Abstract
CD40/CD40 ligand (CD40L) cross-talk plays a key role in B-cell terminal maturation in the germinal centers. Genetic defects affecting CD40 cause a rare form of hyper-immunoglobulin M (IgM) syndrome, a disorder characterized by low or absent serum IgG and IgA, associated with recurrent infections. We previously reported on a few patients with homozygous CD40 mutations resulting in lack or severe reduction of CD40 cell surface expression. Here we characterize the 3 CD40 mutants due to missense mutations or small in-frame deletions, and show that the mutated proteins are synthesized but retained in the endoplasmic reticulum (ER), likely due to protein misfolding. Interestingly, the intracellular behavior and fate differ significantly among the mutants: progressive accumulation of the P2 mutant causes endoplasmic reticulum stress and the activation of an unfolded protein response; the mutant P4 is rather efficiently disposed by the ER-associated degradation pathway, while the P5 mutant partially negotiates transport to the plasma membrane, and is competent for CD40L binding. Interestingly, this latter mutant activates downstream signaling elements when overexpressed in transfected cells. These results give new important insights into the molecular pathogenesis of HIGM disease, and suggest that CD40 deficiency can also be regarded as an ER-storage disease.
Introduction
The hyper-immunoglobulin M (IgM) or HIGM syndrome is a rare immunodeficiency due to genetic defects in CD40L, CD40, or enzymes involved in class-switch recombination (CSR) and somatic hyper mutation (SHM).1,2 and references therein Five patients from 4 unrelated families with CD40 deficiency (also referred as type 3 HIGM) due to homozygous mutations in the CD40 gene (TNFRSF5) have been previously described by our group.3–5 From the immunologic standpoint these patients show normal to increased IgM levels, but impairment of CSR resulting in very low serum IgG and IgA, and low number of switched memory B cells.6 SHM and dendritic cell (DC) maturation have been originally investigated in some patients and found to be defective.3,7 As a consequence, these patients present recurrent bacterial and opportunistic infections similar to that observed for CD40L deficiency.
CD40 (NP_001241.1) is a cysteine-rich type I transmembrane cell surface protein belonging to the tumor necrosis factor receptor (TNF-R) superfamily. Besides B cells, it is also constitutively expressed on the surface of macrophages, monocytes, DCs, and several nonimmune cell types.8–10 The extracellular region of CD40 interacts with CD40L (CD154), a type II transmembrane molecule mainly expressed by activated CD4+ T lymphocytes and platelets. CD40/CD40L interaction causes CD40 trimerization and activation of intracellular signals. These signals promote B lymphocyte proliferation, homotypic cell adhesion, activation of CSR and SHM, and rescue of B cells from apoptosis. Furthermore, CD40-mediated signaling activates monocyte-derived dendritic cells, promoting their maturation and cytokine secretion.7 The signals are mediated through activation of TNF-Receptor Associated Factors (TRAFs), which in turn recruit different kinases leading to activation of nuclear factor (NF)-κB.11 Two distinct and evolutionarily conserved NF-κB signaling pathways have been described downstream of CD40.12 The “canonical” pathway involves NF-κB isoforms, which are normally kept in an inactive form in the cytoplasm by interaction with inhibitory proteins (IκB α, β, ϵ). After stimulation, IκBs are phosphorylated and degraded, thus allowing cytoplasmic NF-κB to translocate to the nucleus and drive expression of target genes. The “noncanonical” pathway does not involve inhibitor degradation but requires proteolytic cleavage of an inactive precursor (p100) into the p52 NF-κB subunit. p52 then forms dimers with other NF-κB factors to transactivate target genes.
Among the CD40 deficient patients, one showed no detectable CD40 protein, while the other patients carried either missense mutations or in-frame deletions that are theoretically permissive for the expression of a mutated protein. As shown for other genetic diseases, the presence of minor in-frame mutations may not impair protein synthesis in the endoplasmic reticulum (ER), but rather cause conformational alterations that are recognized by ER quality control mechanisms leading to retention of the mutant proteins in the ER.13,14 In many cases, the misfolded proteins are targeted, after a lag period, to an ER-associated degradation (ERAD) pathway, which consists of dislocation of the protein to the cytosol, ubiquitination and degradation by proteasomes.15,16 If the protein is not efficiently disposed of by ERAD, it progressively accumulates in the organelle causing ER stress, which may in turn activate a complex protective cell response called unfolded protein response (UPR).17–19 The UPR, through a transient block of protein synthesis and by potentiating the folding and degradation capacity of the ER, may restore homeostasis and favor cell survival. However, a strong or prolonged ER stress may activate maladaptive UPR pathways leading to cell apoptosis.20,21
In the present study we have investigated how the in-frame CD40 mutations affect protein biosynthesis and posttranslational maturation. Using B lymphoblastoid cell lines (BLCLs) derived from the patients as well as human embryonic kidney (HEK)-293 transfectants, we show that CD40 mutants are expressed, but retained in the ER, likely due to protein misfolding. However, the different CD40 mutants have different cellular behavior and elicit different cellular responses.
Methods
Cell lines and transfectants
BLCLs were obtained by Epstein-Barr virus transformation of peripheral blood mononuclear cells from 2 healthy subjects (C, C'), 4 CD40 deficient patients (P1, P2, P4, and P5) and from 2 carriers (P2′ mother and P5′ mother). BLCLs, Daudi, Jurkat, and HEK-293 cell lines were grown in RPMI 1640–10% fetal bovine serum (EuroClone). Primary B and T lymphocytes were separated from peripheral blood mononuclear cells of a normal subject by magnetic beads (Miltenyi Biotech). Control and mutant cDNAs were obtained from BLCLs RNA by reverse transcription, and polymerase chain reaction (PCR) amplification (5′-CACCGCTATGGTTCGTCTGC and 5′-AACTGCCTGTTTGCCCAC). The cDNAs were cloned using a TOPO-TA Cloning Kit (Invitrogen) and inserted into the pcDNA3 expression vector (Invitrogen). Transfection into HEK-293 cells was performed using Lipofectamine 2000 (Invitrogen) and stable transfectants were selected.
