We conducted a retrospective collaborative study to cytogenetically characterize splenic marginal zone lymphoma (SMZL) and ascertain the prognostic value of chromosomal aberrations. Of 330 cases, 72% displayed an aberrant karyotype, 53% were complex, and 29% had a single aberration. The predominant aberrations were gains of 3/3q and 12q, deletions of 7q and 6q and translocations involving 8q/1q/14q. CD5 expression was detected in 39 of 158 cases (25%). The cytogenetic makeup of the CD5+ group differed significantly from that of the CD5− group. Cases with unmutated IGHV were significantly associated with deletions of 7q and TP53. A strong association was noted between usage of the IGVH1-2 and deletion 7q, 14q alterations, and abnormal karyotype. On univariate analysis, patients with more than or equal to 2 aberrations, 14q alterations, and TP53 deletions had the shortest survival; 7q deletion did not affect survival. On multivariate analysis, cytogenetic aberrations did not retain prognostic significance; the parameters negatively affecting survival were hemoglobin and age. In conclusion, the cytogenetic profile of SMZL is distinct from other B-cell lymphomas. Complexity of the karyotype, 14q aberrations, and TP53 deletions are poor prognostic indicators and may be considered together with other clinicobiologic parameters to ascertain the prognosis of SMZL.
Introduction
Splenic marginal zone lymphoma (SMZL) is a rare low-grade B-cell lymphoma considered as a distinct entity in the World Health Organization (WHO) classification.1,2 The diagnosis may be problematic in cases without spleen histology. For this reason, recently, the Splenic Lymphoma Group (SBLG) proposed guidelines for the diagnosis of SMZL and outlined the main clinical, cytogenetic, and biologic features of the disease.3,–5
Cytogenetic data have been reported in several series of SMZL, mainly including patients with active disease that required splenectomy.6,,,,,–12 Approximately 70% to 80% of cases have been reported to exhibit cytogenetic and/or molecular genetic abnormalities, however, without a disease-specific cytogenetic alteration.3 The most frequent cytogenetic abnormalities are deletions at the 7q22-q32 band, followed by gains of chromosome 3/3q. Complex karyotypes are common: the chromosomes most frequently involved are 1, 3, 7, 8, and 14.
The true incidence of cytogenetic abnormalities in SMZL has not been well established. Furthermore, it is still uncertain whether cytogenetic abnormalities have a prognostic impact in SMZL, as is the case for chronic lymphocytic leukemia (CLL). Deletions of chromosomes 7q or 17p have been proposed as adverse prognostic indicators; however, the true prognostic impact of such abnormalities is difficult to assess, given the small size of the respective cohorts.13,–15
In the present study, we describe in detail the cytogenetic findings in a large group of well-characterized SMZLs collected by the SBLG to ascertain: (1) the true cytogenetic profile of SMZL, (2) possible correlations between cytogenetic abnormalities and other disease features, and (3) prognostic implications of cytogenetic findings.
Methods
Patients
A total of 330 patients with a diagnosis of SMZL were included in the study. Cases were referred from the French (E. Callet-Bauchu, C.T., and A.T.-G.), British (D.O. and E.M.), German (R. Siebert and J.D.), Belgian (G.V. and I.W.), and Greek (K.S., A.A., and T.P.) groups and the Spanish Cooperative Group for Hematological Cytogenetics (GCECGH).
For splenectomized cases (n = 143), the diagnosis was established according to the 2008 WHO classification criteria.1,2 For nonsplenectomized cases (n = 187), the diagnosis of SMZL was established according to SBLG guidelines,3 based on a combination of features, including clinical presentation, lymphocyte morphology, immunophenotype, and bone marrow (BM) histology because of the limited number of cases with splenic histology for review (n = 143). The phenotype in patients without available spleen histology was suggestive of SMZL. In particular, it revealed the presence of a clonal B-cell population with low CLL scores, negative for either CD103 or (in all tested cases) CD123; a proportion of cases were CD11c+ and a few weakly expressed CD25; on these grounds, a diagnosis of hairy cell leukemia was excluded.
Table 1 summarizes the main clinical and hematologic features as well as follow-up data for all patients with available information (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Progression was defined as: requirement to treatment because of development or progression of lymphadenopathy, systemic symptoms, or histologic transformation into large B-cell lymphoma.
