Abstract
Autosomal recessive STAT1 deficiency is associated with impaired cellular responses to interferons and susceptibility to intracellular bacterial and viral infections. We report here a new form of partial STAT1 deficiency in 2 siblings presenting mycobacterial and viral diseases. Both carried a homozygous missense mutation replacing a lysine with an asparagine residue at position 201 (K201N) of STAT1. This mutation causes the abnormal splicing out of exon 8 from most STAT1 mRNAs, thereby decreasing (by ∼ 70%) STAT1 protein levels. The mutant STAT1 proteins are not intrinsically deleterious, in terms of tyrosine phosphorylation, dephosphorylation, homodimerization into γ-activating factor and heterotrimerization into ISGF-3, binding to specific DNA elements, and activation of the transcription. Interestingly, the activation of γ-activating factor and ISGF3 was impaired only at early time points in the various cells from patient (within 1 hour of stimulation), whereas sustained impairment occurs in other known forms of complete and partial recessive STAT1 deficiency. Consequently, delayed responses were normal; however, the early induction of interferon-stimulated genes was selectively and severely impaired. Thus, the early cellular responses to human interferons are critically dependent on the amount of STAT1 and are essential for the appropriate control of mycobacterial and viral infections.
Introduction
Interferons (IFNs) are important mediators of immunity.1 STAT1 plays an important role in mediating the physiologic and therapeutic effects of IFNs in humans.2-4 Human STAT1 deficiency was first described in patients with Mendelian susceptibility to mycobacterial diseases (MIM209950),5-7 which is characterized by clinical phenotypes of recurrent and/or disseminated disease caused by weakly virulent mycobacteria in otherwise healthy patients.8,9 Ten known patients with partial dominant STAT1 deficiency have been shown to carry a heterozygous mutation impairing either tyrosine phosphorylation (L706S; Figure 1A)9 or DNA-binding activity (E320Q and Q463H, Figure 1A).8 Cells with these heterozygous alleles displayed an impaired response to IFN-γ in terms of the amount of bioactive γ-activating factor (GAF) produced. The dominant negative effect of the heterozygous mutations identified results from the inability of homodimers containing a mutant STAT1 molecule to enter the nucleus or to bind the consensus GAS element. By contrast, dominant STAT1 deficiency does not impair cellular responses to IFN-α/β. Its clinical penetrance is incomplete, as only 6 patients have developed mycobacterial diseases, heterogeneous in their nature, although all patients were cured with antibiotic treatment.8,9
We also identified 3 patients with complete recessive STAT1 deficiency carrying 2 null alleles, resulting in a complete lack of STAT1 expression and abolition of the STAT1-dependent responses to both IFN-γ and IFN-α/β (through GAF and interferon-stimulated gene F3 [ISGF3]; Figure 1A).10,11 The STAT1-dependent cellular responses to IFN-λ and interleukin-27 (IL-27) are also abolished in these patients. All patients developed severe bacillus Calmette-Guérin (BCG) infection after vaccination and viral infections, and all died between the ages of 11 and 16 months.10,11 Two siblings with partial recessive STAT1 deficiency were recently reported to carry an exonic splicing enhancer mutation (P696S, Figure 1A), leading to aberrant mRNA splicing, resulting in the production of only approximately 10% the normal amount of STAT1 protein. However, the mutant STAT1 protein produced functions normally in terms of phosphorylation, dimerization, and DNA binding. Impaired, but not abolished, responses to IFN-α/β, IFN-γ, IFN-λ, and IL-27 render the patients with this deficiency susceptible to severe intracellular bacterial and multiple viral infections.12 Both early and late IFN responses are impaired in these siblings. We report here the clinical and biologic findings for 2 other siblings from an unrelated consanguineous family with a new form of partial recessive STAT1 deficiency resulting in severe impairment of the early, but not late, responses to IFNs.
Methods
Patients
We investigated 2 siblings (P1 and P2) with mycobacterial diseases from a consanguineous family originating from and living in Saudi Arabia. P1 did not have BCG vaccination and developed disseminated mycobacterial infection at the age of 6 years (positive acid-fast bacillus staining on an axillary lymph node biopsy). He was then treated with intermittent isonicotinic acid hydrazide, rifampicin, pyrasinamide, and ethambutol for 12 months with moderate improvement. One year later, he was referred to King Faisal Specialist Hospital and Research Center with multiple large cervical lymph nodes more than 1 cm in diameter and a 3-cm × 3-cm draining abscess on the scalp. Computed tomography scan showed diffuse neck nodal diseases, bilateral chest nodular opacities, and multiple calvarial lytic lesions with subgaleal inflammatory fluid loculation. Mycobacterium avium was cultured from multiple tissues. The patient's condition improved significantly, with resolution of the scalp lesion and cervical lymph nodes after treatment with cycloserine, ethionamide, ethambutol, and moxifloxacin, according to the results of drug susceptibility tests. At the age of 8 years, P1 developed disseminated varicella, requiring intensive care, which improved after antiviral treatment. He then suffered from Candida parapsillosis sepsis resulting from central venous line contamination, which was resolved by removal of the line and antifungal treatment. At the age of 9 years, P1 presented persistent vomiting and intermittent seizures with progressive spasticity. MRI showed multiple vasogenic edemas involving both cerebral hemispheres and multiple enhancing lesions throughout the brain parenchyma. M avium was again cultured from brain tissue, with the same pattern of drug susceptibility. Linezolid treatment was initiated, together with high-dose IFN-γ. The patient's condition stabilized and the seizures stopped, but he became blind. He remains hospitalized, with severe sequelae. P1 has normal white cell counts with normal CD4+ and CD8+ populations. His immunoglobulin level is normal, with IgG antibodies against cytomegalovirus, Epstein-Barr virus (EBV), and Toxoplasma gondii. P2 was the younger sister of P1. She received BCG vaccination after birth and developed disseminated BCG infection. She died at the age of 3 years, from septic shock. The patients live and were followed up in Saudi Arabia, where their informed consent was obtained in accordance with local regulations, with approval from the Institutional Review Board. The experiments described here were conducted in the United States, according to the local regulations and with approval of the Institutional Review Board of Rockefeller University.
Cells, plasmids, and reagents
EBV-transformed B cells (EBV-B), SV40-transformed fibroblasts (SV40-fibroblasts), 293T cells, and STAT1-deficient U3C cells were cultured as previously described.12 Stimulations were performed with the indicated doses of IFN-γ (Imukin; Boehringer Ingelheim), IFN-α 2b (IntronA, Schering Plough), IL-27 (R&D Systems), and IFN-λ (R&D Systems). Genomic DNA and cDNA were amplified and sequenced with STAT1-specific primers. Primer sequences and polymerase chain reaction (PCR) conditions are available on request. We used 3 different STAT1 expression vectors: pcDNA3.1 STAT1-V5, pcDNA3 STAT1-Flag, and pcDNA6.0 STAT1-myc. The mutated K201N, E320Q, P696S, L706S, Δex8, and F77A+F78A (F77A) STAT1 plasmids were obtained by site-directed mutagenesis with the Quickchange kit (Stratagene). U3C cells were transfected with a calcium phosphate–based transfection kit (Invitrogen). Immunoprecipitation and Western blotting were carried out as previously described,13 with antibodies against Tyr-701-phosphorylated STAT1 (612132, BD Biosciences Transduction Laboratories), the N-terminus of STAT1 (610116, BD Biosciences Transduction Laboratories; H-95, Santa Cruz Biotechnology), the C-terminus of STAT1 (sc-345, Santa Cruz Biotechnology), Tyr-690-STAT2 (sc-21689, Santa Cruz Biotechnology), STAT2 (sc-476, Santa Cruz Biotechnology), α-tubulin (Santa Cruz Biotechnology), V5 (Invitrogen), Myc (clone 4A6, Sigma-Aldrich), and Flag (Sigma-Aldrich). We added 500nM staurosporine (Calbiochem) to cells after 30 minutes of IFN-γ stimulation, for pulse-chase experiments monitoring the kinetics of STAT1 Tyr-701 phosphorylation.