Protein sequence alignment and homology modeling
CD40 sequences from H sapiens (NP_001241.1), M mulatta (XP_001104333.1), E caballus (NP_001075371.1), B taurus (NP_001099081.1), S scrofa (NP_999359.1), C lupus familiaris (NP_001002982.1), and M musculus (NP_035741.2) were aligned with ClustalW (http://www.ebi.ac.uk/Tools/clustalW2/). CD40 structure was predicted with Geno3D (http://geno3d-pbil.ibcp.fr),22 mainly based on similarity with 1JMA,23 and visualized with PyMol Version 0.99 (DeLano Scientific).
Flow cytometry
Cells were stained either with a phycoerythrin (PE)-conjugated anti-CD40 (5C3, BD Biosciences) or with the 626.1 mAb anti-CD40 (kindly provided by Dr. D. Vercelli) followed by a PE-conjugated secondary antibody. Cells were acquired using a FACSCalibur (BD Biosciences) and analyzed by the FlowJo Version 8.8.6 software (TreeStar).
RT-PCR analysis
Total RNA was purified by the RNeasy kit (QIAGEN), treated on-column with DNaseI (QIAGEN), and reverse-transcribed by the ImProm II RT (Promega) with random hexamers. RT-PCR for XBP1 mRNA splicing detection was performed with primers flanking the XBP1 intron (5′-GAAGCCAAGGGGAATGAAGT and 5′-TGGGTCCAAGTTGTCCAGA). Real-time PCR for CD40 mRNA expression analysis was performed using Assays-on-Demand (Applera) designed on exons 1-2 and exons 5-6 junctions. Real-time PCRs for GRp78 (5′-AAAGAAGACGGGCAAAGATG and 5′-GCTTGATGCTGAGAAGACAGG) and ERdj4 (5′-GCCATGAAGTACCACCCTGA and 5′-CCACTACCTCTTTGTCCTTTACC) were performed with the iQ SYBR Green Supermix (Bio-Rad). Relative expressions were calculated by the qBase Version 1.3.5 software.20
Immunoblot
Cells were lysed at room temperature in 150mM NaCl, 50mM Tris-HCl pH7.4, 1% Nonidet P-40 (octylphenoxylpoly-ethoxyethanol), 10mM N-ethylmaleimide (Sigma-Aldrich) and protease inhibitors (Roche). CD40 expression analysis was performed on 11% sodium dodecyl sulfate polyacrylamide gel electrophoresis with a polyclonal rabbit anti-CD40 (H120, Santa Cruz Biotechnology). Deglycosylation of cell lysates was performed with either PNGaseF (Peptide: N-Glycosidase F) or EndoH (Endoglycosidase H; New England Biolabs). When time course deglycosylation was performed, CD40 was previously immunoprecipitated with mouse anti-CD40 (Lob11, Santa Cruz Biotechnology), and treated, for the indicated times, with 0.5 U PNGaseF. Surface biotinylation was performed in Hanks Balanced Salt Solution with 2mM N-Hydroxysuccinimidobiotin (Pierce) for 20 minutes on ice. Biotinylated proteins were immunoprecipitated with Streptavidin-Sepharose (Pierce) for 1 hour at 4°C, and together with leftovers were treated with EndoH. The degradation rate of CD40 mutants was analyzed by incubating cells for 1, 2, 4, and 8 hours with 100 μg/mL cycloheximide (CHX; Sigma-Aldrich). When indicated, 5μM MG132 was added (Sigma-Aldrich). CD40 bands were quantified by densitometric analysis using the ImageQuant Version 5.2 software (Molecular Dynamics). The GRp78 and GRp94 protein expression analysis was performed on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis using a mouse anti-KDEL (lys-asp-glu-leu) antibody (Stressgen). Either rabbit anti-actin or mouse anti-tubulin (Sigma-Aldrich) were used for loading controls.
Confocal microscopy
Cells were fixed and permeabilized in suspension using the Fix and Perm kit (Caltag Laboratories). The following antibodies were used: mouse anti-CD40 (lob11, Santa Cruz Biotechnology, followed by goat anti–mouse AlexaFluor568); rabbit anti-calnexin (USbiological, followed by goat anti–rabbit AlexaFluor488); rabbit anti-giantin AlexaFluor488-conjugate (Abcam); sheep anti-TGN46 (AbD Serotec, followed by donkey anti–sheep AlexaFluor488). For detection of endosomal recycling compartment, cells were incubated with transferrin AlexaFluor488-conjugate (Molecular Probes) for 1 hour at 37°C, before proceeding to CD40 staining. Cells were analyzed with the LSM510META confocal microscope (Zeiss).
Analysis of CD40 signaling
HEK-293 clones were stimulated for either 30 minutes or 5 hours at 37°C with 10 μg/mL recombinant CD40L (Glaxo) or 5 ng/mL recombinant TNFα (R&D Systems). Cells were lysed in a previously reported buffer,24 and sonicated. Lysates were separated on a 4%-12% gradient gel (Invitrogen) and analyzed by immunoblot using a rabbit anti-IκBα (C21, Santa Cruz Biotechnology), a rabbit mAb anti-p100/p52 (18D10, Cell Signaling Technology) and a mouse anti-tubulin (Sigma-Aldrich). The IκBα and p52 bands were quantified as described for immunoblots.
Results
CD40 mutations causing HIGM
Only 5 CD40 deficient patients, from 4 unrelated families, have been reported to date.3–5 Their clinical, molecular and immunologic features, relevant for the present study, are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Briefly, P1 mutation results in skipping of exon 5 in the CD40 mRNA leading to a premature stop codon, while the other patients carry missense mutations or small in-frame deletions, compatible with the expression of a mutant protein.