The study was approved by the local Ethics Review Committee of each Institution, in accordance with the guidelines of the Declaration of Helsinki.
Cytogenetic and FISH studies
Cytogenetic studies were performed in all patients at diagnosis before any treatment or at the time of splenectomy. Chromosome analysis was carried out on lymphoid cells from peripheral blood (n = 222), bone marrow (n = 36), lymph nodes (n = 6), or spleen (n = 66), as previously described.10 Cytogenetic analysis with G- and/or R-banding was performed locally, whereas reports were reviewed centrally by the main investigators (C.B., M.S., and F.S.). Karyotypes were described according to the International System for Human Cytogenetic Nomenclature.16
The following definitions were adopted: (1) complex karyotype: any karyotype presenting 3 or more cytogenetic aberrations or the existence of 2 or more clones; and (2) “2-aberration” group: patients exhibiting 2 aberrations in the same karyotype or 2 karyotypically independent clones with a single change. Single aberrations may include numerical or structural changes; the latter category encompasses derivative chromosomes and translocations.
Fluorescence in situ hybridization (FISH) was performed locally in cases with available fixed cells previously processed for G-banding analysis, as described.10 The DNA probes used for FISH analyses are listed in supplemental Table 1. The following cut-offs were used for chromosome gain, loss, or rearrangement: 5%, 10%, or 5%, respectively.
Cross-species color banding or spectral karyotyping techniques were applied in some cases with a complex karyotype to accurately define the chromosomal aberrations; results from this analysis have been reported previously.10,17 Based on multicolor FISH findings, the chromosomal aberrations of these cases were redefined and compiled with the G-banding karyotype data.
Statistical analysis
The following parameters were analyzed for possible associations with survival: gender, age, performance status, hemoglobin (Hb) level, platelet count, leukocyte and lymphocyte count, serum albumin, serum lactate dehydrogenase (LDH), and β2-microglobulin levels, CD5 expression, splenectomy, and chemotherapy treatment. Regarding cytogenetic and molecular data, the variables analyzed were: complexity of the karyotype (normal karyotype vs sole anomaly vs 2 alterations vs 3 or more alterations), 7q deletions, 14q involvement, 14q32 translocations, 14q deletions, trisomy 18, 6q deletions, gain of chromosome 3/3q by conventional cytogenetics and FISH, and TP53 status by FISH and IGHV mutational status.
Statistical associations were assessed using Pearson's χ2 test and Fisher exact test, depending on sample size. The actuarial survival analysis was performed according to the Kaplan-Meier method, and log-rank test was used for comparison of survival curves between different groups of patients. For each patient, overall survival (OS) was calculated from the date of diagnosis to death (from any cause) or last follow-up. Patients still alive were censored at the last known date of contact. A total of 95% confidence limits for OS were computed using the Kalbfleisch and Prentice method.
For multivariate analyses, only cases with complete data were included (n = 140). Parameters that were statistically significant in the univariate analyses were included in the multivariate regression procedure. Multivariate analysis was performed using the Cox regression method. All statistical analyses were performed with SPSS, version 15 (SPSS).
Results
Cytogenetic and FISH analyses
Numerical and structural aberrations.
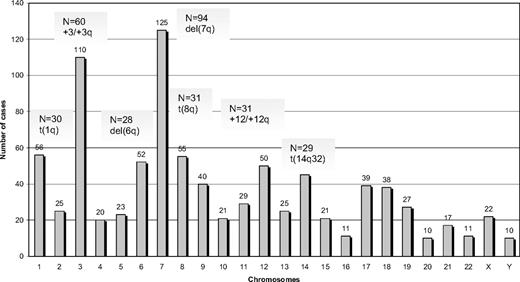
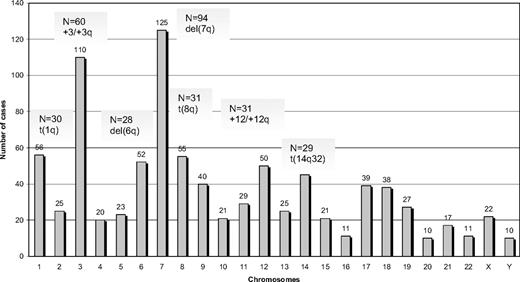
Among 330 SMZL cases analyzed by G- or R-banding cytogenetics, 239 (72%) displayed an aberrant karyotype. Within this subgroup, 68 cases (29%) showed a single cytogenetic aberration (supplemental Table 2); an additional 44 of 239 cases (18%) carried 2 alterations, whereas 127 of 239 cases (53%) showed a complex karyotype (3 or more cytogenetic abnormalities). Cytogenetic results are summarized in Figure 1 and Tables 2 to 4.