Determination of mRNA levels by quantitative real-time PCR
Total RNA was extracted with Trizol reagent (Invitrogen) from EBV-B cells or SV40-fibroblasts left unstimulated or stimulated with 103 IU/mL IFN-α or IFN-γ or with 100 ng/mL IL-27 or 20 ng/mL IFN-λ. The RNA was treated with RNase-free DNase (Roche Diagnostics) and cleaned by passage through an RNAeasy column (QIAGEN). Reverse transcription was then carried out directly with random primers, using the TaqMan Reverse Transcription kit (Applied Biosystems), for the determination of IRF1, IRF8, CCL2, CXCL9, CXCL10, MxA, GBP1, ISG15, SOCS3, FOS, and IFIT1 mRNA levels, using the TaqMan probes delivered by Applied Biosystems for these genes. The results were normalized with respect to the values obtained for the endogenous GUS cDNA. Transcription of the STAT1 sequence carried by the plasmids used for transfection was quantified with a pair of primers binding to the 5′-untranslated region of pcDNA3.1 and exon 1 of STAT1, and the results were normalized with respect to the levels of glyceraldehyde-3-phosphate dehydrogenase mRNA, with the SYBR Green Master Mix (Applied Biosystems). Representative results from 2 or 3 independent experiments were compared in unpaired, 2-tailed Student t tests.
Reporter assay and other
We used 300 ng GAS, IFN-stimulated response element (ISRE), IRF1, IFIT1, IFIT2 firefly luciferase reporter plasmids, separately, to transfect 0.2 million U3C cells. The cells were also transfected with 300 ng wild-type (WT) or mutant STAT1 expression plasmid and 30 ng of the Renilla luciferase plasmid. We then stimulated cells, 24 hours after transfection, with either 103 IU/mL IFN-α or 103 IU/mL IFN-γ. The cells were harvested 24 hours after stimulation. Luciferase levels were measured with the Dual Luciferase assay, according to the manufacturer's instructions (Promega). The methods for exon trapping, electrophoretic mobility shift assay, flow cytometric analysis, and viral assay are described in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
Identification of the K201N STAT1 allele
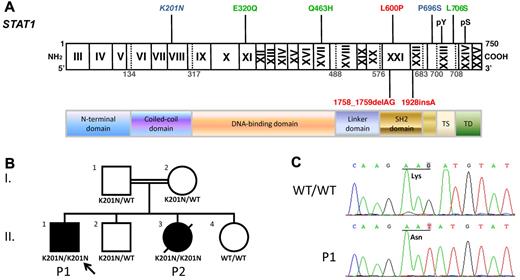
We investigated a male proband (P1) presenting both mycobacterial and viral diseases (“Patients”). Sequencing the exons and flanking intron regions of STAT1, we found a homozygous nucleotide substitution at position 603 in exon 8 (G → T) in genomic DNA from the patient's leukocytes (Figure 1A-C). This substitution replaces a lysine with an asparagine residue (K201N) in the coiled-coil region of STAT1. No other STAT1 mutations were found. Genomic DNA sequencing showed that his younger sister (P2), who had BCG-osis, also carried the homozygous K201N mutation (data not shown). Both parents were healthy and heterozygous for this mutant allele, and the 2 healthy siblings carried at least one WT allele, consistent with autosomal recessive segregation. We excluded the possibility that this mutation was a common or irrelevant polymorphism by sequencing 200 controls from the same ethnic group. In addition, alignment of the human STAT1 sequence with those from 23 animal species present in databases showed the K201 residue to be strictly conserved throughout evolution. These results suggest that the K201N STAT1 allele is responsible for the autosomal recessive STAT1 deficiency in this family.
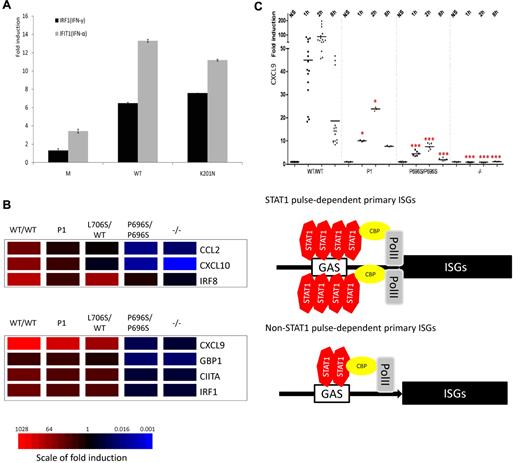
The STAT1 K201N mutation caused susceptibility to mycobacterial and viral infections. (A) The human STAT1 coding region with its known mutations. Coding exons are numbered with Roman numerals and delineated by vertical bars. The N-terminal domain, coiled-coil domain, DNA-binding domain, linker domain, SH2 domain, tail segment domain (TS), and transactivation domain (TD) are indicated, together with their amino acid boundaries. Phosphorylation sites, Tyr 701 (pY) and Ser 727 (pS), are indicated. Mutations in red are recessive mutations associated with complete STAT1 deficiency. Mutations in green are heterozygous mutations associated with partial dominant STAT1 deficiency. Mutations in blue are recessive mutations associated with partial recessive STAT1 deficiency. The mutation reported here is indicated in italics. (B) The pedigree, phenotype, and genotype of the consanguineous kindred from Saudi Arabia. The proband, P1, II.1 is indicated by an arrow, and presented with disseminated M avium infection and disseminated varicella. II.3 had disseminated BCG infection and died of septic shock at the age of 3. These 2 patients are referred to as P1 and P2, respectively. I.1 and I.2 are first cousins. (C) Genomic sequence analysis of exon 8 showing a homozygous G → T mutation in P1, leading to the replacement of a lysine residue by an asparagine residue (K201N/K201N) at position 201 of the protein.
The STAT1 K201N mutation caused susceptibility to mycobacterial and viral infections. (A) The human STAT1 coding region with its known mutations. Coding exons are numbered with Roman numerals and delineated by vertical bars. The N-terminal domain, coiled-coil domain, DNA-binding domain, linker domain, SH2 domain, tail segment domain (TS), and transactivation domain (TD) are indicated, together with their amino acid boundaries. Phosphorylation sites, Tyr 701 (pY) and Ser 727 (pS), are indicated. Mutations in red are recessive mutations associated with complete STAT1 deficiency. Mutations in green are heterozygous mutations associated with partial dominant STAT1 deficiency. Mutations in blue are recessive mutations associated with partial recessive STAT1 deficiency. The mutation reported here is indicated in italics. (B) The pedigree, phenotype, and genotype of the consanguineous kindred from Saudi Arabia. The proband, P1, II.1 is indicated by an arrow, and presented with disseminated M avium infection and disseminated varicella. II.3 had disseminated BCG infection and died of septic shock at the age of 3. These 2 patients are referred to as P1 and P2, respectively. I.1 and I.2 are first cousins. (C) Genomic sequence analysis of exon 8 showing a homozygous G → T mutation in P1, leading to the replacement of a lysine residue by an asparagine residue (K201N/K201N) at position 201 of the protein.
