The transcription factor KLF4 may act both as an oncogene and a tumor suppressor in a tissue-depending manner. In T- and pre-B-cell lymphoma, KLF4 was found to act as tumor suppressor. We found the KLF4 promoter methylated in B-cell lymphoma cell lines and in primary cases of B-cell lymphomas, namely, follicular lymphoma, diffuse large B-cell lymphoma, Burkitt lymphoma, and in classic Hodgkin lymphoma (cHL) cases. Promoter hypermethylation was associated with silencing of KLF4 expression. Conditional overexpression of KLF4 in Burkitt lymphoma cell lines moderately retarded proliferation, via cell-cycle arrest in G0/G1. In the cHL cell lines, KLF4 induced massive cell death that could partially be inhibited with Z-VAD.fmk. A quantitative reverse-transcribed polymerase chain reaction array revealed KLF4 target genes, including the proapoptotic gene BAK1. Using an shRNA-mediated knock-down approach, we found that BAK1 is largely responsible for KLF4-induced apoptosis. In addition, we found that KLF4 negatively regulates CXCL10, CD86, and MSC/ABF-1 genes. These genes are specifically up-regulated in HRS cells of cHL and known to be involved in establishing the cHL phenotype. We conclude that epigenetic silencing of KLF4 in B-cell lymphomas and particularly in cHL may favor lymphoma survival by loosening cell-cycle control and protecting from apoptosis.
Introduction
Krüppel-like factor 4 (KLF4) is a member of the KLF zinc-finger-containing transcription factor family consisting of 16 members.1 KLF4 is most closely related to KLF1 and KLF2. These transcription factors are characterized by 3 highly conserved zinc fingers of the C2H2 type and by a transactivating domain in the N-terminus. KLF4 is also referred to as GKLF (gut KLF), but this name is misleading because KLF4 is also expressed in other tissues and cell types, particularly in B cells.2
Mounting evidence has established KLF4 as a context-dependent oncogene or tumor suppressor.3 As an oncogene, KLF4 was shown to transform rat kidney epithelial cells. In addition, KLF4 overexpression is associated with squamous cell carcinoma in larynx and skin and ductal carcinoma of the breast. The oncogenic properties of KLF4 might be explained by transcriptional repression of p53 and hence bypassing of p53-mediated senescence and apoptosis pathways.3 Furthermore, KLF4 together with Oct3/4, Sox2, and c-Myc is able to reprogram differentiated cells to pluripotent stem cells.4 The tumor suppressor role of KLF4 in gastrointestinal epithelium has been well established by a number of studies of gastric, pancreas, and colorectal carcinogenesis.5,–7 KLF4 is highly expressed in epithelial cells of the gastrointestinal tract and the skin during growth arrest and acts as an inhibitor of cell-cycle progression by transcriptional activation of CDKN1A or CDKN1B and repression of CCNB1 or CCND1.6,7 Although most of the studies on the oncogenic/anticancer effects of KLF4 focused on epithelial tumors, several recent studies indicate the involvement of KLF4 in the regulation of apoptosis, proliferation, and differentiation of B cells and B-cell malignancies. Strong up-regulation of KLF4 expression was detected in multiple myeloma cell lines in the process of apoptosis induced by fibroblast growth factor receptor 3 (FGFR3) silencing.8 KLF4 was shown to be able to suppress ABL-induced transformation of pre-B cells by induction of apoptosis and CDKN1A-mediated cell-cycle arrest in a murine model of B-cell acute lymphoblastic leukemia, suggesting a tumor suppressor role in some B-cell malignancies.2 KLF4 is involved in the regulation of B-cell maturation and antibody responses. KLF4 expression seems to be important for maintenance of B-cell quiescence, and KLF4 was shown to be down-regulated on B-cell activation.9 Remarkably, inhibition of B-cell proliferation by FOXO transcription factors is associated with KLF4 induction.10 Nevertheless, the effect of KLF4 in B-cell proliferation is probably complex. A complete knockdown of KLF4 was shown to hamper B-cell proliferation. Furthermore, in the absence of KLF4, fewer B cells entered S-phase of the cell cycle and completed cell division in response to the engagement of BCR and/or CD40 in vitro. The pro-proliferative effect of KLF4 in B cells was explained by a direct activation of CCND2 transcription.11
We set out to investigate the role of KLF4 in the pathogenesis of lymphomas derived from mature germinal center (GC) B cells. We investigated the methylation status of the KLF4 promoter in primary GC lymphoma cases. In addition, we studied the influence of KLF4 on proliferation and apoptosis in Burkitt lymphoma (BL) and classic Hodgkin lymphoma (cHL) cell lines. Finally, we investigated the mechanism of the observed KLF4-induced apoptosis in cHL cell lines.
Methods
Cell culture and treatments
cHL cell lines (KM-H2, L428, L540, and L1236), primary mediastinal B-cell lymphoma cell line (MedB-1), BL cell lines (Bjab, Raji, Ramos, and Namalwa), and lymphoblastoid cell lines (LCLs; LCL 364921, LCL 131, LCL 141, LCL Meier, and UM1) were cultured in complete RPMI 1640 medium as described earlier.12 KM-H2-KLF4-ER, L428-KLF4-ER, Namalwa-KLF4-ER, and Ramos-KLF4-ER clones, as well as cell lines expressing empty vector were generated by transfection with pcDNA3.1-KLF4ER expression vector (a kind gift from Jonathan Alder13 ) or empty vector pcDNA3.1(+) (Invitrogen). Cells were transfected with Amaxa Nucleofector using nucleofection buffer “V,” program Q-07 for L428, U-01 for KMH2, and T-06 for both Namalwa and Ramos (Lonza). Twenty-four hours later, transfected cells were plated in 96-well plate and positive clones were selected with 1 mg/mL G418 (Geneticin, Invitrogen); 5-aza-2′-deoxycytidine (5-aza-dC) and 4-hydroxytamoxifen (4-OHT) were purchased from Calbiochem, zVAD.fmk from Biotrend Chemicals AG.
Quantitative PCR
Total RNA was isolated from 1 × 106 cells using High Pure RNA Isolation Kit (Roche Diagnostics), and first-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (Promega). Templates were amplified using QuantiTect SYBR Green PCR Kit (QIAGEN) using a LightCycler 480 (Roche Diagnostics). Primers were designed applying Genscript online software (www.genscript.com). Quantitative polymerase chain reaction (PCR) primers synthesized by biomers.net, sequences 5′ to 3′, sense and antisense, annealing temperature 60°C: KLF4: GGCACTACCGTAAACACACG and CTGGCAGTGTGGGTCATATC; RPL13A: CGGACCGTGCGAGGTAT and CACCATCCGCTTTTTCTTGTC; CDKN1A: GCAGACCAGCATGACAGATTT and GGATTAGGGCTTCCTCTTGGA; CDKN1C: GCCAATTTAGAGCCCAAAGA and GTTGCTGCTACATGAACGGT; CCND2: AATGGTCATTTCAGGCACAA and CACATTTGCTGATGGCTTCT; BCL2-antagonist/killer 1 (BAK1): GGTCCTGCTCAACTCTACCC and CCTGAGAGTCCAACTGCAAA; CXCL10: TCTGAGCCTACAGCAGAGGA and CAGCAAATCAGAATGGCAGT; CD86: CTCTTTGTGATGGCCTTCCT and AGCTCACTCAGGCTTTGGTT; CD52: GCCACGAAGATCCTACCAAA and TTCTCTTGCGAGTGATGGTG; MSC/ABF-1: GCTTCCAGTTACATCGCTCA-3′ and 5′-GAATGGCCATGTCAGGTTC-3.