A total of 882 alterations were identified, of which 216 were numerical, whereas the remaining 666 were structural. The mean number of alterations per case was 2.7 (range, 1-25): (1) numerical: 0.6 (range, 0-11); and (2) structural: 2.1 (range, 0-19). Chromosomes most frequently involved (in order of frequency) were: 7, 3, 1, 8, 6, 12, and 14. Chromosomal gains (n = 125) were more frequent than losses (n = 86). The most common numerical aberrations were gain of material from chromosome 3/3q (60 of 239 cases with aberrant karyotype, 25%), trisomy 18 (25 of 239 cases, 10%), and trisomy 12 (18 of 239 cases, 8%).
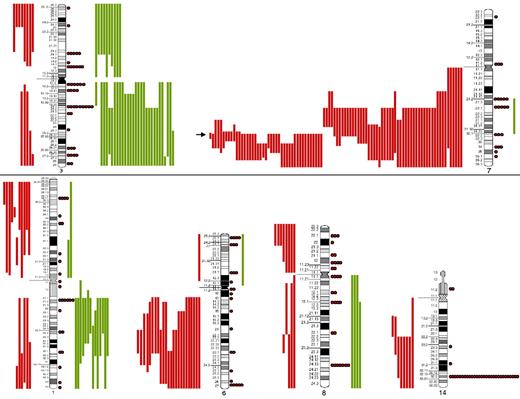
The most frequent chromosomal regions affected by structural abnormalities were 7q (109 cases), 3q (42 cases), 14q (40 cases), 6q (37 cases), 1q (30 cases), and 8q (29 cases; Figure 2); the predominant structural aberration was deletion 7q (94 of 239; 39%). Deletion breakpoints at 7q were heterogeneous, being q21 the most proximal and q36 the most terminal breakpoint. In order of frequency, 7q22 (35 cases) was the most recurrent breakpoint followed by 7q32 (22 cases), 7q31 (16 cases), 7q21 (7 cases), 7q34 (6 cases), and 7q33 (3 cases). In 6 cases, this information was not available. Seventeen of 94 (18%) cases had concomitant deletion 7q and gain of 3q in the same karyotype.
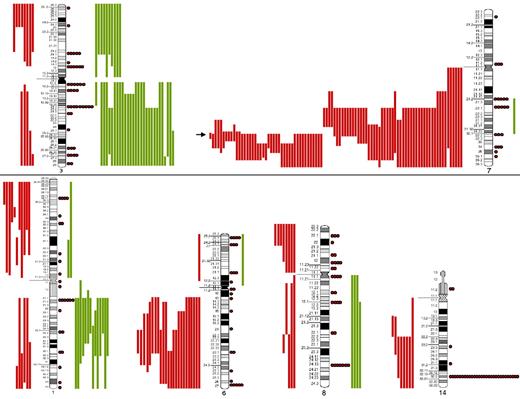
SORIs of the main chromosomes involved in SMZL. Red lines represent losses; green lines, gains; red dots, rearrangements; and arrow, the Smallest Overlapping Region of Imbalance (SORI) 7q32.1-q32.2. The most frequent chromosomal regions affected by structural abnormalities were 7q (109 cases), 3q (42 cases), 14q (40 cases), 6q (37 cases), 1q (30 cases), and 8q (29 cases).
SORIs of the main chromosomes involved in SMZL. Red lines represent losses; green lines, gains; red dots, rearrangements; and arrow, the Smallest Overlapping Region of Imbalance (SORI) 7q32.1-q32.2. The most frequent chromosomal regions affected by structural abnormalities were 7q (109 cases), 3q (42 cases), 14q (40 cases), 6q (37 cases), 1q (30 cases), and 8q (29 cases).
Translocations involving the immunoglobulin gene loci.