The K201N mutation causes abnormal STAT1 mRNA splicing
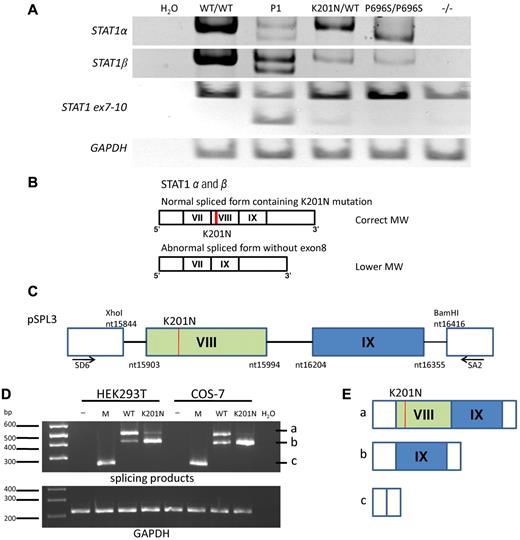
We carried out reverse-transcribed (RT)-PCR to study the STAT1 mRNAs in EBV-B cells and SV40-fibroblasts from a healthy control person (WT/WT), P1 (K201N/K201N), a healthy heterozygous relative (K201N/WT), and previously described patients with partial recessive STAT1 deficiency (P696S/P696S)12 or complete STAT1 deficiency (1928insA/1928insA).10 As expected, 2 different STAT1 transcripts, STAT1α and STAT1β, were amplified from healthy control cells, whereas a product of lower molecular weight (MW) was obtained for STAT1α from P696S/P696S cells (Figure 2A). RT-PCR amplified 2 fragments each for STAT1α and STAT1β from P1 cells: one fragment with the expected MW and another with a lower MW than the WT mRNA (Figure 2A). Both fragments were sequenced, and the smaller fragment was found to lack exon 8, whereas the fragment of normal MW corresponded to the full-length STAT1 gene, including the K201N mutation in exon 8 (Figure 2B). Amplification of the STAT1 mRNA fragment extending from exon 7 to exon 10 confirmed that exon 8 was spliced out by an abnormal splicing process, which was also observed in healthy heterozygous relatives (K201N/WT). Unlike the previously reported P696S mutation, which leads to exon 23 being spliced out of STAT1α, but not out of STAT1β, the K201N mutation impairs the splicing of both STAT1α and STAT1β mRNAs. For confirmation of the role of the K201N mutation in the abnormal splicing of STAT1 mRNA, we transfected HEK293T and COS-7 cells with the exon trapping pSPL314 mock vector, or with pSPL3 containing the STAT1 genomic region, including exons 8 and 9 and their surrounding introns, with or without introducing the K201N mutation (Figure 2C). In both cell types used, the pattern of splicing differed between the WT and K201N STAT1 alleles. A lower MW product, from which exon 8 had been eliminated by splicing (Figure 2D-E), was clearly detectable in cells transfected with the K201N allele and only barely detectable in cells transfected with the WT allele, whereas the normally spliced product of the expected MW was barely detectable in K201N-transfected cells. These results suggest that the K201N mutation is responsible for the splicing out of STAT1 exon 8 in both the STAT1α and STAT1β mRNAs, with residual, leaky splicing in of exon 8 (resulting in ∼ 30% the normal amount of full-length mRNAs, as shown by TA cloning of RT-PCR products, data not shown). Consistent with this hypothesis, in silico analysis predicted the creation of a new binding site for ETR-3 (data not shown) by the K201N mutation, and ETR-3 has been reported to activate exon skipping.15,16
Abnormal mRNA splicing resulting from K201N. (A) RT-PCR of a full-length STAT1α, STAT1β, STAT1 fragment running from exon 7 to exon 10, and glyceraldehyde-3-phosphate dehydrogenase from mRNA extracted from the EBV-B cells of a healthy control (WT/WT), P1, and persons with the following genotypes: K201N/WT, P696S/P696S, and 1928insA/1928insA (−/−). This result is representative of 3 independent experiments. H2O was used as a negative control for PCR. (B) Schematic diagram of the STAT1α and STAT1β mRNA in the cells of P1. The upper band corresponds to the form with an MW identical to that of the WT mRNA, with normal splicing and containing the K201N mutation (red line). The lower band was observed for the cells of P1 and corresponds to forms of the STAT1α and STAT1β mRNAs lacking exon 8. The exons are numbered with Roman numerals. (C) Schematic diagram of the plasmid used for exon trapping. The genomic STAT1 region from nucleotides 15844 to 16416 (NC_000002) was inserted into the pSPL3 plasmid, between the XhoI and BamHI sites, with or without the K201N mutation (line in red). Exon 8 is shown in the green box and exon 9 in the blue box. SD6 and SA2 primer positions are indicated. (D) HEK293T and COS-7 cells were transfected with no vector (−), pSPL3 mock vector (M), pSPL3 vector containing the WT STAT1 gene (WT), and pSPL3 vector containing the K201N-mutated STAT1 gene (K201N). RT-PCR was carried out to amplify the splicing products 24 hours after transfection. (Top panel) PCR with the SD6 and SA2 primers. (Bottom panel) PCR with the glyceraldehyde-3-phosphate dehydrogenase primers. This result is representative of 2 independent experiments. (E) Schematic diagram of the 3 forms of mRNA splicing products: (i) exon 8 and exon 9 plus vector sequence; (ii) exon 9 plus vector sequence; and (iii) 263-bp product corresponding to exonic sequence in the vector.
Abnormal mRNA splicing resulting from K201N. (A) RT-PCR of a full-length STAT1α, STAT1β, STAT1 fragment running from exon 7 to exon 10, and glyceraldehyde-3-phosphate dehydrogenase from mRNA extracted from the EBV-B cells of a healthy control (WT/WT), P1, and persons with the following genotypes: K201N/WT, P696S/P696S, and 1928insA/1928insA (−/−). This result is representative of 3 independent experiments. H2O was used as a negative control for PCR. (B) Schematic diagram of the STAT1α and STAT1β mRNA in the cells of P1. The upper band corresponds to the form with an MW identical to that of the WT mRNA, with normal splicing and containing the K201N mutation (red line). The lower band was observed for the cells of P1 and corresponds to forms of the STAT1α and STAT1β mRNAs lacking exon 8. The exons are numbered with Roman numerals. (C) Schematic diagram of the plasmid used for exon trapping. The genomic STAT1 region from nucleotides 15844 to 16416 (NC_000002) was inserted into the pSPL3 plasmid, between the XhoI and BamHI sites, with or without the K201N mutation (line in red). Exon 8 is shown in the green box and exon 9 in the blue box. SD6 and SA2 primer positions are indicated. (D) HEK293T and COS-7 cells were transfected with no vector (−), pSPL3 mock vector (M), pSPL3 vector containing the WT STAT1 gene (WT), and pSPL3 vector containing the K201N-mutated STAT1 gene (K201N). RT-PCR was carried out to amplify the splicing products 24 hours after transfection. (Top panel) PCR with the SD6 and SA2 primers. (Bottom panel) PCR with the glyceraldehyde-3-phosphate dehydrogenase primers. This result is representative of 2 independent experiments. (E) Schematic diagram of the 3 forms of mRNA splicing products: (i) exon 8 and exon 9 plus vector sequence; (ii) exon 9 plus vector sequence; and (iii) 263-bp product corresponding to exonic sequence in the vector.