Reference genes tested included ACTB, HPRT1, HMBS, and RPL13A. Variation of reference gene expression was analyzed by geNorm software (www.medgen.ugent.be/∼jvdesomp/genorm). RPL13A was the most reliable reference gene and used for calculations. Human apoptosis primer library (Real Time Primers LCC) was used to identify KLF4-target genes.
Human material and laser capture microdissection of the HRS DNA samples
Nine cases of follicular lymphoma (FL), 9 cases of diffuse large B-cell lymphoma (DLBCL), 18 cases of cHL, 5 cases of BL, as well as CD19+ cells obtained from peripheral blood and from tonsils of 13 persons were analyzed. Lymphoma diagnosis was in accordance with the 2008 World Health Organization classification.14 Lymphoma samples were drawn from our archive of fresh and formalin-fixed, paraffin-embedded tissues and pseudonymized to comply with the German law for correct usage of archival tissue for clinical research (Deutsches Ärzteblatt 2003; 100 A1632). Approval for these studies was obtained from the University of Ulm ethics board. For B-cell isolation, we used buffy coats, obtained from the German Red Cross Blood Donation Service. Tonsils were from 3- to 7-year-old patients undergoing tonsillectomy at the Department of Otorhinolaryngology, Head and Neck Surgery, University of Ulm, Ulm, Germany. Written informed consent was obtained. CD19+ cells and B-cell subsets were isolated using microbeads (Miltenyi Biotec). For detailed description of B-cell subset isolation, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Preparations demonstrating more than 90% purity were used for analyses.
For laser capture microdissection, cryopreserved tumor samples were used. According to the morphologic criteria, cHL cases 1, 2, 3, 5, and 9 were diagnosed as nodular sclerosis, 7, 8, and 10 as mixed cellularity forms, and 4 and 6 as lymphocyte-rich subtypes. The 14-μm slices of frozen tumor tissue were mounted onto membrane-covered glass slides (Carl Zeiss Microimaging), fixed with 70% ethanol (EtOH) at −20°C, and stained with hematoxylin for 1 minute, followed by a rinse with DNAse-free water, stained with eosin for 30 seconds, differentiated in 95% EtOH, and dried in 100% EtOH for 1 minute. Microdissection and laser pressure catapulting were performed using the PALM-Microbeam C system (Carl Zeiss) equipped with a Zeiss Axiovert 200M microscope. Hodgkin and Reed-Sternberg (HRS) cells were identified by their characteristic cytomorphology. From each lymphoma specimen, 100 HRS cells and the same amount of bystander and stromal cells were collected.
Quantitative analysis of KLF4 promoter methylation by pyrosequencing and MSP
Genomic DNA was isolated, converted by bisulfite natrium, and amplified with primers 5′-GAGGAGGAAAAGGTTGTAGAGAAGGAAGTTA-TAAGTAAGGA-3′ (forward) and biotin-5′CCCCCACCCCCTCTACTCCCC-3′ (reverse) (annealing temperature 64°C) as described earlier.12 The PCR product was pyrosequenced with the sequencing primer 5′-GGTTGTAGAGAAGGAAGTTAT-3′ on a PSQ 96MA system. To isolate and convert DNA from microdissected samples, we used EZ DNA Methylation-Direct Kit (Zymo Research). Nested PCR was performed by outside primer pair, 5′-GGAGAGAAGAAAGGGAGGGG-3′ and 5′-TAAAAAAATAAAAAACCAAAAAAAC-3′ (annealing temperature 50°C). Further PCR amplification and pyrosequencing were done as described. To detect methylation in the cHL samples, we used the quantitative methylation-sensitive PCR (MSP).15 Converted gDNA was amplified using methylation specific primer pair (mKLF4, 5′ to 3′, sense and antisense, annealing temperature): mKLF4: TATAGTAACGATGGAAGGGAGTTTC and ACGTTCGTTCTCTCTAATCGAA, 56°C. For normalization of DNA input, we used primer set (mACTB) corresponding to the region of ACTB gene free from CpG dinucleotides.15 The methylation ratio was calculated as ECt(mACTB)/ ECt(mKLF4), where E is specific amplification efficiency and Ct is crossing point for mKLF4 and mACTB, respectively.
Immunoblot
Immunoblot was done as described earlier.16 Primary antibodies included rabbit anti-KLF4 (clone H180, 1:500 dilution), goat anti-BAK1 polyclonal antibody (N-20, 1:1000 dilution), and anti–ABF-1 goat antibodies (sc-9556; all from Santa Cruz Biotechnology). Rabbit anticleaved caspase-3 (Asp175) antibody was from Cell Signaling, and rabbit antiactin antibody (A5060, 1:15 000 dilution) was from Sigma-Aldrich. Secondary antibodies included goat anti–rabbit IgG-horseradish peroxidase (sc-2004) and donkey anti–goat IgG-horseradish peroxidase (sc-2020; both Santa Cruz Biotechnology).
Cell proliferation assay, apoptosis, and cell-cycle analysis
For apoptosis analysis, 1 × 105 cells were stained by annexin V (BioVision) and 2 μg/mL propidium iodide (PI; Sigma-Aldrich). Cell death was measured by a Flow Cytometer FACSCalibur (BD Biosciences). To determine cell-cycle distribution, 1 × 106 cells were fixed and stained with 70% cold ethanol and PI. DNA contents were measured by flow cytometry. Data were analyzed using ModFit cell-cycle analysis software (Verity Software House).
shRNA knock-down of BAK1
BAK1 shRNA targeted the sequence 5′-GTACGAAGATTCTTCAAAT-3′ using the pRetro-Super vector (Oligoengine). As a control, we used luciferase mRNA targeting the 5′-GATTATGTCCGGTTATGTA-3 sequence. The vectors were transfected into the KM-H2 and L428 cells by nucleofection. After selection for 5 days with puromycin (0.5 μg/mL for KM-H2 and 1 μg/mL for L428), resistant cells were used for further experiments.
Gene expression profiling
KM-H2-KLF-ER and L428-KLF-ER cells were incubated with 200nM 4-OHT or vehicle (EtOH) for 24 hours. Total RNA was isolated with RNeasy mini kit (QIAGEN). Microarray analyses were performed using 200 ng total RNA as starting material and 5.5 μg ssDNA per hybridization (GeneChip Fluidics Station 450; Affymetrix). Total RNA was amplified and labeled following the Whole Transcript Sense Target Labeling Assay (Affymetrix). Labeled ssDNA was hybridized to Human Gene 1.0 ST Affymetrix GeneChip arrays. Chips were scanned with an Affymetrix GeneChip Scanner 3000 and analyzed using Affymetrix Expression Console Software. Probe level data were obtained using the robust multichip average normalization algorithm. A fold change cutoff of 3 was applied. The analysis of differentially expressed genes was done on a gene-by-gene basis using annotation supplied by GeneSifter microarray data analysis system (www.genesifter.net; VizX Laboratories). For both cell lines, 2 biologic replicas were analyzed. Microarray data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (www.ncbi.nlm.nih.gov/gds) under accession number GSE21296; 12.04.2010.
