We previously reported the adverse prognostic impact of Wilms tumor 1 gene (WT1) mutations in younger adult cytogenetically normal acute myeloid leukemia (CN-AML). Here, we investigated 243 older (≥ 60 years) primary CN-AML patients. WT1 mutated (WT1mut) patients (7%) had FLT3-ITD more frequently (P < .001), lower hemoglobin (P = .01), higher white blood cell count (P = .03) and percentage blood blasts (P = .03), and a shorter overall survival (P = .08) than WT1 wild-type (WT1wt) patients. Comparing older and younger WT1mut patients, they had similar pretreatment characteristics and outcome. By contrast, among WT1wt CN-AML, younger patients had a significantly better outcome. A WT1 mutation-associated gene-expression signature, reported here for the first time, included CD96, a leukemia stem cell-specific marker, and genes involved in gene regulation (eg, MLL, PML, and SNRPN) and in proliferative and metabolic processes (eg, INSR, IRS2, and PRKAA1), supporting the role of mutated WT1 in deregulating multiple homeostatic processes. Our results indicate that WT1mut CN-AML represents a distinct entity with poor treatment response across age groups. This study has been registered at www.clinicaltrials.gov as #NCT00900224.
Introduction
Dysregulation of the Wilms tumor 1 gene (WT1) by mutations and/or overexpression is relatively frequent in acute myeloid leukemia (AML) and might play a role in malignant blast proliferation and impaired differentiation.1
The prognostic impact of WT1 mutations in cytogenetically normal (CN) AML was recently reported for cohorts comprising exclusively2,–4 or almost exclusively5,6 younger (< 60 years) adults. WT1 mutations, clustering in exons 7 and 9, occurred in 9% to 13% of patients2,,,–6 and were associated with lower complete remission (CR) rates,6 higher relapse rates,2,3,6 shorter disease-free survival (DFS)2,6 and overall survival (OS),2,3,6 or had no prognostic impact4 compared with WT1 wild-type (WT1wt). The clinical significance of WT1 mutations in older (≥ 60 years) CN-AML remains to be fully investigated.
We report here the frequency of WT1 mutations and their association with outcome in a relatively large cohort of 243 older primary CN-AML patients intensively treated and characterized comprehensively for clinical and molecular prognosticators. To investigate characteristics of WT1-mutated (WT1mut) CN-AML across age groups, we compared our findings in older patients with those in younger patients. Finally, to gain biologic insights, we derived a signature of genes differentially expressed between WT1mut and WT1wt patients.
Methods
We studied pretreatment bone marrow (n = 205) or blood (n = 38) with more than 20% blasts from primary CN-AML patients 60 years of age and older enrolled in Cancer and Leukemia Group B (CALGB) front-line treatment protocols 85257 (n = 24), 89238 (n = 22), 94209 (n = 6), 972010 (n = 113), or 1020111 (n = 78). For comparative analyses, we used the findings in younger CN-AML patients treated on CALGB 962112 (n = 94) and CALGB 1980813 (n = 113).2 Treatment regimens and definitions of clinical endpoints are detailed in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). No patient received allogeneic stem cell transplantation in first CR. The clinical trials and companion protocols the patients of this study were enrolled in were approved by the Institutional Review Board of every participating institution and the CALGB.
Analysis of WT1 exon 7 and 9 mutations, FLT3-internal tandem duplication (FLT3-ITD), FLT3-tyrosine kinase domain mutations (FLT3-TKD), NPM1 and CEBPA mutations, and mRNA-expression levels of BAALC and ERG were assessed centrally as previously reported.2,14
Patients with and without WT1 mutations were compared for baseline demographic, clinical, and molecular features using the Fisher exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively. Estimated probabilities of DFS and OS were calculated using the Kaplan-Meier method, and the log-rank test evaluated differences between survival distributions. All analyses were performed by the CALGB Statistical Center.
Gene expression of samples from patients younger than 60 years enrolled in CALGB 19808 (n = 94) and patients 60 years of age and older enrolled in CALGB 9720 (n = 71) and 10201 (n = 60) was profiled using the Affymetrix U133 plus 2.0 array (Affymetrix) as reported previously14 and detailed in the supplemental data. The microarray data are available at http://www.ebi.ac.uk/microarray-as/ae/ (accession no. E-TABM-1020). Expression signatures were derived by comparing gene expression between WT1mut (n = 15) and WT1wt (n = 210) patients. Univariable significance levels of .001 for gene-expression profiling were used to determine the probe sets that comprised the signatures.