STAT1 expression and phosphorylation
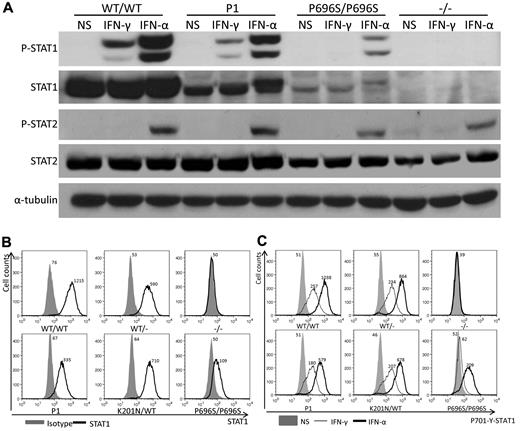
The splicing out of exon 8 (92 nucleotides) induces a frameshift and creates a premature stop codon at position 582, potentially resulting in the production of a truncated STAT1 protein consisting of only the first 194 N-terminal amino acids. To investigate whether such a truncated protein could actually be produced, we generate a plasmid construct containing the STAT1 with exon 8 deleted (Δex8). We transfected 293T and U3C cells with this construct and with a WT construct. We analyzed the results, comparing them with those from P1's EBV-B cells, by Western blotting with a specific antibody recognizing the N-terminal part of STAT1 (amino acids 69-169). A truncated protein was produced in 293T cells but was not detectable in U3C cells or in EBV-B cells from P1 (supplemental Figure 1). To assess the expression of full-length STAT1 protein, we carried out Western blotting on lysates from EBV-B cells and SV40-fibroblasts from P1 (Figure 3A and data not shown), together with cells from one healthy control, and from other patients with partial (P696S/P696S) or complete STAT1 deficiency. Using an antibody recognizing the C-terminal part of the STAT1α isoform, we showed that STAT1 expression was impaired in the cells of P1, which nonetheless produced more of this protein (30% of WT levels) than P696S/P696S cells (10%; Figure 3A). On fluorescence-activated cell sorter (FACS) analysis with an antibody recognizing the N-terminus of STAT1, the specific mean fluorescence intensity of STAT1 was lower in the cells of P1 than in WT/WT, WT/−, and K201N/WT cells but was higher than that in P696S/P696S cells (Figure 3B). Tyrosine 701-phosphorylated STAT1 levels in response to IFN-γ and IFN-α stimulations in vitro were lower in the cells of P1, as demonstrated by both Western blotting and FACS (Figure 3A,C). The different genotypes of cells studied could be ranked in descending order of Tyr-701-phosphorylated STAT1, as follows: WT/WT > K201N/WT > WT/− > K201N/K201N > P696S/P696S > −/− (Figure 3C). The levels of STAT1 phosphorylation were correlated with the amount of STAT1 protein expressed, suggesting that mutant STAT1 proteins underwent normal phosphorylation on the Tyr701 residue. As a control, STAT2 phosphorylation levels in response to IFN-α stimulation were found be similar in all the cells studied (Figure 3A). We then transfected STAT1-deficient U3C cells with the STAT1 K201N allele to compare the stability and phosphorylation of the mutant protein with those of the WT STAT1. The mutant K201N STAT1 was produced in normal amounts and phosphorylated on tyrosine 701 (supplemental Figure 2). The K201N mutation caused the abnormal splicing out of exon 8 in most mRNAs, resulting in the impairment, but not total abolition, of full-length STAT1α and STAT1β isoform expression. The K201N allele results in lower levels of STAT1 through abnormal mRNA splicing, as the K201N missense mutation is not itself intrinsically deleterious for the expression and phosphorylation of STAT1.
STAT1 protein expression is impaired in the cells of P1. EBV-B cells from WT/WT, P1, K201N/WT, 1928insA/WT (WT/−), P696S/P696S, and 1928insA/1928insA (−/−) persons were stimulated with 105 IU/mL IFN-α or IFN-γ or left unstimulated for 30 minutes. (A) Western blotting was carried out with an antibody against Tyr701-phosphorylated STAT1 (P-STAT1), STAT1α (STAT1), Tyr690-phosphorylated STAT2 (P-STAT2), STAT2, and α-tubulin as a reference. (B) FACS analysis of intracellular STAT1 levels with an antibody against the N-terminus of STAT1. Solid area represents isotype control antibody. Solid line indicates STAT1 antibody. (C) FACS analysis of intracellular Tyr701-phosphorylated STAT1 levels in untreated cells (NS) or in cells treated with 105 IU/mL IFN-α or IFN-γ for 30 minutes. Solid area represents untreated cells. Thin line indicates 105 IU/mL IFN-γ stimulation; and thick line, 105 IU/mL IFN-α stimulation. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments.
STAT1 protein expression is impaired in the cells of P1. EBV-B cells from WT/WT, P1, K201N/WT, 1928insA/WT (WT/−), P696S/P696S, and 1928insA/1928insA (−/−) persons were stimulated with 105 IU/mL IFN-α or IFN-γ or left unstimulated for 30 minutes. (A) Western blotting was carried out with an antibody against Tyr701-phosphorylated STAT1 (P-STAT1), STAT1α (STAT1), Tyr690-phosphorylated STAT2 (P-STAT2), STAT2, and α-tubulin as a reference. (B) FACS analysis of intracellular STAT1 levels with an antibody against the N-terminus of STAT1. Solid area represents isotype control antibody. Solid line indicates STAT1 antibody. (C) FACS analysis of intracellular Tyr701-phosphorylated STAT1 levels in untreated cells (NS) or in cells treated with 105 IU/mL IFN-α or IFN-γ for 30 minutes. Solid area represents untreated cells. Thin line indicates 105 IU/mL IFN-γ stimulation; and thick line, 105 IU/mL IFN-α stimulation. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments.
STAT1 dimerization and dephosphorylation
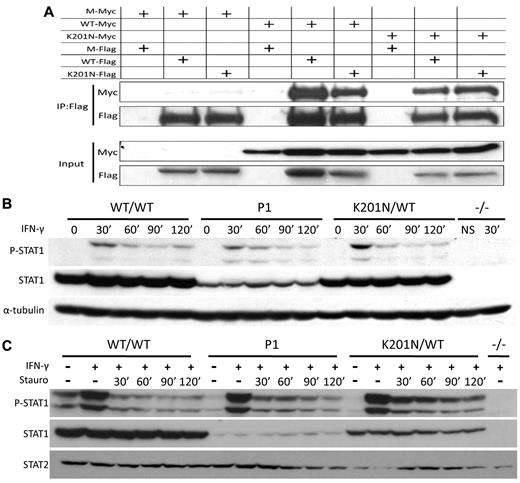
Different mutations in the STAT1 gene cause various functional defects.17 Certain hydrophilic amino acids in the N-terminal domain, coiled-coil domain, and DNA-binding domain play an important role in converting the structure of the molecule from a parallel dimer to an antiparallel dimer. Mutations affecting these hydrophilic amino acids may impair the dimerization of unphosphorylated STAT1, resulting in the prolonged phosphorylation and nuclear retention of STAT1.18-20 We investigated the possible deleterious effects of K201N on this dimerization by modeling K201N STAT1 homodimers, based on the known 3-dimensional structure of WT STAT1 homodimers.20 K201N is not located on the dimer interface (data not shown). We nevertheless investigated the dimerization of unphosphorylated and phosphorylated STAT1 by carrying out coimmunoprecipitation followed by Western blotting in U3C cells transfected with various combinations of WT and K201N STAT1 tagged with either Myc or Flag (Figure 4A). The proteins precipitated with an anti-Flag antibody were subjected to Western blotting with an anti-Myc antibody. For both phosphorylated and unphosphorylated forms of STAT1, the interaction between K201N and WT or K201N was normal (Figure 4A; and data not shown). We then investigated the kinetics of STAT1 activation and dephosphorylation in the cells of P1 and in STAT1-deficient U3C cells transfected with the mutant K201N allele. In EBV-B cells and SV40-fibroblasts from P1, STAT1 phosphorylation was observed at 30 minutes, decreasing thereafter as rapidly as in the control cells, in response to IFN-γ stimulation (Figure 4B and data not shown). With staurosporine, which blocks continuous tyrosine kinase activation, the kinetics of phosphorylation and dephosphorylation were similar in the cells of P1 and in control cells (Figure 4C). Moreover, in U3C cells transfected with K201N, STAT1 phosphorylation and dephosphorylation kinetics were similar to those of the WT protein. This was not the case after transfection with the F77A STAT1 allele,18 which displays prolonged STAT1 phosphorylation (supplemental Figures 2-3). Thus, the K201N missense mutation does not affect the phosphorylation, homodimerization, or dephosphorylation of STAT1.