A total of 29 of 239 cases with aberrant karyotype (12%) carried a translocation involving the chromosome 14q32 region (IGH gene locus), in 2 cases as a single anomaly [both with t(6;14)(p21;q32)]. In 13 of 29 cases with a rearranged chromosome 14q32, the partner chromosome could not be defined (supplemental Table 3). Among the remaining cases, the following chromosomal aberrations were identified: t(14;19)(q32;q13) (4 cases), t(6;14)(p21;q32) (3 cases), t(9;14)(p13;q32) (4 cases), and t(1;14)(q21;q32) (2 cases); translocations t(1;14)(p34;q32), t(1;14)(p22;q32), t(9:14)(p11.3;q32), t(11;14)(q21;q32), and t(12;14)(q23;q32) and duplicationdup(14)(q32.1q32.3) were detected in 1 case each. Three of 4 patients with t(14;19) had additional 14q32 alterations. Translocations t(11;14)(q13;q32), t(14;18)(q32;q21), and t(3;14)(q27;q32) as well as translocations typical of mucosa-associated lymphoid tissue (MALT) lymphomas were not detected.
All cases with t(9;14)(p13;q32) were CD5−. Spleen histology information was available in 3 cases: interestingly, 2 of 3 showed a diffuse pattern of infiltration. Finally, FISH analysis was performed in 3 of 4 cases with t(9;14)(p13;q32) and confirmed the involvement of the PAX5 gene.
Translocations involving the immunoglobulin kappa (IGK) or lambda (IGL) light chain gene loci at chromosomes 2p and 22q, respectively, were detected in 6 cases, as follows: t(2;7)(p11-12;q22) (5 cases; 1 confirmed by FISH also involving the CDK6 gene) and t(8;22)(q24;q11) (1 case).
Comparison of classic karyotype and FISH results.
A comparison of classic karyotype and FISH results is given in supplemental Table 4. Notably, no differences were observed with regard to the frequency of detection of either trisomy 3/3q or IGH translocations by conventional cytogenetic or FISH analysis. In contrast, a higher proportion of cases with deletion 17p was detected by FISH versus conventional karyotype (18% vs 8.7%, respectively). Among cases with 11q deletion (n = 16), FISH analysis did not provide evidence of involvement of the ATM gene. Regarding deletions 7q or 13q as well as trisomy 12, only a few cases were analyzed by FISH using relevant probes, thus precluding any meaningful comparisons with the results from classic cytogenetic analysis.
Cytogenetic profiling of CD5+ cases
Among 158 cases with available data, 39 (25%) were CD5+. Table 4 compares clinical features and cytogenetic findings in CD5+ and CD5− cases. The CD5+ subgroup did not differ from the CD5− subgroup with regard to age, gender, β2-microglobulin and LDH levels, hematologic values, the presence of villous lymphocytes, and IGHV gene mutational status.
Interestingly, however, despite the relative homogeneity of the 2 groups defined by CD5 expression, their cytogenetic makeup differed. In particular, trisomy 3/3q, deletion 6q, and trisomy 18 were significantly more frequent in the CD5+ group (P = .045, P = .004, and P = .038, respectively), whereas a single abnormality was more common in the CD5− group (P = .040). Complex karyotypes were more frequent among CD5+ cases; however, the difference was not statistically significant (P = .073). CD5 expression was not associated with trisomy 12, 14q32 translocations, or TP53 deletions. In contrast, deletion 7q as a single abnormality was less frequent in CD5+ versus CD5− cases (1 of 13 cases, 7.7%; vs 13 of 47 cases, 27.7%; P = .264).
Immunoglobulin gene repertoire, mutational analysis, and associations with cytogenetic findings
Sequence information about the clonotypic IGHV-D-J rearrangements was available in 100 cases of the present study. The most frequent IGHV gene was IGHV1-2 (32 cases, 32%), followed by IGHV4-34 (11 cases, 11%) and IGHV3-23 (10 cases, 10%). After the 98% identity cut-off value, 59 of 100 cases (59%) from the present series carried IGHV genes with less than 98% identity to germline and were defined as “mutated,” whereas the remainder (41 of 100 cases, 41%) carried “unmutated” IGHV genes; within the latter subgroup, only 14 cases carried IGHV genes with 100% germline identity. At the individual gene level, the distribution of rearrangements of IGHV genes according to mutation status varied significantly; in particular, IGHV1-2 rearrangements were more frequently unmutated compared with other IGHV genes (43.9% vs 23.7%; P = .033).