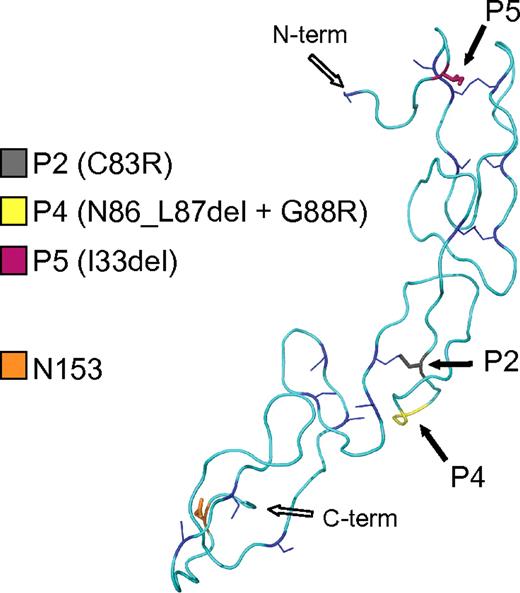
By sequence alignment of the CD40 protein of 7 mammalian species, we observed that C83, mutated in P2, as well as N86 and G88, mutated in P4, are invariant among mammals. Instead, the residue deleted in P5 (I33) is not conserved (supplemental Figure 1). We evaluated the effects of the mutations on a structural model of the CD40 extracellular domain based on homology to the ectodomain of Herpesvirus Entry Mediator A receptor, a member of the TNFR superfamily (Figure 1).23 The C83R mutation in P2 affects a cysteine involved in a conserved disulphide bridge in the second cysteine-rich region. By destroying this structurally important intramolecular bond, the C83R mutation is predicted to lead to a major conformational change. In the P4 mutant, the deleted residues and the substituted glycine are 3 amino acids away from C83. The deletion may perturb the formation of the conserved disulphide bridge formed by C83 by modifying the orientation of the cysteines relative to each other due to a shortened loop. The I33 deletion of the P5 mutant occurs in the first cysteine-rich region, and may shorten the distance between the cysteines at position 26 and 37, presumably preventing disulphide bridge formation and resulting in a distorted structure.
CD40 mutations causing HIGM. Structural model of the CD40 extracellular domain (amino acid 26 to 157) showing the positions of the P2, P4, and P5 mutations (solid black arrows) and the position of 1 of the 2 N-glycosylation sites (orange box). The cysteine residues and the conserved disulphide bridges are shown in blue.
CD40 mutations causing HIGM. Structural model of the CD40 extracellular domain (amino acid 26 to 157) showing the positions of the P2, P4, and P5 mutations (solid black arrows) and the position of 1 of the 2 N-glycosylation sites (orange box). The cysteine residues and the conserved disulphide bridges are shown in blue.
Expression analysis of CD40 mutants
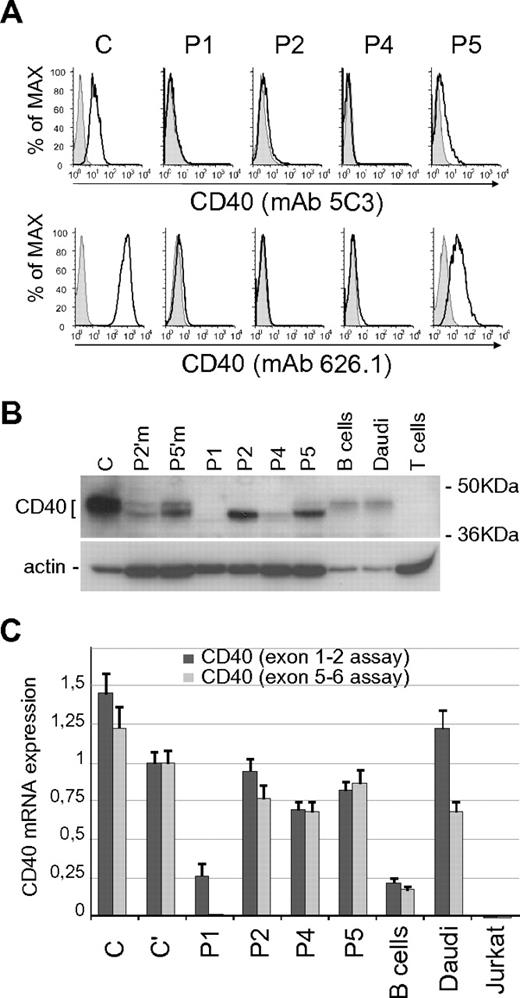
We investigated the expression of CD40 mutants in BLCLs derived from patients. By flow cytometric analysis, P1, P2, and P4 BLCLs lack CD40 surface expression, whereas P5 BLCL shows a residual CD40 (Figure 2A). The analysis was performed using 2 monoclonal antibodies recognizing different epitopes, mAb 5C3 and mAb 626.1, the latter binding at the CD40L binding site.25,26 Notably, a similar pattern of CD40 expression was observed in primary B cells obtained from patients (supplemental Figure 2).3–5 We then analyzed by immunoblot the electrophoretic profile of the CD40 mutants, in comparison with wild-type CD40 from control BLCLs (C), purified B cells, and Daudi cells. Comparison was also made with CD40 from the cells of 2 heterozygous carriers (Figure 2B). Our data confirmed previous results showing that the P1 mutant was not expressed, and the P2 mutant had a molecular weight lower than normal.3 Data also showed that P4 mutant resembled P2, although its expression level was significantly reduced, and the electrophoretic profile of the P5 mutant showed both a lower sized band and a faint band with an apparently normal molecular weight (Figure 2B).