K201N mutation does not impair STAT1 dephosphorylation and homodimerization. (A) K201N does not impair homodimerization. U3C cells were transfected with a combination of mock (M), WT, and K201N-mutated STAT1 plasmids tagged with either Flag or Myc. Proteins were extracted 48 hours after transfection and subjected to coimmunoprecipitation and Western blotting. (Bottom panel) Western blotting for the detection of Myc or Flag expression in input with a specific antibody. (Top panel) Immunoprecipitation with an anti-Flag antibody, followed by Western blotting with an anti-Myc or anti-Flag antibody. (B) STAT1 phosphorylation kinetics. EBV-B cells from a healthy control (WT/WT), P1, a subject heterozygous for the K201N allele (K201N/WT), and a patient with complete STAT1 deficiency (1928insA/1928insA, −/−) were stimulated with 105 IFN-γ for the time indicated (in minutes). Western blotting was carried out with an antibody against Tyr701-phosphorylated STAT1 (P-STAT1), or STAT1α (STAT1), or with an antibody against α-tubulin, as a reference. (C) Pulse-chase experiment. EBV-B cells from a healthy control (WT/WT), P1, a subject heterozygous for the K201N allele (K201N/WT), and a patient with complete STAT1 deficiency (1928insA/1928insA, −/−) were stimulated with 105 IU/mL IFN-γ for 30 minutes, and staurosporine was added and the mixture incubated for the time indicated. Western blot shows Tyr701-phosphorylated STAT1 and STAT1, with STAT2 as a reference.
K201N mutation does not impair STAT1 dephosphorylation and homodimerization. (A) K201N does not impair homodimerization. U3C cells were transfected with a combination of mock (M), WT, and K201N-mutated STAT1 plasmids tagged with either Flag or Myc. Proteins were extracted 48 hours after transfection and subjected to coimmunoprecipitation and Western blotting. (Bottom panel) Western blotting for the detection of Myc or Flag expression in input with a specific antibody. (Top panel) Immunoprecipitation with an anti-Flag antibody, followed by Western blotting with an anti-Myc or anti-Flag antibody. (B) STAT1 phosphorylation kinetics. EBV-B cells from a healthy control (WT/WT), P1, a subject heterozygous for the K201N allele (K201N/WT), and a patient with complete STAT1 deficiency (1928insA/1928insA, −/−) were stimulated with 105 IFN-γ for the time indicated (in minutes). Western blotting was carried out with an antibody against Tyr701-phosphorylated STAT1 (P-STAT1), or STAT1α (STAT1), or with an antibody against α-tubulin, as a reference. (C) Pulse-chase experiment. EBV-B cells from a healthy control (WT/WT), P1, a subject heterozygous for the K201N allele (K201N/WT), and a patient with complete STAT1 deficiency (1928insA/1928insA, −/−) were stimulated with 105 IU/mL IFN-γ for 30 minutes, and staurosporine was added and the mixture incubated for the time indicated. Western blot shows Tyr701-phosphorylated STAT1 and STAT1, with STAT2 as a reference.
Nuclear translocation and DNA-binding activity of the mutant STAT1 protein
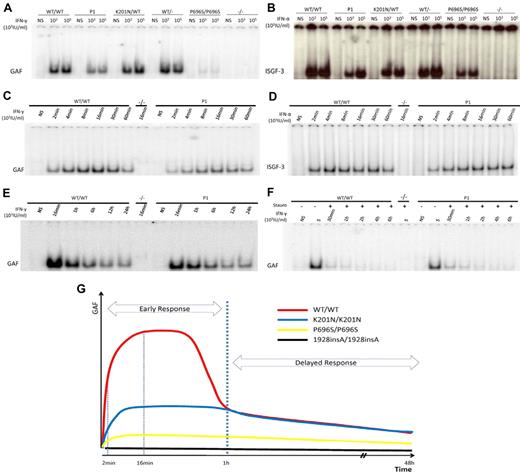
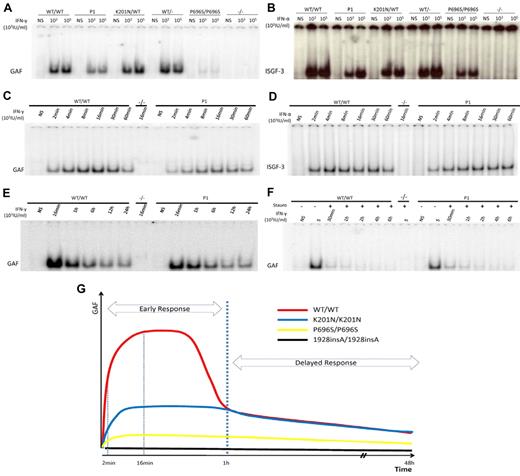
The phosphorylation of STAT1 triggers its translocation into the nucleus in the form of GAF dimers or ISGF3 trimers, which bind to consensus promoter sequences and initiate the transcription of downstream ISGs. Immunofluorescence analyses showed that STAT1 was constitutively expressed in the cytoplasm of control fibroblasts and in the fibroblasts of P1 but was not detectable in fibroblasts from patients with partial (P696S/P696S) or complete STAT1 deficiency. In response to IFN-γ or IFN-α stimulation, STAT1 was translocated from the cytoplasm to the nucleus, as normal, in the fibroblasts of P1 (supplemental Figure 4). We quantified the DNA-binding activity of mutant STAT1-containing GAF and ISGF-3 complexes, by carrying out electrophoretic mobility shift assays with GAS and ISRE probes in cells from P1, and comparing the results obtained with those for healthy controls and cells from patients with other forms of STAT1 deficiency. The cells of P1 displayed levels of GAF binding activity 36% (± 5%) of those from normal controls after 16 minutes of stimulation with 103 IU/mL or 105 IU/mL IFN-γ. This level of activity was higher than that of P696S/P696S mutant (∼ 8%; Figure 5A) and L706S/WT mutant (impairing phosphorylation and, thereby, DNA binding, ∼ 20%) cells, but lower than that of E320Q/WT mutant (impairing DNA binding, ∼ 40%) cells (data not shown). After 16 minutes of stimulation with 103 IU/mL or 105 IU/mL IFN-α, P1 ISGF-3 DNA-binding activity was determined and was shown to be 38% (± 14%) or 62.7% (± 11%) that of normal controls. These levels were similar to those of the P696S/P696S mutant after stimulation with 103 IU/mL IFN-α but higher than those observed after stimulation with 105 IU/mL (Figure 5B). We also evaluated the DNA-binding activity of K201N STAT1 after its overexpression in U3C cells. Unlike L706S and E320Q mutant proteins, K201N STAT1 displayed normal DNA-binding activity, such as P696S STAT1 (supplemental Figure 5 and data not shown). Furthermore, transcripts lacking exon 8 had no dominant-negative effect on WT STAT1 GAF-DNA binding activities in U3C cells in response to IFN-γ (supplemental Figure 6).