At cohort level, significant differences were not identified with regard to clinical features or outcome between cases with mutated versus unmutated IGHV genes. However, 7q and TP53 deletions were significantly more frequent in the latter group (P = .006 and P = .045, respectively). At the individual IGHV gene level, IGHV1-2 usage was associated with a lymphocyte count lower than 5 × 109/L (P = .024) and Hb level less than 12g/dL (P = .01). Notably, significant differences were noted with regard to cytogenetic profiles between cases expressing the IGHV1-2 versus other IGHV genes. In particular, 7q deletions, alterations at 14q, and abnormal karyotypes were significantly more frequent in the IGHV1-2 group (P = .002, P = .011, and P = .049, respectively; supplemental Table 5).
Prognostic factors for survival
The results of the univariate and multivariate analysis identifying significant prognostic factors for OS are summarized in Table 5. On univariate analysis, age more than 65 years, Hb level less than 12 g/dL, and serum β2-microglobulin level more than 3 mg/L were associated with a shorter survival (median 6.4 vs 14.5 years, P < .001; median 8.5 vs 14 years, P = .004; and median 8.3 vs 14.5 years, P = .008, respectively). Patients with a lymphocyte count less than 4 × 109/L and a platelet count less than 100 × 109 had a poor prognosis and shorter survival (7.2 vs 11.6 years, P = .027 and 6 vs 10.8 years, P = .092, respectively; supplemental Figure 2). CD5 expression was not found to affect survival (P = .898).
A total of 74 patients were stratified according to the Intergruppo Italiano Linfomi Score18 in 3 prognostic groups: low-risk (n = 26), intermediate-risk (n = 24), and high-risk (n = 24; Table 1). The median survival times for each risk group did not differ significantly (14.5, 8.2, and 7.2 years, respectively; P = .256).
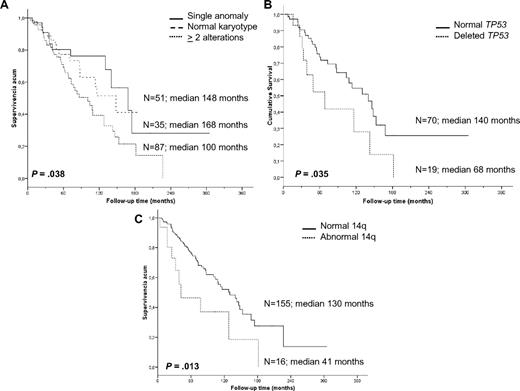
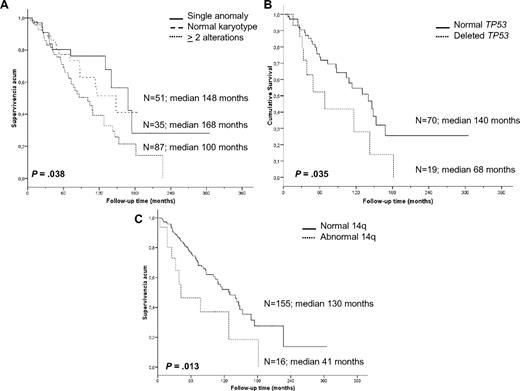
Cytogenetic findings had an impact on OS. Patients with a single anomaly had a better survival than those with 2 or more aberrations (median 14 vs 8.4 years, P = .038); however, no differences were observed between patients with 2, 3, or more chromosomal alterations. Furthermore, patients with 2 or more cytogenetic aberrations had a higher risk of progression than patients with normal karyotype or a single anomaly (53.3% vs 37%, respectively, P = .041); the latter 2 groups experienced similar survival (P = .619). Moreover, we studied the association between progression and the most frequent genomic aberrations detected by classic cytogenetic and/or FISH analysis (7q−, +3/+3q, and 14q aberrations; 6q−, 1q+, and 8q alterations; and TP53 deletions); however, no significant associations were identified, probably because of the heterogeneity of the karyotypes.
