
The recent progress made in understanding the pathophysiology of the 5q− syndrome highlights the fact that the precise mode of action of lenalidomide in the 5q− syndrome remains uncertain.82 Lenalidomide has a wide range of potential properties ranging from inhibition of tumor necrosis factor-α and interleukin-6 to angiogenesis and stimulation of T cells and natural killer cells via induction of interleukin-2 and interferon-γ.80,81,89
The molecular targets of lenalidomide have been investigated by studying its in vitro effects on growth, maturation, and global gene expression in differentiating erythroblasts from patients with MDS with del(5)(q31). Lenalidomide inhibited growth of del(5q) erythroblasts but did not affect normal cells.70 Treatment with lenalidomide significantly influenced the pattern of gene expression in del(5q) intermediate erythroblasts, with VSIG4, PPIC, TPBG, activin A, and SPARC genes up-regulated by more than 2-fold in all samples.70 Up-regulation of the tumor suppressor gene SPARC is of particular interest because it is located at 5q32 within the CDR of the 5q− syndrome.22 The principal function of SPARC is the regulation of extracellular matrix interactions. SPARC has antiproliferative, anti-adhesive, and anti-angiogenic properties,90,91 all recognized effects of immunomodulatory drugs. The up-regulation of SPARC is one possible mode of action of lenalidomide. Interestingly, marked up-regulation of SPARC has been demonstrated in several non-Hodgkin lymphoma cell lines after treatment with lenalidomide.92
Another intriguing link between the mode of action of lenalidomide and genes on 5q was provided by Wei et al,93 who showed that lenalidomide inhibits 2 phosphatases that regulate the cell cycle, Cdc25C and PP2Acalpha. The genes for these phosphatases are located on 5q and are deleted in the majority of patients with 5q− syndrome. Haploinsufficiency for Cdc25C and PP2Acalpha gives rise to an enhanced sensitivity for lenalidomide in this model.93
Future
It is probable that further mouse models will be created which will prove insightful as to the genes and combination of genes important in the 5q− syndrome. A full understanding of the p53 mechanism may lead to new therapeutic approaches for the disease.
The relationship of the 5q− syndrome to other MDS and AML bearing 5q deletions will be pursued and may depend on genes localized outside 5q. Whole genome sequencing of the 5q− syndrome and related disorders will be critical here. In addition, a full genomic study of the 5q− syndrome and other forms of refractory anemia by whole genome sequencing and related technologies would be informative as to whether similar or different mutations are present across a spectrum of early MDS which result in clonal expansion.
The mode of action of lenalidomide is uncertain, but animal models of the 5q− syndrome will help unravel the complexities of its action and facilitate the development of other drugs. The 5q− syndrome has begun to deliver its long held promise to provide insights into the pathophysiology of MDS.
Note added in proof:
Following the review of this article, Tehranchi et al99 published a study demonstrating that malignant stem cells persist in del(5q) MDS in lenalidomide-induced remission.
Acknowledgments
We are grateful to Professor Douglas Higgs (Weatherall Institute of Molecular Medicine, Oxford) for critical reading of the manuscript and to Leukaemia & Lymphoma Research (UK) for support.
Authorship
Contribution: J.B., A.P., A.N.J.M., and J.S.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Jacqueline Boultwood, University Reader at University of Oxford, Co-director of the LLR Molecular Haematology Unit, Nuffield Department of Clinical Laboratory Sciences, John Radcliffe Hospital, Headington, Oxford OX3 9DU, United Kingdom; e-mail: jacqueline.boultwood@ndcls.ox.ac.uk.