Results
KLF4 is epigenetically silenced in B-cell lymphomas
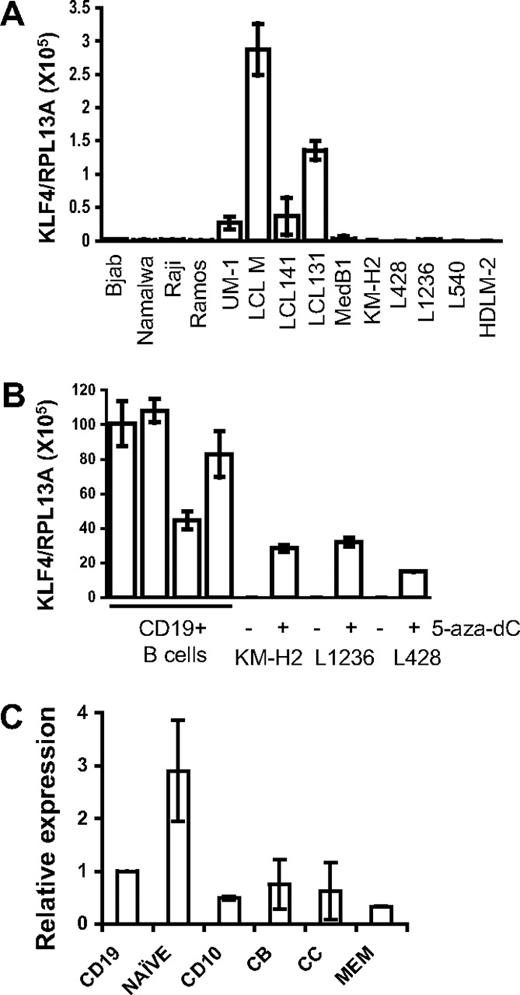
KLF4 is often epigenetically silenced in tumors. To investigate whether KLF4 is down-regulated in B-cell lymphomas, we studied its expression in LCL, BL, primary mediastinal B-cell lymphoma (PMBL), and cHL cell lines using quantitative PCR. We found that in LCLs, which represent human B cells immortalized by infection with Epstein-Barr virus, expression of KLF4 is much higher than in B-cell lymphoma cell lines (Figure 1A). The lowest levels of KLF4 mRNA were detected in cHL cell lines. We then treated the 3 cHL cell lines KM-H2, L1236, and L428 with the DNA methyltransferase I inhibitor 5-aza-dC to test whether promoter hypermethylation is involved in KLF4 silencing. 5-aza-dC treatment restored KLF4 expression in cHL cell lines to levels comparable with those of KLF4 expression in tonsillar CD19+ cells (Figure 1B). The majority of B-cell lymphomas, including BL, FL, and cHL, are thought to arise from GC B cells. Although tonsillar CD19+ cells represent mainly GC B cells, naive cells are also abundant in the tonsils.17,18 High expression of KLF4 in naive cells compared with memory B cells has been reported.9 Thus, we investigated whether low expression of KLF4 in the B lymphomas simply reflects physiologically low levels of KLF4 in their GC progenitors (Figure 1C). When we define KLF4 expression levels in tonsillar CD19+ cells as 100%, then naive cells express 2.9-fold higher levels, whereas CD10+ GCC, CB, CC, and memory cells express 49%, 78%, 54%, and 33%, respectively. Thus, KLF4 expression in GC cells, including CB and CC, is only 2-fold less than in CD19+ cells (ie, still much higher than in B-cell lymphoma cell lines and LCLs). We also surveyed the recent microarray databases19 (GSE10821 and GSE10831, Gene Expression Omnibus database) with the help of Genesifter software (Geospiza). This analysis also revealed comparable relative expression levels of KLF4 in the 3 main B-cell populations: naive B cells, GC (CD10+) cells, and memory B cells (data not shown).
KLF4 is down-regulated in B-cell lymphomas. (A) KLF4 is not expressed in B-lymphoma cell lines. Expression of KLF4 in B-cell lymphoma and in LCL cell lines was analyzed by quantitative PCR. The results are represented as mean value plus or minus SD of target gene (KLF4) to reference gene (RPL13A) ratio. (B) Reactivation of KLF4 expression in cHL cell lines by treatment with 5-aza-dC. cHL cell lines were incubated with 2μM 5-aza-dC for 24 hours; then the medium was replaced. After an additional 96 hours of incubation, cells were harvested and used for mRNA extraction. Experiments were repeated at least twice with similar results (A-B). KLF4 expression was analyzed by quantitative PCR. (C) Differential expression of KLF4 in the tonsillar B-cell subsets. CD19+ cells (n = 9) and B-cell subsets: naive B cells, GC CD10+ B cells (GCC), centroblasts CD77+ (CB), centrocytes CD77−/CD10+ (CC), and memory B cells (n = 3 or 4) were isolated from human tonsils. Expression was determined by quantitative PCR and calculated in relation to internal control gene RPL13A by the comparative Ct method. Mean of CD19+ values was used as comparator. Data are represented as mean ± SD.
KLF4 is down-regulated in B-cell lymphomas. (A) KLF4 is not expressed in B-lymphoma cell lines. Expression of KLF4 in B-cell lymphoma and in LCL cell lines was analyzed by quantitative PCR. The results are represented as mean value plus or minus SD of target gene (KLF4) to reference gene (RPL13A) ratio. (B) Reactivation of KLF4 expression in cHL cell lines by treatment with 5-aza-dC. cHL cell lines were incubated with 2μM 5-aza-dC for 24 hours; then the medium was replaced. After an additional 96 hours of incubation, cells were harvested and used for mRNA extraction. Experiments were repeated at least twice with similar results (A-B). KLF4 expression was analyzed by quantitative PCR. (C) Differential expression of KLF4 in the tonsillar B-cell subsets. CD19+ cells (n = 9) and B-cell subsets: naive B cells, GC CD10+ B cells (GCC), centroblasts CD77+ (CB), centrocytes CD77−/CD10+ (CC), and memory B cells (n = 3 or 4) were isolated from human tonsils. Expression was determined by quantitative PCR and calculated in relation to internal control gene RPL13A by the comparative Ct method. Mean of CD19+ values was used as comparator. Data are represented as mean ± SD.