In contrast, a significantly shorter survival was noted for patients with recurrent 14q abnormalities (3.5 vs 10.8 years, P = .013), which mostly accounted for the markedly inferior prognosis of cases with 14q deletion. Indeed, the OS of such cases was only 2.6 years compared with patients with 14q32 translocations who had an OS of 10.6 years (P = .007). Deletion of 7q (9 vs 10.8 years, P = .613), gain of 3q by cytogenetics and FISH (9 vs 10.6 years, P = .379), trisomy 18 (10.8 vs 10.8 years, P = .960), and 6q deletion (7.1 vs 10.8 years, P = .307) did not have any influence on survival. Finally, the presence of TP53 deletions was an unfavorable indicator (median OS 5.6 vs 11.6 years, P = .035; Figure 3).
Kaplan-Meier survival curves of the cytogenetic findings with prognostic impact. (A) Complexity of karyotype. (B) TP53 status by FISH. (C) 14q aberrations.
Kaplan-Meier survival curves of the cytogenetic findings with prognostic impact. (A) Complexity of karyotype. (B) TP53 status by FISH. (C) 14q aberrations.
On multivariate analysis, the only parameters that maintained a negative prognostic influence on OS were hemoglobin less than 12 g/dL and age more than 65 years (Table 5).
Discussion
Cytogenetic findings
We document the chromosomal alterations identified in a series of 330 SMZL patients, by far the largest to date, with the use of conventional karyotype and FISH. Previous studies on smaller patient cohorts have reported clonal chromosomal abnormalities, with an incidence ranging from 43% to 87%.6,,–9,10,–12 These discrepancies could be attributed to small size of most series as well as different inclusion criteria, given that some studies included patients with or without lymphocytosis or were performed after treatment. In addition, at least a subset of cases included in our analysis may represent diffuse red pulp small B-cell lymphoma, a provisional entity recently included in the last review of the WHO classification. Because of the low number of cases with splenic histology available, the cases with diffuse pattern of splenic infiltration were classified according to SBLG.3
In the present series, the overall incidence of chromosomal abnormalities was 72%, mainly (53%) in the form of a complex karyotpe. Most frequently involved (in order of frequency) were chromosomes 7, 3, 1, 8, 6, 12 and 14. In agreement with previous studies,6,,–9,10,–12 albeit on significantly smaller series, deletion 7q21q36, gain 3/3q, 14q32 translocations and deletion 6q were the predominant structural alterations. The most frequent numerical abnormalities were whole or partial trisomy 3, trisomy 18, and trisomy 12 (in the latter 2 cases, usually associated with gain of chromosome 310,12,19,–21 ; supplemental Table 6).
Trisomy 3 is a common abnormality in marginal zone B-cell lymphomas, in particular nodal and extranodal MALT lymphomas.8,10,19,21 It has been suggested that a gene dosage effect for genes localized on 3q rather than a specific gene disruption could be involved in the development and/or disease progression of marginal zone B-cell lymphomas.22 However, unlike other marginal cell lymphomas, in which whole trisomy 3 is the most frequent gain, in SMZL the gain specifically occurs at 3q23 and is more frequently associated with complex karyotypes. Taking into account the variety of breakpoints and partners involved in 3q rearrangements, these structural aberrations probably represent secondary events.
Deletion of 7q was the most frequent abnormality in the present series, detected in 39% of patients, a higher incidence than previously reported (17%-20%)6,10 but similar to that documented by Mateo et al who evaluated loss of heterozygosity at chromosome 7q.24 In our study, deletion 7q was found to span from 7q21 to 7q36; 7q22 and 7q32 were the most common breakpoints, whereas 7q32.1-q32.2 was the smallest overlapping region of imbalance as previously reported by conventional cytogenetics, loss of heterozygosity, FISH, and comparative genomic hybridization (CGH).19,23,25 A recent study by array CGH26 in SMZL has demonstrated a high incidence of major imbalances, including, among others, deletions at 7q (7q22-q36). The authors suggested that the SHH (7q36.2) and POT1 (7q32.32) genes could have a potentially relevant role in the pathogenesis of SMZL.26 Of note, in 22 cases of our series, deletion 7q was found as a single anomaly, further supporting the notion that, unlike other aberrations, it may represent a primary pathogenic event in SMZL.