Expression analysis of CD40 mutants. (A) Surface expression of CD40 in BLCLs from control (C) and from P1, P2, P4, and P5 patients, analyzed by flow cytometry with mAb 5C3-PE conjugated (top panels) and mAb 626.1 (bottom panels). Negative staining is shown as shaded peaks. (B) Total expression of CD40 analyzed by immunoblot with a polyclonal anti-CD40 antibody (H120), comparing CD40 expression in controls (C; primary B cells; Daudi cell line), heterozygous carriers (P2′m, P2′ mother; P5′m, P5′ mother) and mutants (P1; P2; P4; P5). T cells were used as negative control. Immunoblot for β-actin is shown as loading control. (C) CD40 mRNA expression in controls (C and C'; primary B cells; Daudi and Jurkat cell lines) and in patients' BLCLs, analyzed by real-time PCR using 2 Taqman assays designed on exons 1-2 (dark gray) and exons 5-6 (light gray) junctions. Expression values, relative to β-actin, are normalized to the expression observed in (C). Each analysis was performed with triplicate samples, and data are shown as means ± SD.
Expression analysis of CD40 mutants. (A) Surface expression of CD40 in BLCLs from control (C) and from P1, P2, P4, and P5 patients, analyzed by flow cytometry with mAb 5C3-PE conjugated (top panels) and mAb 626.1 (bottom panels). Negative staining is shown as shaded peaks. (B) Total expression of CD40 analyzed by immunoblot with a polyclonal anti-CD40 antibody (H120), comparing CD40 expression in controls (C; primary B cells; Daudi cell line), heterozygous carriers (P2′m, P2′ mother; P5′m, P5′ mother) and mutants (P1; P2; P4; P5). T cells were used as negative control. Immunoblot for β-actin is shown as loading control. (C) CD40 mRNA expression in controls (C and C'; primary B cells; Daudi and Jurkat cell lines) and in patients' BLCLs, analyzed by real-time PCR using 2 Taqman assays designed on exons 1-2 (dark gray) and exons 5-6 (light gray) junctions. Expression values, relative to β-actin, are normalized to the expression observed in (C). Each analysis was performed with triplicate samples, and data are shown as means ± SD.
We performed CD40 mRNA analysis by real-time PCR assays (Figure 2C). For this purpose, since different CD40 mRNA isoforms are generated by alternative splicing,27,28 we used 2 different assays designed on the exons 1-2 and exons 5-6 junctions. In particular, the use of the exon 5-6 assay was meant to exclude the exon 5 skipping isoform, previously identified in P1 mRNA,3 as well as the isoform with skipping of both exons 5 and 6. Levels of CD40 mRNA were similar in BLCLs from patient P2, P4, P5, and controls. Reduced expression of CD40 mRNA was observed in BLCL from patient P1 using the exon 1-2 assay, and no transcript was detected using the exon 5-6 junction probe, thus demonstrating the absence of full-length CD40 mRNA.
N-glycosylation pattern of CD40 mutants
Based on the lack of CD40 cell surface expression and on CD40 protein modeling, we hypothesized that P2, P4, and P5 mutants were retained in the ER. We studied their N-linked glycan maturation based on the assumption that the N-glycan processing proceeds in a precise order along the exocytic pathway and reflects the intracellular localization of the glycoprotein. If the protein is trapped in the ER, the N-glycans remain in the high-mannose state, while the protein transported to plasma membrane bears high molecular weight complex sugars.
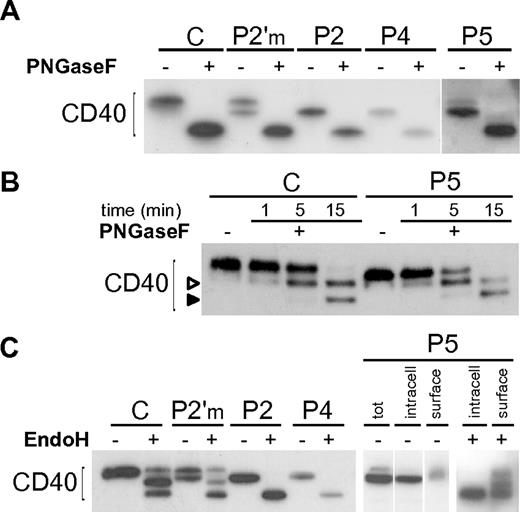
We first performed a complete deglycosylation of the CD40 mutants with PNGaseF, a N-glycosidase that cleaves N-linked sugars, and showed that their lower molecular weight was due to differences in the N-glycan moieties (Figure 3A). By a time-course digestion with PNGaseF we showed that the 2 predicted glycosylation sites (N153 and N180) were used in both wild-type and mutants (Figure 3B), demonstrating that the different glycosylation of mutants was not due to the loss of a glycosylation site, but rather to a different sugar maturation state. Thus we tested their susceptibility to EndoH, an N-glycanase that cleaves only high-mannose or hybrid N-glycans, but not complex oligosaccharides. As hypothesized, P2 and P4 mutants were both fully susceptible to EndoH treatment (Figure 3C), unlike wild-type CD40 displaying a composite pattern probably due to the existence of a plasma membrane fraction bearing hybrid sugars. A partial susceptibility to EndoH could also be seen in purified B cells from a healthy donor (supplemental Figure 3A). As shown in Figure 2B, the CD40 P5 mutant appeared as a doublet, likely representing an ER-retained form and a less abundant plasma membrane form. We separated cell surface proteins from the intracellular proteins by surface biotinylation, prior to EndoH treatment, and demonstrated that the lower band of P5 mutant was intracellular and fully susceptible to EndoH, while the upper band, in the surface fraction, resembled control CD40 (Figure 3C).