Results and discussion
Pretreatment characteristics and outcome associated with WT1 mutations in older patients
Sixteen (7%) patients harbored WT1 mutations: 10 patients had single exon 7 mutations, 4 had single exon 9 mutations, and 2 had 2 concurrent mutations (supplemental Table 1).
WT1mut patients had lower hemoglobin (P = .01), higher white blood cell count (P = .03) and percentage of blood blasts (P = .03), and FLT3-ITD more frequently (P < .001) than WT1wt patients (Table 1). All except one WT1mut patient also had either NPM1 or CEBPA mutations, or FLT3-ITD (supplemental Figure 1).
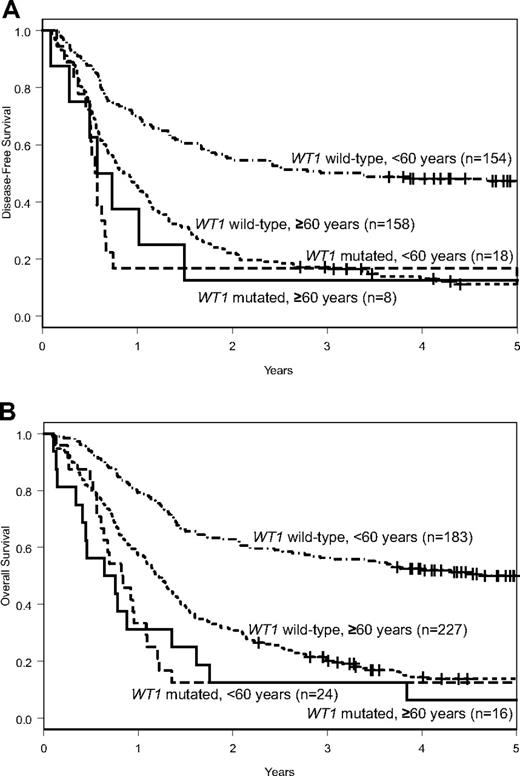
WT1mut patients had fewer CRs (50% vs 70%, P = .16) than WT1wt patients (Table 1). Although DFS was similar (P = .59; 3-year rates, 13% vs 17%; Figure 1A), WT1mut patients tended to have a shorter OS (P = .08; 3-year rates, 13% vs 20%; Figure 1B) than WT1wt patients (Table 1).
Outcomes of patients with CN-AML according to WT1 mutation status. DFS (A) and OS (B) of patients aged 60 years or older and of those younger than 60 years according to WT1 mutation status.
Outcomes of patients with CN-AML according to WT1 mutation status. DFS (A) and OS (B) of patients aged 60 years or older and of those younger than 60 years according to WT1 mutation status.
Comparison of WT1 mutations in older and younger CN-AML patients
To evaluate the clinical impact of WT1 mutations across different age groups, we compared the older patients with 207 younger adults with CN-AML, 24 (12%) of whom carried mutations in WT1 exon 7 and/or 9.2
The frequency of WT1 mutations tended to be lower in older than younger patients (7% vs 12%; P = .07), whereas mutation types and their localization were similar (supplemental Table 2). There were also no significant differences between younger and older WT1mut patients with regard to pretreatment clinical and molecular characteristics. Older WT1mut patients had fewer CRs (50% vs 75%, P = .18) than younger WT1mut patients, but, despite differences in postremission treatment intensity, there were no differences in DFS (P = .66; 3-year rates, 13% vs 17%), or OS (P = .68; 3-year rates, 13% vs 13%; Table 1; Figure 1). These results were confirmed in multivariable models (supplemental Table 3). In contrast, older WT1wt patients had significantly worse CR rates (70% vs 84%, P < .001), DFS (P < .001; 3-year rates, 17% vs 50%), and OS (P < .001; 3-year rates, 20% vs 56%) than younger WT1wt patients (Table 1; Figure 1). Similar pretreatment characteristics and poor outcome results indicate that WT1mut CN-AML constitutes a relatively uniform molecular subset of CN-AML across age groups. The less evident adverse prognostic impact of WT1 mutations in older patients is probably the result of the poor outcome of the respective comparison group of older WT1wt patients. In contrast to younger WT1wt patients, there is only a minor outcome advantage to being WT1wt in the older age group.