Impaired GAF and ISGF-3 DNA binding in the cells of P1 early in stimulation. EBV-B cells from WT/WT, P1, K201N/WT, WT/−, P696S/P696S, and −/− subjects were stimulated with the indicated doses of IFN-γ (A) or IFN-α (B) for 15 minutes. Nuclear proteins were extracted to determine GAF (A,C) and ISGF-3 (B,D) levels, with the corresponding probe. Kinetics of GAF (C,E-F) or ISGF-3 (D) DNA-binding activity after IFN-γ and IFN-α, respectively, as determined with stimulation for the times indicated. (F) Pulse-chase experiment with the addition of staurosporine after 15 minutes of IFN-γ stimulation. The results are representative of 3 independent experiments. (G) Schematic diagram of GAF DNA-binding kinetics in healthy control, P1, P696/P696S, and 1928insA/1928insA cells. The x-axis indicates duration of IFN-γ stimulation; and the y-axis, GAF DNA-binding activities.
Impaired GAF and ISGF-3 DNA binding in the cells of P1 early in stimulation. EBV-B cells from WT/WT, P1, K201N/WT, WT/−, P696S/P696S, and −/− subjects were stimulated with the indicated doses of IFN-γ (A) or IFN-α (B) for 15 minutes. Nuclear proteins were extracted to determine GAF (A,C) and ISGF-3 (B,D) levels, with the corresponding probe. Kinetics of GAF (C,E-F) or ISGF-3 (D) DNA-binding activity after IFN-γ and IFN-α, respectively, as determined with stimulation for the times indicated. (F) Pulse-chase experiment with the addition of staurosporine after 15 minutes of IFN-γ stimulation. The results are representative of 3 independent experiments. (G) Schematic diagram of GAF DNA-binding kinetics in healthy control, P1, P696/P696S, and 1928insA/1928insA cells. The x-axis indicates duration of IFN-γ stimulation; and the y-axis, GAF DNA-binding activities.
Kinetics of DNA-binding by STAT1-containing complexes in the cells of P1
As the patients had low levels of STAT1 in their cells and as some mutations in the STAT1 coiled-coil region are known to lead to the accumulation of activated GAF in the nucleus,19 we examined the kinetics of GAF DNA-binding activity in EBV-B cells and SV40-fibroblasts from P1 and controls on stimulation with IFN-α or IFN-γ, with or without staurosporine. In healthy control cells, GAF-binding activity was detected after 2 minutes of stimulation, gradually increasing thereafter to reach a peak at 16 minutes, and began to decrease again after 30 minutes. The activated GAF did not disappear until 48 hours of stimulation (Figure 5C,E and data not shown). With staurosporine, GAF binding rapidly decreased, becoming undetectable at 2 hours (Figure 5F). Comparisons with healthy controls showed that the cells of P1 had defects in GAF-binding activity only at early time points (levels 20%-40% those of controls). However, whereas GAF-binding in control cells decreased after 30 minutes, GAF levels decreased at different rates in cells of P1 and in control cells, resulting in similar levels of GAF-binding in the cells from P1 and control cells after 1 hour of stimulation. In the presence of staurosporine, cells from P1 displayed lower levels of DNA-binding activity than control cells at the peak of activation and similar levels of DNA-binding during deactivation (Figure 5F). The kinetics of ISGF-3 DNA-binding activity after IFN-α stimulation followed a pattern similar to that observed after IFN-γ stimulation in the cells of P1 (Figure 5D). These results suggest that the K201N mutation impaired the early response to IFN stimulation in terms of GAF and ISGF-3 activation, as the normal peaks of activation were not reached, but that the late phase of activated GAF and ISGF-3–binding activity was normal (Figure 5G).
Transcriptional activation and downstream gene induction
We investigated whether the K201N mutation impaired the transcriptional activity of STAT1, by transfecting STAT1-deficient U3C cells with a WT or mutant STAT1-encoding plasmid, together with reporter plasmids, with expression of the firefly luciferase under the control of GAS, ISRE, IRF1, IFIT1, or IFIT2 promoters. Cells were also cotransfected with a Renilla luciferase reporter as a reference. After IFN-γ stimulation, the K201N mutant STAT1 induced the transcription of reporter genes under the control of GAS, IRF1, or IFIT2 promoters, with the level of induction attained similar to that for the WT (Figure 6A; and data not shown). On IFN-α stimulation, reporter genes under the control of ISRE, IFIT1, or IFIT2 promoters were also induced to similar levels by WT and K201N STAT1 mutant proteins (Figure 6A; and data not shown). The P696S STAT1 mutant also displayed normal transcriptional activity, compared with the L706S mutant, which was a loss-of-function mutant in these reporter assays (data not shown). We then studied the downstream induction of ISGs by quantitative PCR, in EBV-B cells and SV40-fibroblasts from 5 healthy controls, K201N/K201N, L706S/WT, P696S/P696S, and 1928insA/1928insA cells, on stimulation with IFN-γ or IFN-α. Comparisons with healthy controls showed the induction of one group of genes to be decreased in the K201N/K201N cells after 2 hours of stimulation with IFN-γ. This group of genes included the CCL2, IRF8, and CXCL10 genes. However, another group of genes, including CXCL9, CIITA, GBP1, and IRF-1, was induced well in cells from P1 (Figure 6B). The lowest levels of expression of these target genes were observed in the P696S/P696S and 1928insA/1928insA fibroblasts. In cells from a patient with dominant STAT1 deficiency (L706S/WT), induction of the CCL2, CXCL9, and CXCL10 genes show some decreases. For the IFN-α–induced genes, we found that the induction of CXCL9, GBP1, IFIT1, ISG15, and MxA was STAT1-dependent, whereas the induction of SOCS3 and FOS was STAT1-independent; CXCL9, GBP1, and IFIT1 were early-response genes for which induction typically peaked 2 hours after stimulation (Figure 6C; supplemental Figure 7A-B; and data not shown). In addition, MxA and ISG15 displayed a delayed mRNA induction peak at approximately 8 hours. EBV-B cells from P1 contained significantly smaller amounts of CXCL9 and GBP1 mRNA than control cells after 2 hours of induction, but these levels remained higher than those in P696S/P696S cells and cells from a patient with complete STAT1 deficiency (Figure 6C). However, the induction of MxA, ISG15, and IFIT1 was not impaired in cells from P1 (supplemental Figure 7A-B and data not shown).
Selective impairment of downstream gene induction in the cells of P1. (A) The K201N allele has normal transcriptional activity. U3C cells were transfected with M, WT, and K201N mutant alleles, with firefly luciferase under IRF1 or IFIT1 promoters and a Renilla luciferase control, and stimulated with 103 IU/mL IFN-γ or IFN-α for 24 hours. (B) Hotmap of 2 categories of ISGs quantified by quantitative PCR in SV40-fibroblasts from P1, with comparison of the results obtained for WT/WT, L706S/WT, P696S/P696S, and −/− subjects. The induction of CCL2, CXCL10, and IRF8 was significantly impaired in the cells of P1, as shown by comparison with 5 healthy controls. These genes are identified as “STAT1 pulse-dependent primary ISGs”; CXCL9, GBP1, CIITA, and IRF1 displayed a similar pattern in the cells of P1 and controls and are designated as “non-STAT1 pulse-dependent primary ISGs.” (C) After stimulation with 103 IU/mL IFN-α, CXCL9 induction was significantly impaired in EBV-B cells from P1, at 1-hour and 2-hour time points. However, the level of induction of CXCL9 in the cells of P1 remained higher than those in P696S/P696S and −/− EBV-B cells. The results are representative of 3 independent experiments. One red asterisk indicates P < .05; and 3 red asterisks indicate P < .005.