The low KLF4 expression in primary FL and DLBCL samples compared with GC B cells is further corroborated by a previous study2 and by microarray data20 mined from the Oncomine database (www.oncomine.org, 25.03.2010; supplemental Figure 1). In addition, low expression of KLF4 in FL samples in comparison with normal lymph node was recently reported.21
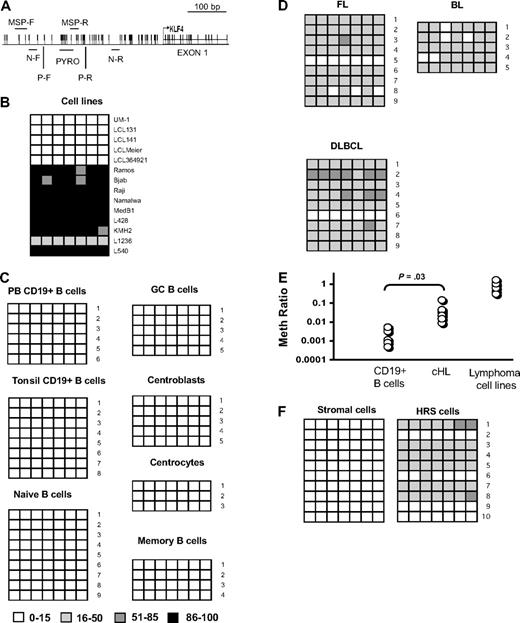
To directly demonstrate promoter methylation, we measured by pyrosequencing the methylation status of 7 sequential CpG dinucleotides, located in a CpG island in the promoter region of KLF4 gene (Figure 2A). In 5 LCLs, expressing low but detectable levels of KLF4, the methylation rate of individual CpGs never exceeded 15%. In contrast, in 4 BL cell lines, 1 PMBL cell line, and in 3 cHL cell lines, the methylation rate varied between 86% and 100%. Only in the L1236 cHL cell line, the methylation rate was between 16% and 50% (Figure 2B). Thus, low methylation of KLF4 promoter correlated with high KLF4 expression and vice versa. To clarify the role of hypermethylation to KLF4 expression in GC cells compared with naive cells, we analyzed KLF4 promoter methylation in 9 samples of naive B cells, 5 samples of GC CD10+ cells (composing CB and CC), 5 samples of CB, 3 samples of CC, and 4 samples of memory cells. In addition, we investigated 8 samples of the CD19+ tonsillar B cells, composing mainly GC cells, as well as 6 samples of peripheral blood CD19+ cells (Figure 2C). We did not find hypermethylation of KLF4 promoter in any of the investigated primary samples. Therefore, promoter methylation is not involved in regulation of KLF4 expression in activated B cells. Of note, in 5 LCLs sharing an activated B-cell phenotype and expressing much lower levels of KLF4 than tonsillar CD19+ or GC cells (Figure 1A,C), KLF4 promoter hypermethylation was not detected either (Figure 2B).
KLF4 promoter is hypermethylated in B-cell lymphomas. (A) Region of KLF4 promoter analyzed by pyrosequencing (PYRO) from −226 to −252 base pairs from start of transcription. MSP-F and MSP-R indicate the positions of methylation-specific primers for Q-MSP; N-F and N-R, locations of outer primers for nested PCR; and P-F and P-R, locations of the pyrosequencing amplification primers. The transcription start site is indicated by an arrow. (B) Pyrosequencing analysis of KLF4 promoter methylation in LCL and lymphoma cell lines. The methylation status of 7 CpG dinucleotides was analyzed. The ratio of methylated CpG is indicated by color coding. (C) Pyrosequencing analysis of KLF4 promoter methylation in peripheral blood and tonsillar CD19+ cells as well as in tonsillar B-cell subsets. (D) Pyrosequencing analysis of KLF4 promoter methylation in the primary cases of FL, DLBCL, and BL. (E) Methylation status of cHL samples. Bisulfite-converted genomic DNA from 7 samples of normal CD19+ cells isolated from peripheral blood, 8 samples of primary cHL cases, and from B-cell lymphoma cell lines L428, KM-H2, L540, Namalwa, Raji, and Ramos were analyzed for the presence of methylated KLF4 alleles by Q-MSP. Data represent the ratio of methylated to unmethylated KLF4 alleles in the sample. The ACTB template, which is amplified independently of methylation, is used as a reference. CD19+ normal B cells represent hypomethylated control, and B-lymphoma cell lines are used as hypermethylated control. KLF4 promoter methylation level in cHL samples is significantly higher than in normal CD19+ cells (2-sided t test). (F) Genomic DNA was isolated from 100 microdissected HRS or stromal and bystander cells isolated from 10 cHL samples each. Bisulfite-converted samples were amplified by nested PCR with primers located as shown in panel A, and methylation status of 7 individual CpGs was measured as described in panel B.
KLF4 promoter is hypermethylated in B-cell lymphomas. (A) Region of KLF4 promoter analyzed by pyrosequencing (PYRO) from −226 to −252 base pairs from start of transcription. MSP-F and MSP-R indicate the positions of methylation-specific primers for Q-MSP; N-F and N-R, locations of outer primers for nested PCR; and P-F and P-R, locations of the pyrosequencing amplification primers. The transcription start site is indicated by an arrow. (B) Pyrosequencing analysis of KLF4 promoter methylation in LCL and lymphoma cell lines. The methylation status of 7 CpG dinucleotides was analyzed. The ratio of methylated CpG is indicated by color coding. (C) Pyrosequencing analysis of KLF4 promoter methylation in peripheral blood and tonsillar CD19+ cells as well as in tonsillar B-cell subsets. (D) Pyrosequencing analysis of KLF4 promoter methylation in the primary cases of FL, DLBCL, and BL. (E) Methylation status of cHL samples. Bisulfite-converted genomic DNA from 7 samples of normal CD19+ cells isolated from peripheral blood, 8 samples of primary cHL cases, and from B-cell lymphoma cell lines L428, KM-H2, L540, Namalwa, Raji, and Ramos were analyzed for the presence of methylated KLF4 alleles by Q-MSP. Data represent the ratio of methylated to unmethylated KLF4 alleles in the sample. The ACTB template, which is amplified independently of methylation, is used as a reference. CD19+ normal B cells represent hypomethylated control, and B-lymphoma cell lines are used as hypermethylated control. KLF4 promoter methylation level in cHL samples is significantly higher than in normal CD19+ cells (2-sided t test). (F) Genomic DNA was isolated from 100 microdissected HRS or stromal and bystander cells isolated from 10 cHL samples each. Bisulfite-converted samples were amplified by nested PCR with primers located as shown in panel A, and methylation status of 7 individual CpGs was measured as described in panel B.
To exclude the possibility that KLF4 promoter hypermethylation in B-cell lymphoma cell lines was established during in vitro culturing, we investigated KLF4 promoter methylation in the samples of primary B-cell lymphoma cases. In 8 of 9 cases of FL, 8 of 9 cases of DLBCL, and in 5 of 5 cases of BL, methylation of promoter CpGs varied between 16% and 85% (Figure 2D). The somewhat lower methylation ratio in primary tumors compared with cell lines may be the result of the ongoing KLF4 promoter methylation process in lymphoma cells or by contamination with stromal cells, the proportion of which was estimated to be approximately 20% in the non-Hodgkin lymphoma samples investigated. In cHL samples, the proportion of tumor HRS cells varies between 1% and 4%. To assess methylation in samples with the expected low frequency of methylated alleles, we used a quantitative MSP approach (Figure 2E). We found that the methylation levels in primary cHL samples were significantly higher than in normal CD19+ cells and correlated well with the estimated proportion of HRS in cHL tumors. To exclude the possibility that KLF4 promoter methylation detected with the MSP was derived from stromal cells, we used laser capture microdissection to isolate approximately 100 HRS cells and the same number of stromal and bystander cells from each of 10 cHL samples. Using a nested PCR approach followed by pyrosequencing, we found hypermethylated CpGs in 7 of 10 cHL samples (Figure 2F). The methylation status of the KLF4 promoter in stromal and bystander cells of cHL did not exceed levels observed in the normal CD19+ cells and B-cell subsets. Therefore, we conclude that KLF4 promoter methylation is a common and specific event in B-lymphomagenesis.