N-glycosylation of CD40 mutants. (A) Immunoblot showing treatment of Control (C), an heterozygous carrier (P2′m) and CD40 mutants with PNGaseF. (B) A time-course treatment with PNGaseF was done for C and one of the mutants (P5), showing the appearance of a mono-glycosylated intermediate (empty arrowhead), and a complete deglycosylated product (solid arrowhead). (C) Immunoblot showing treatment of C and CD40 mutants with EndoH. P5 (right end panels) was analyzed after separation of cell surface biotinylated proteins from intracellular proteins. Vertical lines have been inserted to indicate repositioned gel lanes.
N-glycosylation of CD40 mutants. (A) Immunoblot showing treatment of Control (C), an heterozygous carrier (P2′m) and CD40 mutants with PNGaseF. (B) A time-course treatment with PNGaseF was done for C and one of the mutants (P5), showing the appearance of a mono-glycosylated intermediate (empty arrowhead), and a complete deglycosylated product (solid arrowhead). (C) Immunoblot showing treatment of C and CD40 mutants with EndoH. P5 (right end panels) was analyzed after separation of cell surface biotinylated proteins from intracellular proteins. Vertical lines have been inserted to indicate repositioned gel lanes.
Finally, we evaluated the N-glycan processing of CD40 by a pulse chase experiment. Wild-type CD40 cotranslationally acquired high-mannose sugar chains which slowly mature to higher molecular weight complex sugars, reflecting a correct transport to the cell surface. In contrast, even after 12 hours of chase, the CD40 mutants appeared in the high-mannose glycosylation state, not distinguishable from that observed in cells treated with drugs that prevent the ER to Golgi protein transport (supplemental Figure 3B-C). On the whole, these results demonstrate that the CD40 mutants are actively kept within the ER, apart from a fraction of P5 which is correctly targeted to the plasma membrane.
Localization of CD40 mutants
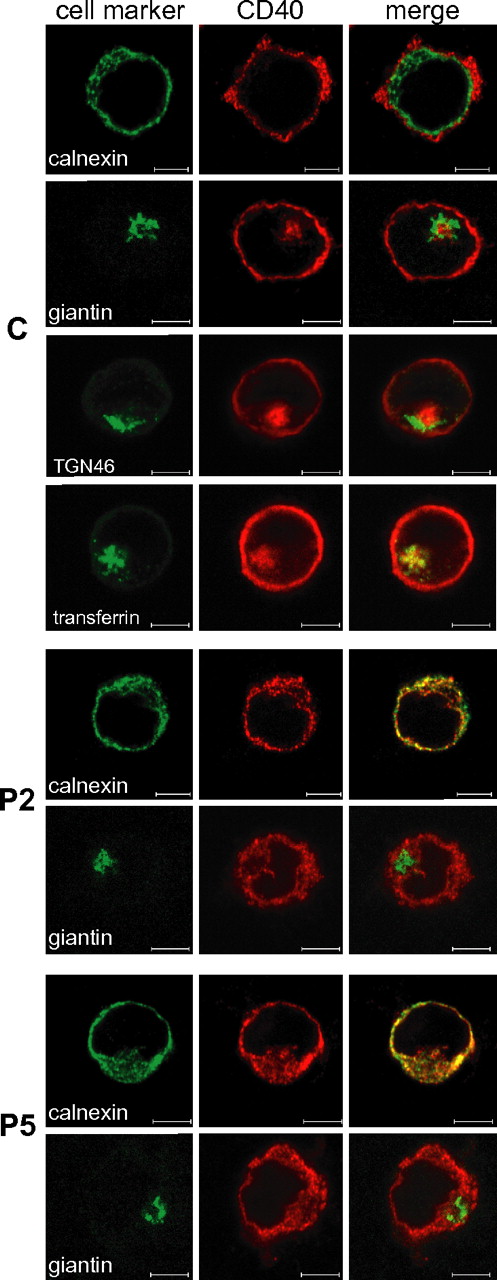
We used confocal microscopy to define the subcellular localization of CD40 mutants (Figure 4), which were observed to colocalize with calnexin (an ER marker), confirming their retention in the ER. No colocalization was observed neither with giantin (a cis-medial Golgi marker), or with trans-Golgi network marker TGN46 (supplemental Figure 4). As expected, wild-type CD40 mainly localized on the cell surface but interestingly a fraction was also localized in the recycling endosomal compartment, as shown by superimposition of CD40 signal and fluorescently labeled transferrin, internalized via its receptor (Figure 4). In agreement with their ER retention, CD40 mutants were not detected in the recycling endosomal compartment (supplemental Figure 4).
Localization of CD40 mutants in the ER. Intracellular double staining of CD40 (AlexaFluor568) with either calnexin, an ER marker (AlexaFluor488), or giantin, a Golgi Apparatus marker (AlexaFluor488-conjugate), in BLCLs from control (C) and from P2 and P5 patients. A further double staining was performed on C either with TGN46, a Trans Golgi Network marker (AlexaFluor488), or after cell incubation with fluorescently labeled transferrin (AlexaFluor488-conjugate) that localizes to the recycling endosomal compartment. Colocalization is visible as yellow staining in the merged micrographs. Scale bars, 5 μm. Analysis was performed by the LSM510META confocal microscope (Zeiss) with a EC Plan-Neofluar 100×/1.30 oil objective and a scan zoom of 1-1.5 ×. Digital images were analyzed with the LSM Image Browser (Zeiss) and processed with Adobe Photoshop 6.0 (Adobe Systems).
Localization of CD40 mutants in the ER. Intracellular double staining of CD40 (AlexaFluor568) with either calnexin, an ER marker (AlexaFluor488), or giantin, a Golgi Apparatus marker (AlexaFluor488-conjugate), in BLCLs from control (C) and from P2 and P5 patients. A further double staining was performed on C either with TGN46, a Trans Golgi Network marker (AlexaFluor488), or after cell incubation with fluorescently labeled transferrin (AlexaFluor488-conjugate) that localizes to the recycling endosomal compartment. Colocalization is visible as yellow staining in the merged micrographs. Scale bars, 5 μm. Analysis was performed by the LSM510META confocal microscope (Zeiss) with a EC Plan-Neofluar 100×/1.30 oil objective and a scan zoom of 1-1.5 ×. Digital images were analyzed with the LSM Image Browser (Zeiss) and processed with Adobe Photoshop 6.0 (Adobe Systems).