Selective impairment of downstream gene induction in the cells of P1. (A) The K201N allele has normal transcriptional activity. U3C cells were transfected with M, WT, and K201N mutant alleles, with firefly luciferase under IRF1 or IFIT1 promoters and a Renilla luciferase control, and stimulated with 103 IU/mL IFN-γ or IFN-α for 24 hours. (B) Hotmap of 2 categories of ISGs quantified by quantitative PCR in SV40-fibroblasts from P1, with comparison of the results obtained for WT/WT, L706S/WT, P696S/P696S, and −/− subjects. The induction of CCL2, CXCL10, and IRF8 was significantly impaired in the cells of P1, as shown by comparison with 5 healthy controls. These genes are identified as “STAT1 pulse-dependent primary ISGs”; CXCL9, GBP1, CIITA, and IRF1 displayed a similar pattern in the cells of P1 and controls and are designated as “non-STAT1 pulse-dependent primary ISGs.” (C) After stimulation with 103 IU/mL IFN-α, CXCL9 induction was significantly impaired in EBV-B cells from P1, at 1-hour and 2-hour time points. However, the level of induction of CXCL9 in the cells of P1 remained higher than those in P696S/P696S and −/− EBV-B cells. The results are representative of 3 independent experiments. One red asterisk indicates P < .05; and 3 red asterisks indicate P < .005.
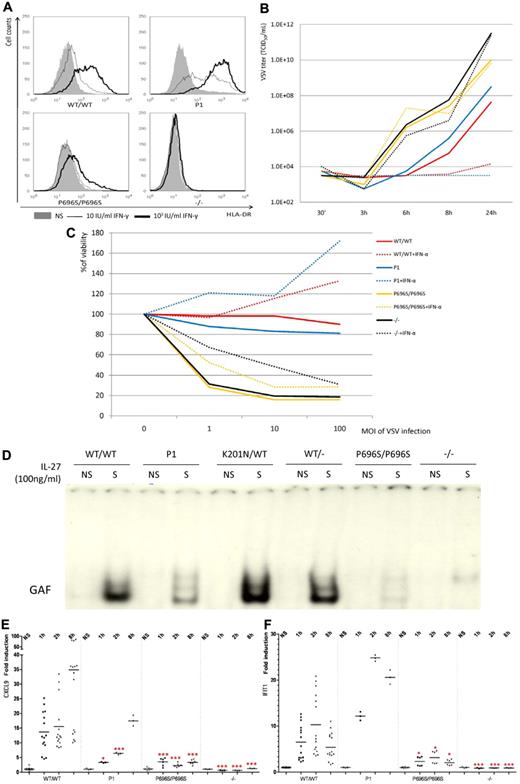
We then studied the late response in cells from P1. HLA-DR displayed a classic delayed response to IFN-γ stimulation in fibroblasts.13 SV40-fibroblasts from P1 showed normal HLA-DR induction with either 10 IU/mL or 103 IU/mL IFN-γ, whereas HLA-DR could not be up-regulated in the fibroblasts from a patient with complete STAT1 deficiency and only residual levels of induction were observed in P696S/P696S fibroblasts (Figure 7A). We then assessed the delayed response by determining the protection against viral challenge conferred by IFN-α in the fibroblasts. Prior treatment with IFN-α for 18 hours decreased vesicular stomatitis virus (VSV) load by a factor of 106 with respect to untreated cells, demonstrating the effective protection of the cells of P1 by IFN-α (Figure 7B). Prior treatment with IFN-α also protected the cells of P1 against VSV-induced cell death, as observed in the healthy control (Figure 7C). Thus, impairment of the early response is not sufficient to abolish the protection mediated by IFN-α.
Delayed interferon responses, IL-27 and IFN-λ responses in the cells of P1. (A) HLA-DR induction in SV-40 fibroblasts of WT/WT, P1, P696S/P696S, and −/− subjects after 48 hours of stimulation with the indicated dose of IFN-γ. HLA-DR was quantified by FACS analysis. Solid area represents no stimulation; thin solid line, 10 IU/mL IFN-γ; and thick solid line, 103 IU/mL IFN-γ. (B) VSV viral titer after VSV challenge. The SV40-fibroblasts of P1 controlled VSV replication after prior treatment of IFN-α for 18 hours. Bold line indicates without IFN-α; and dashed line, with 105 IU/mL IFN-α. (C) Cell viability after VSV challenge. The viability of SV40-fibroblasts from P1 was similar to that of healthy control cells after prior treatment with 105 IU/mL IFN-α for 18 hours. The results are representative of 2 independent experiments. (D) GAF DNA-binding activity was impaired in the EBV-B cells of P1, as shown by comparison with the cells of healthy controls after stimulation with 100 ng/mL IL-27 for 15 minutes. (E) On stimulation with 100 ng/mL IL-27, CXCL9 induction was significantly impaired in the EBV-B cells of P1 at the 1-hour and 2-hour time points, as shown by comparison with healthy controls. However, CXCL9 induction was nonetheless stronger in the cells of P1 than in P696S/P696S and −/− EBV-B cells. (F) After stimulation with 20 ng/mL IFN-λ, IFIT1 induction was normal in the EBV-B cells of P1. This level of induction was greater than that observed in P696S/P696S and −/− EBV-B cells. The results are representative of 3 independent experiments. One red asterisk indicates P < .05; and 3 red asterisks indicate P < .005.
Delayed interferon responses, IL-27 and IFN-λ responses in the cells of P1. (A) HLA-DR induction in SV-40 fibroblasts of WT/WT, P1, P696S/P696S, and −/− subjects after 48 hours of stimulation with the indicated dose of IFN-γ. HLA-DR was quantified by FACS analysis. Solid area represents no stimulation; thin solid line, 10 IU/mL IFN-γ; and thick solid line, 103 IU/mL IFN-γ. (B) VSV viral titer after VSV challenge. The SV40-fibroblasts of P1 controlled VSV replication after prior treatment of IFN-α for 18 hours. Bold line indicates without IFN-α; and dashed line, with 105 IU/mL IFN-α. (C) Cell viability after VSV challenge. The viability of SV40-fibroblasts from P1 was similar to that of healthy control cells after prior treatment with 105 IU/mL IFN-α for 18 hours. The results are representative of 2 independent experiments. (D) GAF DNA-binding activity was impaired in the EBV-B cells of P1, as shown by comparison with the cells of healthy controls after stimulation with 100 ng/mL IL-27 for 15 minutes. (E) On stimulation with 100 ng/mL IL-27, CXCL9 induction was significantly impaired in the EBV-B cells of P1 at the 1-hour and 2-hour time points, as shown by comparison with healthy controls. However, CXCL9 induction was nonetheless stronger in the cells of P1 than in P696S/P696S and −/− EBV-B cells. (F) After stimulation with 20 ng/mL IFN-λ, IFIT1 induction was normal in the EBV-B cells of P1. This level of induction was greater than that observed in P696S/P696S and −/− EBV-B cells. The results are representative of 3 independent experiments. One red asterisk indicates P < .05; and 3 red asterisks indicate P < .005.