KLF4 induces cell-cycle arrest in BL cell lines
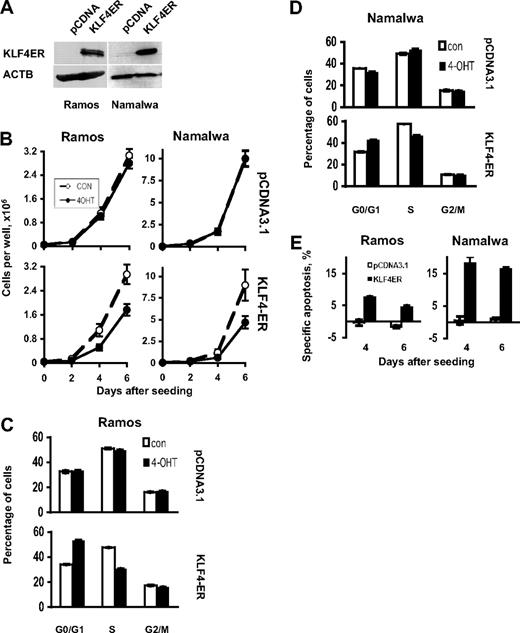
The epigenetic silencing of KLF4 in B-cell lymphoma suggests a tumor suppressor activity of this gene in B-cell lymphomagenesis. To test this hypothesis, we generated BL and cHL cell lines, stably expressing a KLF4-ER fusion gene. This construct contains the full-length KLF4 coding sequence fused in frame with a mutated estrogen receptor (ER) ligand-binding domain. This KLF4-ER fusion protein is selectively activated by 4-OHT.13 First, we investigated the effect of KLF4 in BL cell lines. Expression of the KLF4-ER protein in the Namalwa and Ramos clones was shown by immunoblot (Figure 3A). Activation of KLF4 attenuated the growth of both BL cell lines (Figure 3B). Growth inhibition was associated with an accumulation of cells in G0/G1 phase accompanied by a proportional decrease of cells in S-phase in both cell lines (Figure 3C-D). In addition, KLF4 activation increased cell death in both BL cell lines (Figure 3E). Thus, KLF4 affects growth of BL cell lines by influencing cell-cycle progression and survival.
KLF4 inhibits growth of BL cell lines. (A) Expression of KLF4-ER protein in stable clones of BL cell lines Ramos and Namalwa was measured by immunobloting using anti-KLF4 antibody. Anti-ACTB antibody was used as a loading control. (B) Cell lines were plated in 6-well plates at a density of 0.5 × 105 cells per well in 3 mL of complete culture medium (day 0). 4-OHT was added at concentration 200nM at the days 0, 2, and 4. Cells were counted with a hemocytometer. Cell viability was controlled by trypan blue staining. All experiments were repeated at least 3 times. The data represent mean ± SD of the most representative experiment. (C-D) Cell-cycle analysis. Cells were seeded at the density of 1 × 106/10 mL of the complete medium and were treated with 200nM 4-OHT at the same day; in 48 hours, cells were harvested and cell-cycle distribution was measured by PI staining as described in “Cell proliferation assay, apoptosis, and cell-cycle analysis.” Data are mean ± SD obtained in 3 independent experiments. (E) Cells were seeded at concentration of 1 × 106 cells per 10 mL of complete medium. The 4-OHT was added at the same concentration and schedule as for cell proliferation experiment and cell-cycle experiments. Cell death was measured by annexin V/PI staining. The results are represented as specific apoptosis (SA): SA (%) = 100(AE − AC)/(1 − AC), where AE equals percentage of apoptotic cells in the experimental (+4-OHT) group and AC equals percentage of apoptotic cells in the control (not treated) group. Data are mean ± SD of 1 most representative of at least 3 independent experiments.
KLF4 inhibits growth of BL cell lines. (A) Expression of KLF4-ER protein in stable clones of BL cell lines Ramos and Namalwa was measured by immunobloting using anti-KLF4 antibody. Anti-ACTB antibody was used as a loading control. (B) Cell lines were plated in 6-well plates at a density of 0.5 × 105 cells per well in 3 mL of complete culture medium (day 0). 4-OHT was added at concentration 200nM at the days 0, 2, and 4. Cells were counted with a hemocytometer. Cell viability was controlled by trypan blue staining. All experiments were repeated at least 3 times. The data represent mean ± SD of the most representative experiment. (C-D) Cell-cycle analysis. Cells were seeded at the density of 1 × 106/10 mL of the complete medium and were treated with 200nM 4-OHT at the same day; in 48 hours, cells were harvested and cell-cycle distribution was measured by PI staining as described in “Cell proliferation assay, apoptosis, and cell-cycle analysis.” Data are mean ± SD obtained in 3 independent experiments. (E) Cells were seeded at concentration of 1 × 106 cells per 10 mL of complete medium. The 4-OHT was added at the same concentration and schedule as for cell proliferation experiment and cell-cycle experiments. Cell death was measured by annexin V/PI staining. The results are represented as specific apoptosis (SA): SA (%) = 100(AE − AC)/(1 − AC), where AE equals percentage of apoptotic cells in the experimental (+4-OHT) group and AC equals percentage of apoptotic cells in the control (not treated) group. Data are mean ± SD of 1 most representative of at least 3 independent experiments.
KLF4 causes apoptosis in cHL cell lines
To investigate potential tumor suppressor effects of KLF4 in cHL cell lines, we generated KM-H2 and L428 cell lines stably expressing the KLF4-ER fusion protein (Figure 4A). Activation of KLF4 rapidly decreased viability of both cHL cell lines, and the effect was much more striking than for the BL cell lines (Figure 4B). We again first analyzed whether KLF4 affected cell-cycle progression and found a significant G0/G1-arrest specifically in KM-H2 cells, but not in L428 cells (Figure 4C-D). Combining annexin V and PI staining, we found that the decrease in the viable cell numbers was mainly the result of induction of cell death (Figure 4E). Although KLF4 induced massive apoptosis in both cell lines, the kinetics of this cell death was slightly different. In KM-H2 cells, specific apoptosis slowly increased from 15.5% on day 2 of 4-OHT treatment to 90.5% on day 6. In L428, 45% of cells were apoptotic already 1 day after the treatment, and on day 2 specific apoptosis reached 86.6% (Figure 4E). 4-OHT did not induce apoptosis in cells transfected with empty vector. To investigate the involvement of caspase activation in KLF4-induced cell death, we first tested the ability of Z-VAD.fmk to inhibit KLF4-induced CASP3 cleavage in the L428 cell line (supplemental Figure 2A). Z-VAD.fmk is a broad caspase inhibitor that was previously shown to inhibit CD95 ligation-induced apoptosis in L428 and other cHL cell lines at a concentration range of 10 to 100μM.22 Of note, KM-H2 cells express very low levels of CASP3, and Z-VAD.fmk does not inhibit apoptosis in this cell line.22,23 We found that CASP3 activation in the L428 cell line was inhibited by Z-VAD.fmk at concentrations of 40μM and 160μM, respectively. Dose dependence of the antiapoptotic effect of Z-VAD.fmk was investigated at concentrations from 2 to 320μM (supplemental Figure 2B). Z-VAD.fmk only slightly inhibited apoptosis in both cell lines in the 20 to 320μM concentration range. Finally, we investigated the antiapoptotic activity of the Z-VAD.fmk at a concentration of 320μM at different time points after KLF4 activation. Z-VAD.fmk only marginally inhibited KLF4-induced apoptosis in both cell lines (Figure 4F). Thus, KLF4 activation in cHL cell lines induces mainly caspase-independent cell death.