Role of ERAD in the turnover of CD40 mutants
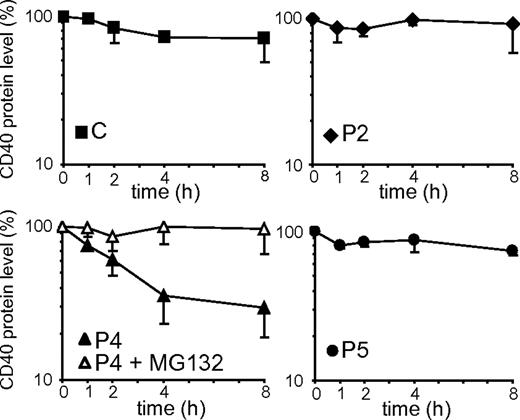
Misfolded proteins retained in the ER can be delivered to degradation by quality control mechanisms. The turnover rate of the CD40 mutants was investigated by culturing BLCLs with CHX, an inhibitor of protein synthesis. The CD40 expression values obtained by densitometric analysis of immunoblots are shown in Figure 5. We observed that control CD40, as well as P2 and P5 mutants, remained relatively stable over the 8 hours of treatment. In contrast, the P4 mutant had a faster degradation rate, with a half-life of approximately 3 hours. Treatment with MG132, an inhibitor of proteasomal degradation, prevented degradation of P4 CD40, showing that its degradation is mediated by proteasomes. Since no significant difference was seen in the mRNA expression level between P4 and the other CD40 variants (Figure 2C), the lower steady state level of P4 mutant could be explained by the faster turnover rate of the protein.
Degradation rates of CD40 mutants. The graphs represent the decay of control (C) and CD40 mutants in a time-course treatment of BLCLs with CHX. The protein expression values were determined by densitometric analysis of anti-CD40 immunoblots and were expressed as percentages of the CD40 expression in untreated cells (0 hour). In the graph representing P4, the empty symbols show CD40 decay in the presence of both CHX and MG132. Each cell line was analyzed in at least 2 independent experiments, and data are given as means ± SD.
Degradation rates of CD40 mutants. The graphs represent the decay of control (C) and CD40 mutants in a time-course treatment of BLCLs with CHX. The protein expression values were determined by densitometric analysis of anti-CD40 immunoblots and were expressed as percentages of the CD40 expression in untreated cells (0 hour). In the graph representing P4, the empty symbols show CD40 decay in the presence of both CHX and MG132. Each cell line was analyzed in at least 2 independent experiments, and data are given as means ± SD.
Analysis of UPR activation by CD40 mutants
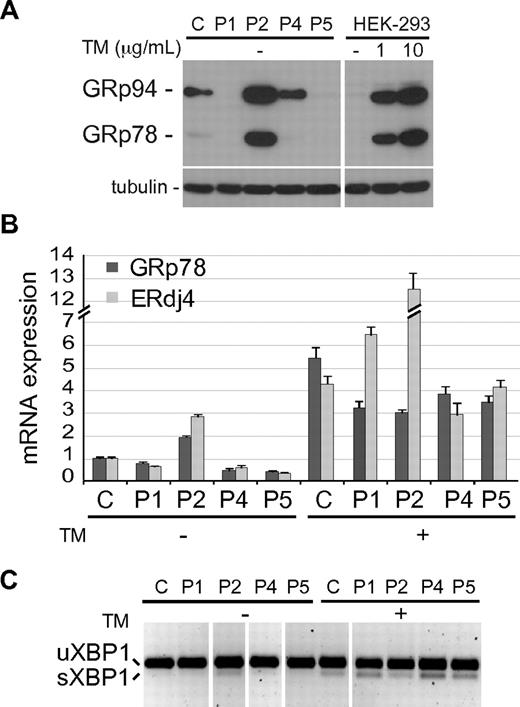
The accumulation of unfolded proteins in the ER may activate a UPR, a complex cell stress response aimed at restoring the ER homeostasis. Up-regulation of the ER chaperone GRp78/BiP is one of the best indicators of UPR activation in an ER undergoing chronic stress, such as that caused by the retention of a mutated protein that is expressed constitutively.29 We hence analyzed GRp78 protein expression in BLCLs from different patients by immunoblot and a remarkable increase was observed in P2 cells, comparable with that observed in HEK-293 cells treated with Tunicamycin (TM), an ER stress inductor (Figure 6A). Interestingly, the ER chaperone GRp94, another UPR-induced protein, was also significantly up-regulated in cells expressing the P2 mutant, although a lower induction was also observed in cells expressing normal CD40 or P4 mutant. Induction of GRp78, as well as that of ERdJ4 (a cochaperone of GRp78) mRNAs was also observed by real-time PCR analysis in the P2 BLCLs (Figure 6B). Furthermore, the spliced mRNA form of XBP1, a key transcription factor mediating UPR, was detected in P2 cells by PCR (Figure 6C).30 Taken together, these results show that accumulation in the ER of P2, but not of the other CD40 mutants, activates a UPR.
UPR induction in P2 BLCL. (A) Immunoblot showing the expression level of GRp78 and GRp94 in control (C) and patient's BLCLs. HEK-293 cells, untreated (-) or treated for 6 hours with 2 different doses of Tunicamycin (TM) are shown as control of UPR induction. Tubulin was used as loading control. (B) Real-time PCR analysis of GRp78 and ERdJ4 mRNA expression, in C and mutant BLCLs, untreated or treated with TM. Expression values, relative to β-actin, are normalized to the expression observed in C. The experiment shown is representative of 2 independent experiments with triplicate samples, and data are given as means ± SD. (C) RT-PCR analysis of XBP1 mRNA, spliced (sXBP1) and unspliced (uXBP1). The experiment shown is representative of 2 independent experiments. Vertical lines have been inserted to indicate repositioned gel lanes.