A significant minority of cases (35 of 239, 15%) carried abnormalities affecting the immunoglobulin (IG) loci, IGH (14q32), IGK (2p12), or IGL (22q11). However, recurrent translocations found in follicular lymphoma, mantle cell lymphoma (MCL), or MALT lymphoma were not identified; only 1 case carried t(8;22)(q24;q11), typical of Burkitt lymphoma. A previous FISH study on 220 paraffin-embedded SMZL specimens documented IG translocations with different partner chromosomes in 6% of cases, with just a single case exhibiting coexistence of this type of aberration with deletion 7q.27 This is in contrast to our series, where a concurrent IG translocation was found in 17 of 35 cases (49%) carrying deletion 7q. This discrepancy may result from different methodologies and/or the status of the specimens analyzed (fresh vs paraffin-embedded).
Our study confirms the low incidence of certain 14q32 translocations, for instance, t(6;14)(p21;q32) and t(9;14)(p13;q32).11,28,29 Translocation t(14;19)(q32;q13), a rare abnormality in B-cell disorders, documented in cases with “atypical” CLL, small B-cell unclassifiable CD5+ leukemia, and various other B-cell malignancies,30,–32 was also rare (4 of 330 cases; 1.2%). Furthermore, in all cases of the present series carrying t(14;19)(q32;q13), this aberration was associated with del(7q) and a complex karyotype, in keeping with a previous report.30
Eight cases exhibited the del14q abnormality, which has been reported in low-grade B-cell malignancies, particularly CLL,33 and often associated with trisomy 12 and unmutated IGHV genes. In the present series, no case with 14q deletion was associated with trisomy 12, as in a previous report.33 Our findings suggest that 14q deletions are recurrent but infrequent in SMZL.
Comparison of CD5+ versus CD5− SMZL
The incidence of CD5 expression in the present series was similar to previous reports.4 It has been suggested that CD5 expression may portend a more aggressive clinical course in SMZL.34 Nevertheless, our data do not support this suggestion, as there were no differences in survival or most other clinicobiologic parameters between CD5+ versus CD5− cases. Similar results were reported by Baseggio et al in a series of 24 CD5+ SMZLs, in which no differences in outcome were found.35
Notably, however, we document here, for the first time, that CD5+ cases have a cytogenetic make-up different from that of CD5− cases, with a higher frequency of trisomy 3/3q, trisomy 18, and deletion 6q and a lower incidence of deletion 7q as a single anomaly, in the latter case contrasting data recently reported by Baseggio et al.35 In addition, in contrast to previous studies by others and our group that have documented a higher incidence of TP53 deletion in CD5+ SMZL,36,37 in this much larger series, TP53 deletion was not associated with CD5 expression.
The possibility that the CD5+ cases from our series were indeed t(11;14)(q13;q32), cyclin D1− MCL cannot be formally excluded. However, in our cases, the absence of generalized lymphadenopathy at diagnosis and the indolent clinical course are in contrast to what has been recently reported for cyclin D1− MCL38 and argue against this possibility. In addition, all CD5+ cases from our series were negative for t(11;14)(q13;q32), thereby excluding the diagnosis of indolent MCL, because, to the best of our knowledge, all indolent MCLs display this aberration.39
The different cytogenetic makeup of the 2 subgroups of SMZL defined by CD5 expression reported here are notable yet admittedly difficult to interpret, especially given the relative homogeneity of both subgroups regarding almost all other clinicobiologic parameters. Therefore, future, prospective studies are strongly warranted to confirm these findings and explore their potential prognostic significance.
IGHV gene repertoire and mutational status
In keeping with previous reports,14,40,41 our study documents a highly restricted IGHV gene repertoire, with biased usage of the IGHV1-2 gene. After the 98% identity cut-off value, which is widely used in CLL to make the clinically relevant distinction between “mutated” and “unmutated” cases,42,–44 59 of 100 sequences (59%) from our series were defined as “mutated,” whereas the remainder (41 of 100 sequences; 41%) had “unmutated” IGHV genes.
The prognostic implications of IGHV gene mutational status in SMZL are still contested.15,40,45,46 Some studies have reported an association between unmutated IGHV genes and a high and intermediate risk according to the Intergruppo Italiano Linfomi score,45 del(7q), an adverse clinical course15,47 and 17q deletion.47 In keeping with previous reports,15 we observed an association between 7q deletion and unmutated IGHV genes; however, these parameters did not have an impact in OS. Nevertheless, given the retrospective nature of our study and the heterogeneity of therapeutic regimens, this issue merits further evaluation in a prospective fashion.