KLF4 induces cell death in cHL cell lines. (A) Expression of KLF4-ER fusion protein in KMH2-KLF4-ER and L428-KLF4-ER as well as in KM-H2-pcDNA and L428-pcDNA clones was investigated as described for Figure 2 legend. (B) KLF4 activation suppresses growth of cHL cell lines. The experiments were done as described for Figure 2B legend, except that cells were seeded at density 1 × 105/well because of the relatively slow growth kinetic of cHL cell lines. (C-D) Cell-cycle distribution was measured as indicated for Figure 2C and D. (E) KLF4 induces apoptosis in cHL cell lines (Figure 2E). (F) Caspase inhibition does not completely protect cHL cell lines from KLF4-induced apoptosis. A total of 2 × 105 cells/well of 6-well plate were seeded in 3 mL of the medium. Z-VAD.fmk was added at the day of seeding at concentration 320μM followed by 200nM of 4-OHT 2 hours later. Cell death is given as percentage of dead cells. Data are mean ± SD of 3 independent experiments.
KLF4 induces cell death in cHL cell lines. (A) Expression of KLF4-ER fusion protein in KMH2-KLF4-ER and L428-KLF4-ER as well as in KM-H2-pcDNA and L428-pcDNA clones was investigated as described for Figure 2 legend. (B) KLF4 activation suppresses growth of cHL cell lines. The experiments were done as described for Figure 2B legend, except that cells were seeded at density 1 × 105/well because of the relatively slow growth kinetic of cHL cell lines. (C-D) Cell-cycle distribution was measured as indicated for Figure 2C and D. (E) KLF4 induces apoptosis in cHL cell lines (Figure 2E). (F) Caspase inhibition does not completely protect cHL cell lines from KLF4-induced apoptosis. A total of 2 × 105 cells/well of 6-well plate were seeded in 3 mL of the medium. Z-VAD.fmk was added at the day of seeding at concentration 320μM followed by 200nM of 4-OHT 2 hours later. Cell death is given as percentage of dead cells. Data are mean ± SD of 3 independent experiments.
KLF4 activates genes involved in regulation of cell-cycle progression and apoptosis
Because cHL cell lines were much more sensitive to KLF4 activation than BL cells, we investigated the molecular aspects of KLF4 action in cHL cell lines. First, we validated the functional activity of the construct by testing induction of known KLF4 target genes, including CDKN1A/p21, CDKN1C/p57, and CCND2/cyclin D22,6,24,25 using quantitative PCR. KLF4 activation increased expression of CDKN1A and CCND2 mRNA in both cell lines, whereas CDKN1C was strongly up-regulated specifically in KM-H2 cells (Figure 5). The most prominent tumor-suppressive effect of KLF4 in cHL-cell lines we observed was induction of apoptosis. We therefore examined the expression level of 88 genes involved in apoptosis pathways, using the Human Apoptosis Primer Library. Fourteen genes were modulated by KLF4 activation in both L428 and KMH2 cells by more than 2-fold (Table 1). Among the up-regulated genes were caspases 4 and 5, which are not classified as initiator or effector caspases, and their involvement in apoptosis has rarely been reported. CASP7, although being an effector caspase, is mainly regulated at the posttranslational level,26 and its activation is inhibited by zVAD-fmk. Therefore, it was unlikely that CASP7 played a dominant role in KLF4-induced apoptosis in cHL cells. We found a moderate up-regulation of several genes involved in the extrinsic apoptosis pathway, CASP8 and FADD-like apoptosis regulator (c-FLIP), tumor necrosis factor receptor superfamily members 10a (TRAILR1) and 10c (TRAILR3).
Validation of functional activity of KLF4 constructs in cHL cell lines by induction of specific target genes. A total of 2 × 106 KMH2-KLF4-ER and L428-KLF4-ER cells were seeded in 10 mL of complete culture medium and treated with 200nM of 4-OHT at the same day. Twenty-four hours later, cells were harvested, and expression of CDKN1A, CCND2, and CDKN1C was assessed by quantitative PCR. Data represent relative mRNA expression (mean ± SD) calculated by the comparative Ct method. The relevant nontreated controls were used as comparators. RPL13A was used as reference gene. All measurements were made in triplicate.
Validation of functional activity of KLF4 constructs in cHL cell lines by induction of specific target genes. A total of 2 × 106 KMH2-KLF4-ER and L428-KLF4-ER cells were seeded in 10 mL of complete culture medium and treated with 200nM of 4-OHT at the same day. Twenty-four hours later, cells were harvested, and expression of CDKN1A, CCND2, and CDKN1C was assessed by quantitative PCR. Data represent relative mRNA expression (mean ± SD) calculated by the comparative Ct method. The relevant nontreated controls were used as comparators. RPL13A was used as reference gene. All measurements were made in triplicate.
Among the genes up-regulated by KLF4 activation, we also found 2 genes encoding mitochondrial proapoptotic proteins: second mitochondria-derived activator of caspases/direct inhibitor of apoptosis-binding protein with low pI (Smac/DIABLO) and BAK1. Because of the level of its up-regulation (13- to 23-fold), we focused on BAK1. BAK1 is a multidomain, proapoptotic member of the BCL-2 family and an essential gateway to the mitochondrion-mediated cell death program. Triggered by various proteins, such as PUMA and NOXA, oligomerization of BAK1 and BAX in the mitochondrial outer membrane forms channels that cause release of mitochondrial proteins, such as cytochrome c, SMAC/Diablo, apoptosis-inducing factor, HtrA2/Omi, and endonuclease G, which may induce caspase activation but may also lead to caspase-independent cell death.27 Given that BAK1 is upstream of Smac/DIABLO in the mitochondrial apoptotic pathway and exhibits the highest up-regulation among the 14 genes, we further investigated the role of BAK1 in KLF4-induced apoptosis in cHL cell lines.
BAK1 is responsible for KLF4-induced cell death
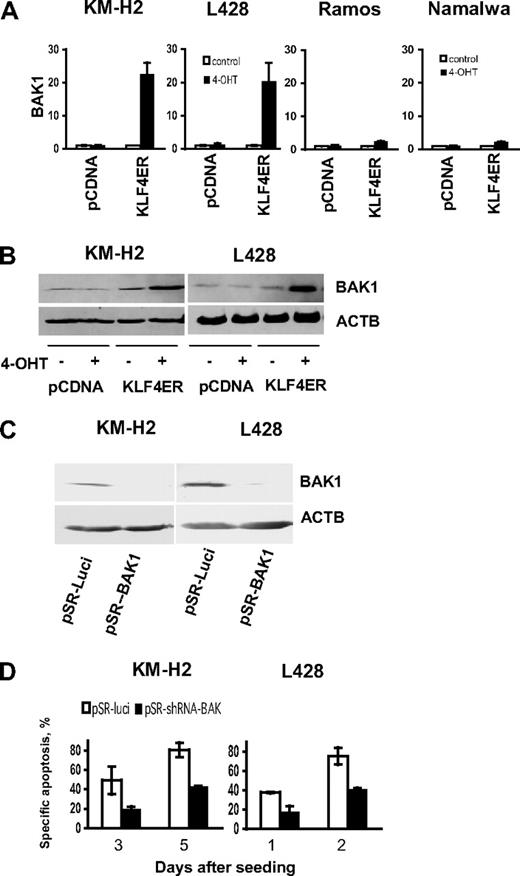
First, we verified KLF4-induced BAK1 mRNA up-regulation in cHL cell lines and compared it with BAK1 expression induced in BL cells, using BAK1-specific primers by quantitative PCR (Figure 6A). We found that, 24 hours after KLF4 activation, BAK1 mRNA levels were increased more than 20-fold in both cHL cell lines, which is in good correlation with results obtained using the Human Apoptosis Primer Library (Table 1). Interestingly, in BL cell lines, KLF4 activation induced only 2-fold up-regulation of BAK1, which correlated with the significantly lower levels of KLF4-induced apoptosis in Namalwa and Ramos cell lines. BAK1 up-regulation in cHL cell lines was observed also at the protein level (Figure 6B). To clarify the role of BAK1 in KLF4-induced apoptosis, we silenced BAK1 in cHL cell lines using a specific shRNA construct. As control, we used vector expressing a shRNA specific for luciferase mRNA. The BAK1 specific shRNA reduced BAK1 protein expression in L428-KLF4-ER and KM-H2-KLF4-ER cell lines, whereas the control shRNA did not (Figure 6C). The transient overexpression of pRS-shBAK1 in L428-KLF4-ER and KM-H2-KLF4-ER significantly decreased KLF4-induced apoptosis (Figure 6D). Thus, we show, for the first time, that KLF4-mediated apoptosis in cHL-cell lines depends on induction of BAK1 expression.