UPR induction in P2 BLCL. (A) Immunoblot showing the expression level of GRp78 and GRp94 in control (C) and patient's BLCLs. HEK-293 cells, untreated (-) or treated for 6 hours with 2 different doses of Tunicamycin (TM) are shown as control of UPR induction. Tubulin was used as loading control. (B) Real-time PCR analysis of GRp78 and ERdJ4 mRNA expression, in C and mutant BLCLs, untreated or treated with TM. Expression values, relative to β-actin, are normalized to the expression observed in C. The experiment shown is representative of 2 independent experiments with triplicate samples, and data are given as means ± SD. (C) RT-PCR analysis of XBP1 mRNA, spliced (sXBP1) and unspliced (uXBP1). The experiment shown is representative of 2 independent experiments. Vertical lines have been inserted to indicate repositioned gel lanes.
Analysis of P5 expression and signaling in HEK transfectants
To investigate whether the P5 mutation, partially permissive for CD40 surface expression, could be also permissive for signal transduction, we analyzed the 2 major NF-κB pathways triggered by CD40 engagement: the “canonical” pathway that involves degradation of IκBs (that takes place within minutes) and the “noncanonical” pathway involving the processing of p100 to p52 (that requires several hours). We first assessed the ability of P5 mutant to bind recombinant monomeric CD40L in the BLCLs from the P5 patient. However, to investigate CD40 signaling, the BLCL-based system is inadequate, due to an overlap between the CD40 signaling pathway and the one used by the Epstein-Barr virus latent membrane protein 1.31 Therefore, we established a model based on stable transfectants in HEK-293 cells expressing control CD40 (HEK-C) and P5 mutant. We isolated 3 clones expressing P5 mutant CD40 protein based on the levels of CD40 surface expression. Of these, clone HEK-P5-cl1 expressed P5 CD40 mutant at levels comparable with HEK-C; clone HEK-P5-cl2 had intermediate (30% of HEK-C) levels of expression of P5 CD40 mutant, whereas clone HEK-P5-cl3 expressed low (4% of HEK-C) levels of P5 CD40 mutant protein (supplemental Figure 5A). The low levels of expression observed in HEK-P5-cl3 clone recapitulate the levels of expression detected on P5 BLCLs compared with control BLCLs. By immunoblot analysis, P5 transfectants showed a mixed electrophoretic profile and a susceptibility to PNGaseF and EndoH comparable with that observed in P5 BLCLs (supplemental Figure 5B).
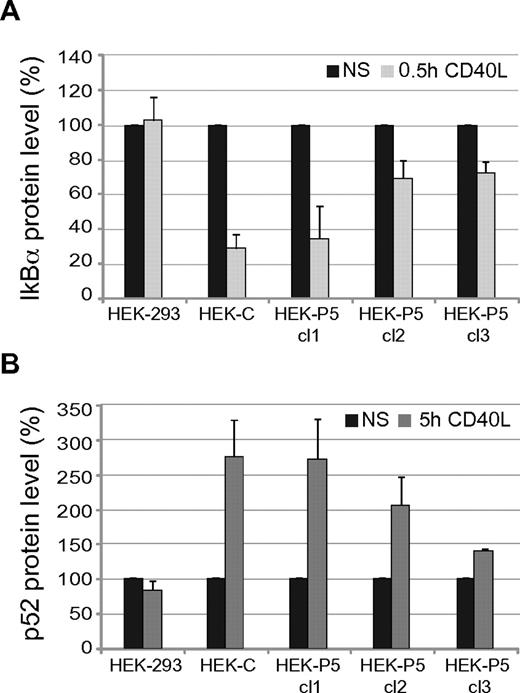
We studied CD40-mediated signal transduction by analyzing both NF-κB pathways in HEK-293 transfectants treated with soluble CD40L for 30 minutes or 5 hours. To this aim we performed immunoblots and determined by densitometric analysis the expression levels of IκBα and p52 (Figure 7A and B, respectively, and supplemental Figure 6). Stimulation with CD40L activated both NF-κB pathways in cells expressing CD40, while it was ineffective in the HEK-293 parental cells. In particular, HEK-C stimulation led to almost complete IκBα degradation after 30 minutes and induction of p52 formation after 5 hours. Stimulation with CD40L of clones expressing P5 mutant also activated both NF-κB pathways; interestingly, the extent of signaling correlated with the amount of CD40 expression on the cell surface. These data clearly demonstrate that the fraction of P5 transported to the cell surface is competent for signaling.
IκBα degradation and p52 production in P5 mutant. Graphs showing IκBα degradation (panel A) and p52 formation (panel B) after stimulation with CD40L of HEK-293 cells (HEK-293), a clone expressing control CD40 (HEK-C), and 3 clones showing decreasing expression of P5 mutant (HEK-P5 cl1, cl2, cl3). Stimulation induced IκBα degradation and p52 formation only in CD40 expressing cells depending on CD40 surface expression levels. The protein expression values were determined by densitometric analysis of immunoblots from 2 independent experiments and were expressed as percentages (means ± SD) of the values in nonstimulated cells (NS).
IκBα degradation and p52 production in P5 mutant. Graphs showing IκBα degradation (panel A) and p52 formation (panel B) after stimulation with CD40L of HEK-293 cells (HEK-293), a clone expressing control CD40 (HEK-C), and 3 clones showing decreasing expression of P5 mutant (HEK-P5 cl1, cl2, cl3). Stimulation induced IκBα degradation and p52 formation only in CD40 expressing cells depending on CD40 surface expression levels. The protein expression values were determined by densitometric analysis of immunoblots from 2 independent experiments and were expressed as percentages (means ± SD) of the values in nonstimulated cells (NS).