Although WT1 mutations were significantly associated with FLT3-ITD in both younger (P = .02) and older (P < .001) patients, we observed no interaction between WT1 mutation and FLT3-ITD status for DFS (P = .89) or OS (P = .74) in the combined cohort of younger and older patients. Gaidzik et al4 recently reported a worse survival of FLT3-ITD-positive/WT1mut patients compared with FLT3-ITD–negative/WT1mut patients in 56 younger adults with CN-AML. However, we found no significant differences in CR rates, DFS, or OS between FLT3-ITD-positive/WT1mut and FLT3-ITD–negative/WT1mut patients in our cohort after controlling for age group.
Genome-wide gene-expression analysis
A WT1mut-associated gene-expression signature, derived from younger and older patients, was composed of 193 probe sets differentially expressed between WT1mut and WT1wt patients. A total of 115 probe sets (representing 74 named genes) were up-regulated and 78 probe sets (40 named genes) were down-regulated in WT1mut patients (supplemental Table 4).
The most up-regulated gene associated with WT1 mutations was GTSF1, which, like WT1, is involved in germ cell development.16 Also up-regulated in WT1mut patients were CD96, encoding a leukemia stem cell–specific marker physiologically expressed on NK cells and T cells,17,18 and MLL and PML, recurrently involved in chromosomal translocations in AML and participating in gene regulation processes, such as chromatin remodeling,19,20 and MACC1, whose expression predicts metastasis formation in colon cancer.21
Among the most down-regulated genes in WT1mut patients were the imprinted SNRPN and SNURF, involved in pre-mRNA splicing,22 and the functionally not characterized FCHO2 and MTX3, located in a 5q chromosomal region frequently deleted in myelodysplastic syndromes and AML.23
The WT1mut signature also included genes involved in metabolic and proliferative regulation. Down-regulated were the genes of the insulin receptor, transcriptionally repressed by WT1,24 and the PI3K activating insulin receptor substrate 2 (IRS2),25 and up-regulated was PRKAA1, encoding a catalytic subunit of the adenosine monophosphate-activated kinase.26
Thus, WT1 mutated CN-AML is seemingly characterized by deregulation of a heterogeneous group of genes involved in biologic activities, such as gene regulation, cell proliferation, and metabolic homeostasis.
In conclusion, WT1 mutations are generally adverse prognostic factors in primary CN-AML, although their impact in older CN-AML is partially obscured by the overall poor outcome of this age group. Future studies are needed to corroborate our results. The WT1mut CN-AML–associated gene-expression signature, first described here, may provide biologic insights into this molecular subset of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Donna Bucci of the Cancer and Leukemia Group B Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center (Columbus, OH) for sample processing and storage services, Lisa J. Sterling and Dr Colin G. Edwards for data management, and Dr Klaus H. Metzeler for fruitful discussions.
This work was supported in part by National Cancer Institute (grants CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657), the Coleman Leukemia Research Foundation, and the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.).
National Institutes of Health
Authorship
Contribution: H.B., G.M., K. Maharry, M.D.R., K. Mrózek, and C.D.B. contributed to the design and analysis of the study and the writing of the manuscript; H.B., S.P.W., P.P., Y.-Z.W., and S.S. carried out laboratory-based research; K. Maharry, M.D.R., D.M., and K.B.H. performed statistical analyses; G.M., B.L.P., T.H.C., J.E.K., M.W., A.J.C., J.O.M., M.R.B., M.A.C., R.A.L., and C.D.B. were involved directly or indirectly in the care of patients and/or sample procurement; and all authors agreed on the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of CALGB participants appears in the supplemental Appendix.
Correspondence: Clara D. Bloomfield, The Ohio State University, Comprehensive Cancer Center, 1216 James Cancer Hospital, 300 W. 10th Ave, Columbus, OH 43210; e-mail: clara.bloomfield@osumc.edu; or Guido Marcucci, The Ohio State University, Comprehensive Cancer Center, 809C Biomedical Research Tower, 460 W. 12th Ave, Columbus, OH 43210; e-mail: guido.marcucci@osumc.edu.