BAK1 activation is responsible for KLF4-induced apoptosis in cHL cell lines. (A) KLF4 induces BAK1 mRNA expression. A total of 2 × 106 KMH2, L428, Namalwa, and Ramos cells stably expressing KLF4-ER fusion protein, or empty vector were seeded in 10 mL of complete culture medium and treated with 200nM of 4-OHT at the same day. Twenty-four hours later, cells were harvested and BAK1 expression was assessed by quantitative PCR. Data are relative mRNA expression (mean ± SD) calculated by the comparative Ct method. The relevant nontreated controls were used as comparators. RPL13A was used as reference gene. All experiments were performed in triplicate. (B) KLF4 activation induces BAK1 protein expression in cHL cell lines. Western immunoblots of transfected cells with or without 4-OHT are shown. ACTB was used as loading control. (C-D) Silencing of BAK1 expression by shRNA prevents KLF4 induced cell death. (C) KM-H2-KLF4-ER or L428-KLF4-ER cells were transiently transfected with pRS-luci and pRS-shBAK1 vectors followed by 5-day selection with 0.5 μg/mL (KM-H2) and 1 μg/mL (L428) of puromycin. BAK1 expression was determined by immunoblot. (D) KM-H2-KLF4-ER and L428-KLF4-ER were transfected with pRS-BAK1 or pRS-luci and were selected for 5 days with puromycin. Then cells were recovered for 1 day in the complete culture medium without puromycin followed by treatment with 200nM of 4-OHT (day 0). Apoptosis was measured on the indicated time points. Data are mean ± SD obtained in 2 independent experiments.
BAK1 activation is responsible for KLF4-induced apoptosis in cHL cell lines. (A) KLF4 induces BAK1 mRNA expression. A total of 2 × 106 KMH2, L428, Namalwa, and Ramos cells stably expressing KLF4-ER fusion protein, or empty vector were seeded in 10 mL of complete culture medium and treated with 200nM of 4-OHT at the same day. Twenty-four hours later, cells were harvested and BAK1 expression was assessed by quantitative PCR. Data are relative mRNA expression (mean ± SD) calculated by the comparative Ct method. The relevant nontreated controls were used as comparators. RPL13A was used as reference gene. All experiments were performed in triplicate. (B) KLF4 activation induces BAK1 protein expression in cHL cell lines. Western immunoblots of transfected cells with or without 4-OHT are shown. ACTB was used as loading control. (C-D) Silencing of BAK1 expression by shRNA prevents KLF4 induced cell death. (C) KM-H2-KLF4-ER or L428-KLF4-ER cells were transiently transfected with pRS-luci and pRS-shBAK1 vectors followed by 5-day selection with 0.5 μg/mL (KM-H2) and 1 μg/mL (L428) of puromycin. BAK1 expression was determined by immunoblot. (D) KM-H2-KLF4-ER and L428-KLF4-ER were transfected with pRS-BAK1 or pRS-luci and were selected for 5 days with puromycin. Then cells were recovered for 1 day in the complete culture medium without puromycin followed by treatment with 200nM of 4-OHT (day 0). Apoptosis was measured on the indicated time points. Data are mean ± SD obtained in 2 independent experiments.
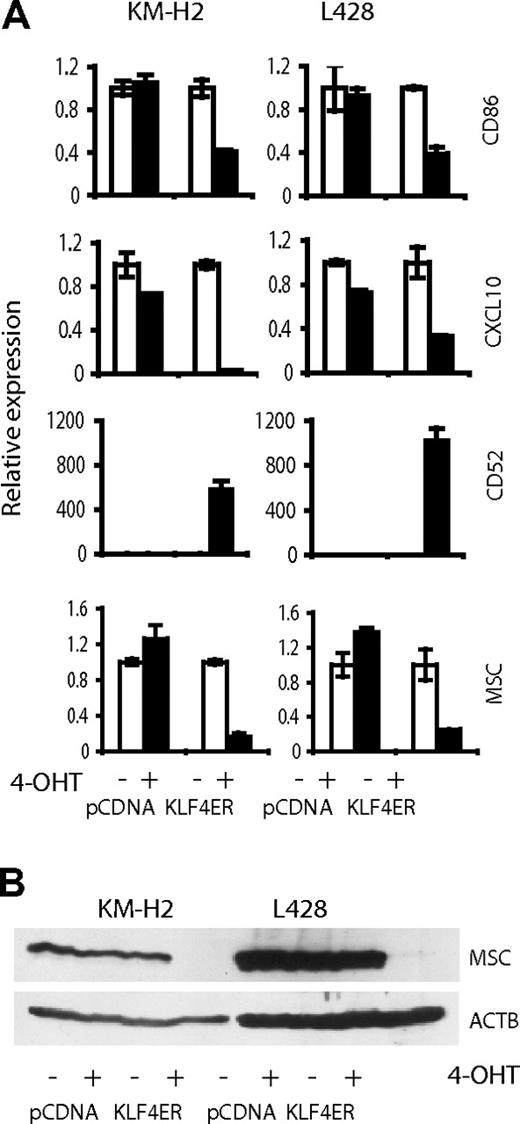
KLF4 down-regulation may be involved in establishing specific phenotype of HRS cells
The characteristic repression of the B-cell phenotype in cHL is thought to be a result of transcription factor network deregulation because of the overexpression of repressor proteins ID2 and ABF-1.28 KLF4 is a versatile transcription factor, participating in regulation of differentiation processes in various tissues.7,29 Epigenetic silencing of KLF4 in cHL hints that KLF4 is involved in the complex mechanism of extinguishing of B-cell phenotype in cHL. To clarify this issue, we investigated transcriptome changes induced by KLF4 activation in KM-H2 and L428. KLF4 activation led to more than 3-fold up-regulation of 59 genes and to more than 3-fold down-regulation of 10 genes in both cell lines (supplemental Table 1). We searched for genes potentially involved in cHL pathogenesis. Among the KLF4-induced genes, only CD52 is differentially expressed in cHL.30 Among the down-regulated genes, CD86/B7-2, CXCL10, and MSC/ABF-1 could be critical targets explaining the cHL phenotype. CD86 and CXCL10 are important for establishing a specific cellular milieu for attraction of infiltrating immune cells into cHL. MSC/ABF-1 together with ID2 is involved in extinguishing the B-cell phenotype in HRS cells.28 Using quantitative PCR, we validated expression changes for the selected genes (Figure 7A). Because of the special role of MSC/ABF-1 in silencing of B-cell phenotype in cHL, we used immunoblot to investigate MSC/ABF-1 expression on the protein level. In both cell lines, KLF4 completely inhibited MSC/ABF-1 protein expression (Figure 7B).