P1's response to IL-27 and IFN-λ
It has been shown that IL-27 activates STAT1, leading to GAF production. The EBV-B cells of P1 produced smaller amounts of GAF than those of healthy controls (Figure 7D). We then studied the transcriptional activation of downstream genes, by quantitative PCR. The transcriptional activation of CXCL9 and CXCL10 was significantly impaired in the cells of P1 after only 1 or 2 hours of stimulation with IL-27, but this activation was similar to that of controls after 8 hours of stimulation (Figure 7E; supplemental Figure 8). By contrast, the induction of IRF1 and GBP1 was not impaired even at early time points in P1 cells, consistent with the selective impairment of induction of early ISGs in P1 cells, but not in P696S/P696S cells or in complete STAT1-deficient cells (data now shown). Moreover, on IFN-λ stimulation, the cells of P1 displayed normal induction of IFIT1 and ISG15 at various time points (Figure 7F; supplemental Figure 9), whereas P696S/P696S cells and complete STAT1-deficient cells displayed an impaired response. The reason for these differences in cellular phenotype between P1 and P696S/P696S cells may be the lower level of IFN-λ receptor expression in EBV-B cells, which could not phosphorylate large amounts of STAT1. In conclusion, the cells of P1 displayed defects in their response to IL-27 stimulation, but not in their response to IFN-λ.
Discussion
We report here 2 siblings with a new form of partial recessive STAT1 deficiency. They are homozygous for a nucleotide substitution leading to both an amino acid substitution (K201N) and a frameshift splice defect of exon 8. Their cells contain approximately 30% the normal amount of STAT1 protein, exclusively in the full-length K201N STAT1α and β forms. Partial STAT1 deficiency was first described in a kindred with the P696S mutation.12 These 2 different exonic missense mutations, K201N and P696S, probably lead to abnormal RNA splicing by introducing exonic enhancer effects through different mechanisms. Despite the location of K201N substitution in the coiled-coil domain, STAT1 dephosphorylation and dimerization were normal, consistent with the presence of the K201N residue on the outside of the homodimer interface.18-20 Mutations of some of the lysine residues of STAT1 mimicking the acetylated form impair the nuclear translocation of STAT1.21,22 However, K201N-mutated STAT1 was translocated from the cytoplasm to the nucleus. The K201N STAT1 protein, like the P696S STAT1 allele, was intrinsically fully functional in terms of phosphorylation, dimerization, translocation, dephosphorylation, DNA binding, and transcriptional activation. However, K201N, like the P696S allele, caused an aberrant mRNA splicing, decreasing the amounts of STAT1 protein in the cells. Cells from patients with the P696S/P696S genotype, however, contain smaller amount of STAT1 (∼ 10%) than cells from patients with the K201N/K201N genotype (∼ 30%), resulting in patients with the K201N mutation having a less severe cellular phenotype on IFN stimulation.
However, the most interesting observation revealed by this experiment of Nature is that the disease-causing K201N allele impairs early, but not late cellular responses to IFNs,23,24 unlike the P696S allele, which impairs both early and late responses to IFNs. The correct duration and magnitude of IFN-induced transcriptional activation are essential to ensure optimal IFN activity, so the regulation of STAT1 activation has been investigated at multiple levels.25 On IFN stimulation, STAT1 undergoes rapid and transient tyrosine phosphorylation, peaking after approximately 16 minutes and gradually decreasing thereafter. In the positive regulation of STAT1, IFN production,26,27 receptor densities,28 kinase activities,29,30 and nuclear factors are tightly regulated to control the physiologic response to IFN stimulation. For STAT1 deactivation, tyrosine phosphatase activity,31,32 the export of STAT1 from the nucleus,33 the SOCS and PIAS protein families,34 and the methylation and acetylation of STAT121,22,35 have been associated with control of the parabolic response to IFN stimulation. Two phases of the cellular response to IFNs can be defined in terms of STAT1 activation: an early response characterized by peak levels of activated STAT1 in the nucleus and a delayed response, in which activated STAT1 is present at low levels in the nucleus. However, little is known about the physiologic importance of these 2 separate phases. We demonstrate here the importance of the early peak response. We provide here the first evidence that impairment of the early human IFN response may cause clinical susceptibility to mycobacterial and viral infections.
Another major finding of this study is the dissociation of DNA-binding activity and the induction of target genes after stimulation with IFNs. The impairment of early transcriptional factor activation does not cause a decrease in mRNA induction for all downstream ISGs. Careful investigation of ISG induction, by quantitative PCR, identified 2 groups of downstream genes in K201N/K201N cells. One group of genes was found to display impaired induction in K201N/K201N cells, consistent with a dependence on the early peak of GAF activation, whereas the genes of the second group were induced normally. The mechanisms of inducible mRNA transcription in response to stimulation have been widely studied recently and seem to involve several sequential processes, from chromatin remodeling and histone modification to the binding of transcription factors to DNA and RNA polymerase II recruitment.36 There may be various reasons for the existence of the 2 groups of induced genes identified in K201N/K201N cells. First, the promoter structures may account for the differences in induction pattern. These effects may result from the presence of cis-elements, CpG islands, and differences in G plus C content.37-39 Second, the 2 categories of inducible ISGs in K201N/K201N cells may differ in terms of the roles played by several cotranscriptional factors40-42 and histone regulation proteins, such as HDAC1, CBP, and BRG1.43-45 Third, the threshold level of STAT1, which binds to the inducible gene loci, required to induce efficient transcription may differ for different genes.46,47 Further careful investigation is required to resolve this point. In any case, the genes not induced in K201N/K201N cells, such as IRF8 in response to IFN-γ48,49 and CXCL9 in response to IFN-α,50 are the best candidate genes for patients with unexplained mycobacterial and viral diseases, respectively.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Levy, Stephane Smale, Bertrand Boisson, Sun Xiao-Jian, Minji Byun, and Melina Herman for helpful discussions and critical reading; David Levy (pIFIT2), John Hiscott (pIFIT1), and Georg Kochs (pIRF1) for providing luciferase reporter plasmids; Ron Liebman, Tatiana Kochetkov, Erin Kirk, Yelena Nemirovskaya, Lucile Jannière, Martine Courat, and Tony Leclerc for technical and secretarial assistance; and all members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions.
X.-F.K. is supported by a Choh-Hao Li Memorial Fund Scholar award and the Shanghai Educational Development Foundation. The Laboratory of Human Genetics of Infectious Diseases is supported by grants from the Rockefeller University Center for Clinical and Translational Science (grant 5UL1RR024143), the Rockefeller University, the Bill and Melinda Gates Foundation, the St Giles Foundation, the Jeffrey Modell Foundation and Talecris Biotherapeutics, National Institute of Allergy and Infectious Diseases (grant 1R01AI089970), the Schlumberger Foundation, the BNP-Paribas Foundation, the Institut Universitaire de France, and the European Union (grant QLK2-CT-2002-0046).
National Institutes of Health
Authorship
Contribution: X.-F.K., S.A.-M., J.-L.C., and S.B.-D. were responsible for the conception and design of the experiments; X.-F.K. and M.C. performed the experiments; X.-F.K., M.C., M.A., J.-L.C., and S.B.-D. analyzed the data; S.A.-H., R.H., and S.A.-M. treated the patients; L. Alsina, T.Z., J.B., J.F., M.A., C.P., V.B., A.K., D.B., X.-X.Z., L. Abel, and D.C. contributed reagents/materials/analysis tools and edited the paper; X.-F.K., J.-L.C., and S.B.-D. wrote the paper; and J.-L.C. supervised all work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Laurent Casanova, St Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, 1230 York Ave, New York, NY 10065; e-mail: jean-laurent.casanova@rockefeller.edu.
References
Author notes
M.C. and S.A.-H. contributed equally to this study and are considered co–second authors.
S.A.-M., J.-L.C., and S.B.-D. contributed equally to this study and are considered co–senior authors.