Discussion
We have focused the present study on the molecular behavior of the CD40 mutants bearing either missense mutations or small in-frame deletions, permissive for protein synthesis. We demonstrate that these CD40 protein mutants are retained in the ER, likely due to specific conformational alterations in the CD40 protein. Both P2 and P4 mutations affect highly conserved amino acids in the second cysteine-rich domain of the molecule, impairing the formation of a conserved disulphide bridge, according to structural modeling. The I33 residue deleted in P5 mutant is not conserved and likely introduces a local perturbation in the first cysteine-rich domain, although the alteration is partially permissive for folding in a native-like conformation as demonstrated by the residual surface expression detected on primary B cells and BLCLs from P5 patient.
On the basis of ER retention of some CD40 mutants, we propose that CD40 deficiency can be included among ER-storage diseases.32–34 These diseases include cystic fibrosis, alpha-antitrypsin deficiency, neurodegenerative disorders, and several endocrinopathies. Interestingly, mutations of a receptor belonging to the same CD40 superfamily, the TNFR1 protein, that cause the TNF receptor associated periodic syndrome, have been also associated to protein misfolding and ER retention.35 A common trait of ER-storage diseases is the loss-of-function due to protein mislocalization. However, the mutated proteins may have different fates; in most cases they are efficiently disposed of by ERAD, but, if the degradation process is insufficient, the protein accumulates and may trigger a UPR that protects the cell by enhancing the ER folding and degradation capacity. Instead, in a subgroup of ER-storage disorders, the intracellular accumulation of the mutated protein results in severe ER stress, that may compromise cell function or lead to cell death. Important differences among CD40 mutants have been disclosed with regard to their turnover rate and cell stress responses. P2 mutant appears stable, progressively accumulates in the ER, and triggers a UPR as assessed by GRp78 overexpression. Instead, P4 mutant is degraded by the ERAD pathway, with a half-life of approximately 3 hours. This faster degradation may explain the very low amount of the P4 mutant detected in BLCLs derived from the patient. On the other hand, the P5 mutant is neither degraded nor seems to activate a canonical UPR, despite its ER accumulation. A very intriguing feature of this mutant is the residual CD40 expression at the plasma membrane. Based on structural data, the region affected by the mutation is not directly involved in binding to CD40L.36 Consistently, we have demonstrated that P5 BLCL and P5 HEK-293 transfectants can bind recombinant CD40L. Moreover, upon stimulation with CD40L, HEK-293 cells expressing the P5 mutant can activate 2 independent NF-κB signaling pathways. Interestingly, we demonstrated that the signal transduction capacity increases progressively with the increase of the surface expression of the receptor. Based on these results we suggest that the impaired CD40-mediated B cell activation observed in patient P5 may be the consequence of the very low surface expression of CD40 rather than of an intrinsic functional defect of the P5 mutant. It should be noted that the 50% expression of CD40 observed in the heterozygous carriers is sufficient to induce a normal immunologic response. On the other hand, the antibody deficiency observed in P5 patients was as severe as in the other patients.
Novel therapeutic strategies aimed at promoting folding in the ER and transport along the exocytic pathway have been attempted for other ER-storage diseases. Transport of the cystic fibrosis transmembrane conductance regulator ΔF508 mutant, the most common mutant causing cystic fibrosis, has been facilitated by treating cells at low temperature or with chemical chaperones such as osmolytes and 4-phenyl butyric acid.37 Recently, the use of modifiers of N-glycosylation such as kifunensine was also proposed as a strategy to restore surface expression of INFγR mutants retained in the ER and causing the atypical mycobacterial disease.38 Similar approaches would be feasible to promote cell surface expression of selected CD40 mutants retained in the ER. The P5 mutant represents an interesting model in this respect, since it appears to be competent for signaling once expressed at higher levels. Preliminary data on BLCLs from patient P5 indicate that exposure of these cells at low temperature promotes transport of the receptor to the cell surface (supplemental Figure 7). This suggests that treatment with chemical chaperones may in principle rescue surface expression of this mutant.
Taken together our results show that CD40 deficiency can be associated to trapping of the mutated proteins in the ER, and show that this primary immunodeficiency can also be regarded as an ER-storage disease. Since the CD40 mutants undergo different intracellular fate, detailed analysis of individual mutants is required to fully understand the pathogenesis of the disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Roberto Sitia, Paolo Dellabona, and Raif Geha for suggestions and discussions, Donata Vercelli and Maria Luisa Malosio for providing reagents, and Luisa Massardi and Valentina Albertini for technical help.
This work was supported by European Union EURO-POLICY-PID SP23-CT-2005-006411 to L.D.N. and S.G., University of Brescia and Fondazione Angelo Nocivelli to S.G., Italian Telethon Foundation GSP02476 to S.F., and Academy of Finland, the Medical Research Fund of Tampere University Hospital, and Sigrid Juselius Foundation to M.V.
Authorship
Contribution: G.L., S.F., A.M.F., and S.G. designed research; G.L., S.C., and A.M.F. performed research; M.V. elaborated structural model; N.K., A.P., and L.D.N. provided patient's clinical data; L.S. provided comments and support; G.L., A.M.F., and S.G. wrote the manuscript; and L.D.N. reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna Maria Fra, General Pathology and Immunology, Dept of Biomedical Sciences and Biotechnology, University of Brescia, Vle Europa, 11-25123 Brescia, Italy; e-mail: fra@med.unibs.it; or Silvia Giliani, A. Nocivelli Institute for Molecular Medicine and Pediatric Clinic, University of Brescia, Ple Spedali Civili, 1-25123 Brescia, Italy; e-mail: giliani@med.unibs.it.
References
Author notes
A.M.F. and S.G. share the senior authorship.