Validation of array-based gene expression profiles. (A) Quantitative PCR. (B) Immunoblot. Expression of CD86, CXCL10, CD52, and MSC was determined in KM-H2 and L428 cells, expressing KLF4-ER or empty vector, treated and not treated with 200nM of 4-OHT. After 24 hours of incubation with 4-OHT, cells were harvested. mRNA expression was determined by quantitative PCR and calculated in relation to internal control gene RPL13A by the comparative Ct method. Mean value of the group expressing empty vector and not activated with 4-OHT was used as comparator. Data are mean ± SD.
Validation of array-based gene expression profiles. (A) Quantitative PCR. (B) Immunoblot. Expression of CD86, CXCL10, CD52, and MSC was determined in KM-H2 and L428 cells, expressing KLF4-ER or empty vector, treated and not treated with 200nM of 4-OHT. After 24 hours of incubation with 4-OHT, cells were harvested. mRNA expression was determined by quantitative PCR and calculated in relation to internal control gene RPL13A by the comparative Ct method. Mean value of the group expressing empty vector and not activated with 4-OHT was used as comparator. Data are mean ± SD.
Discussion
We have shown that KLF4 is hypermethylated in primary cases of B-cell lymphoma and this hypermethylation of the KLF4 promoter is associated with gene silencing. Reexpression of KLF4 leads to growth retardation in BL cell lines and induces apoptosis in cHL cell lines. Activation of BAK1 transcription plays a major role in KLF4-induced apoptosis in cHL cell lines.
KLF4 promoter hypermethylation has been shown for a variety of epithelial tumors where KLF4 typically functions as a tumor suppressor.1,31 In gastric carcinoma, KLF4 is frequently hypermethylated.1 Inactivation of KLF4 by methylation was also observed in adult T-cell leukemia.32 Epigenetic abnormalities are common in B-cell lymphomas12,33,,–36 ; in accordance with this, KLF4 promoter hypermethylation was detected in all tested FL lymphoma21 and in DLBCL37 samples. We extend these findings and provide evidence of promoter methylation also in HRS cells of cHL. We show that KLF4 promoter hypermethylation is a common event in GC B-cell lymphomas. It is probable that low transcriptional activity of the KLF4 gene precedes the hypermethylation of the KLF4 promoter in cHL. Notch1, a known repressor of KLF4, is highly expressed in cHL.38 In contrast, the transcription factor PU.1, a transcriptional activator of KLF4,39 is epigenetically silenced in cHL.12,16 The observed heterogeneity of methylation patterns among the CpGs of the same tumor as well between tumor samples may indicate that KLF4 methylation is gradually acquired in the process of lymphomagenesis. Inheritance of the hypermethylation pattern and its maintenance indicates a negative influence of KLF4 on B-cell growth and survival. Indeed, we found that KLF4 activation moderately delays growth of BL cell lines and induces massive cell death in cHL cell lines. HRS cells of cHL are generally thought to be derived from preapoptotic GC B cells38 ; hence, the transforming events must not only rescue HRS cells from apoptosis but also endow them with the ability of uncontrolled proliferation. Numerous antiapoptotic pathways are typically activated in HRS cells. Death receptor-induced apoptosis is inhibited by c-FLIP overexpression40 and up-regulation of XIAP suppresses caspase activation.41 Constitutively activated in cHL NF-κB leads to high expression of Bcl-xL42 along with defective BAX activation.43 Therefore, to induce apoptosis in HRS cells, KLF4 must corrupt the multicomponent antiapoptotic machinery. The ability of KLF4 to induce apoptosis was demonstrated in adult T-cell leukemia32 and in a mouse model of pre-B cell leukemia.2 In pre-B cell leukemia, KLF4 induced caspase-independent cell death, which could be attenuated by BCL2L1/Bcl-xL overexpression.2 Similarly, in our experiments, KLF4-induced cell death in the human cHL cell lines also was not inhibited by caspase inhibitor Z-VAD.fmk. KLF4-induced activation of BAK1 is reminiscent to KLF4-induced mitochondrial damage described in the pre-B cell lymphoma.2 In healthy cells, BAK1 is specifically sequestrated through interaction with MCL1 or BCL2L1/BCL-xL and induces apoptosis only if freed from both.44 Thus, we assume that KLF4 induces apoptosis by stimulating the overproduction of BAK1 in amounts, exceeding the sequestration capacity of BCLxL. Remarkably, the caspase-independent character of KLF4-induced cell death also hints at the BAK1 involvement. BAX and BAK1 were shown to promote caspase-independent apoptosis.45,46
The additional KLF4-target genes revealed by gene expression profiling include several interesting genes potentially involved in cHL lymphomagenesis. We found that KLF4 negatively regulated CD86 and CXCL10. Therefore, KLF4 down-regulation may favor increasing production of the receptors and cytokines (eg, CD86 and CXCL10), facilitating attraction of the bystander T lymphocytes to HRS cells of cHL.47,48 Given that HRS cell survival largely depends on the extracellular signals provided by the inflammatory milieu, this factor may also be important for cHL pathogenesis.36 Furthermore, the strong inhibition of the repressor MSC/ABF-1 by KLF4 is of significant interest. MSC/ABF-1 is highly expressed in LCL, PMBL, and cHL.28,33 Of note, the MSC/ABF-1 promoter is often hypermethylated in FL, DLBCL, and BL.33 This may explain the absence of MSC/ABF-1 expression in these tumors despite low levels of KLF4. MSC/ABF-1 forms heterodimers with E-box transcription factors E47/E12 suppressing their transactivating potential.49 E47/E12 proteins are principal regulators of B-cell maturation, activating their target genes IGH, CD79A, and AICDA.28 E47/E12 control B-cell proliferation by activating the target gene p21waf/cip1.50 Inactivation of E47/E12 by MSC/ABF-1 and ID2 was suggested to be responsible for silencing of the B-cell-specific expression program and increased proliferation potential of cHL.28 It is possible that down-regulation of KLF4 is a prerequisite for MSC/ABF-1 expression.
In conclusion, our data provide new insight into the mechanisms of lymphomagenesis, especially in cHL. Epigenetic silencing of KLF4 might be necessary to circumvent the apoptotic barriers faced by crippled GC B cells in the process to become HRS cells. Particularly, epigenetic mechanisms, including promoter methylation of KLF4, might be operative in cHL pathogenesis, and its reversible nature makes it an attractive therapeutic target.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anita Kick for excellent technical assistance and Jonathan Alder for donation of the pcDNA3.1-KLF4ER expression vector.
This work was supported by Deutsche Krebshilfe eV (grant 107547; T.W., A.U.) and the Fonds der Chemischen Industrie. H.G. was supported by the China Scholarship Council.
Authorship
Contribution: H.G., L.X., and L.F. performed research; F.L. and P.M. contributed vital reagents and analyzed data; and T.W. and A.U. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Wirth, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, D-89069 Ulm, Germany; e-mail: thomas.wirth@uni-ulm.de.