Interestingly, we identified a significantly higher frequency of del(7q) in cases expressing the IGHV1-2 versus all other IGHV genes (65.6% vs 32.4%, P = .002), independently of the mutational status. This biased cytogenetic profile, reported here for the first time, recalls the situation in CLL, where different groups have recently published evidence pointing to recurrent, “stereotyped” cytogenetic findings in different groups defined by IGHV gene usage and molecular features, for example, exclusive del(13q) in cases expressing stereotypical IGHV4-34 gene rearrangements assigned to subset 4,48,49 t(14;19)(q32;q13) and concurrent trisomy 12 in cases expressing stereotypical IGHV4-39 gene rearrangements assigned to subset 8.30,48,50 Although the underlying pathogenetic mechanisms are elusive and will probably prove hard to decipher, the identified association is too striking to ignore and might be considered as evidence of genetic evolution along distinctive, perhaps B-cell receptor-mediated, pathways for subgroups of SMZL cases characterized by distinctive B-cell receptor structures.
Prognosis
SMZL usually runs an indolent clinical course, but some patients manifest with a more aggressive disease. Clinical and cytogenetic parameters with prognostic value are not well established. Only a few studies, including small numbers of cases and analyzing selected parameters, have been reported (supplemental Table 7). To the best of our knowledge, this is the first report including a large number of SMZL cases and also attempting a comprehensive assessment of the prognostic implications of various clinicobiologic parameters, with a special emphasis on cytogenetic profiles.
Considering clinical variables, univariate associations for shorter OS were identified for age more than 65 years, Hb less than 12 g/dL, and serum β2-microglobulin more than 3 mg/L, in keeping with previous reports.45,46,50,,–53 Hb less than 12 g/dL and age more than 65 years were the only parameters that maintained prognostic significance with regard to OS on multivariate analysis, in concordance with previous reports by Parry-Jones et al53 and Arcaini et al.18 However, we could not confirm the prognostic impact of serum LDH and albumin, as proposed by the Interruppo Italiano Linfomi, probably because the number of cases with this information was relatively small. The finding that only non–disease-specific parameters (ie, age and Hb) retained prognostic significance for OS on multivariate analysis might be taken to imply that death was the result of incidental causes, at least in a proportion of cases; alternatively, the evaluated biomarkers could be predictive of progression but not response to therapy. These issues will require clarification in large prospective studies.
The heterogeneity of chromosomal aberrations and the high incidence of complex karyotypes in SMZL hinder the assessment of the prognostic impact of individual abnormalities. However, our data indicate that the presence of 2 or more aberrations correlates with a shorter survival. Interestingly, patients with normal karyotype showed an intermediate prognosis. Such cases may carry other abnormalities that are undetectable by G/R-banding or FISH; in these cases, the application of single nucleotide polymorphism/CGH arrays might be useful to find gains and losses.
Our findings do not support results from previous small studies that reported an adverse prognostic impact of del(7q)15 and/or associations between trisomy 3, 6q deletion, 7q deletion, 8p deletion, and trisomy 12.12 Instead, we provide, for the first time, evidence to suggest that abnormalities at 14q, especially deletion 14q, may confer an inferior prognosis. However, given that the number of cases with deletion 14q was small, this observation will require validation in larger series, especially as this aberration was usually detected in the context of a complex karyotype, probably accounting for the unfavorable prognosis.
In conclusion, although our study was retrospective and multicentric and had limitations inherent to such analysis (including missing information and nonhomogeneous treatment), it provides a comprehensive overview of the cytogenetic profile of SMZL, identifies undescribed and potentially prognostically relevant cytogenetic subgroups, and highlights the need to perform cytogenetics for diagnostic and prognostic stratification in these patients. Furthermore, the present study reinforces the clinical and biologic heterogeneity of SMZL, which creates the need for future prospective analyses in large series to find powerful prognostic markers specifically associated with this entity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Eveline Callet-Bauchu (Unité de Cytogénétique et Biologie Moléculaire des Hemopathies Malignes, Service d'Hématologie Biologique, Center de Biologie Sud, Lyon, France), Dr José Ángel Martínez Climent (Centro de Investigación de Navarra, Pamplona, Spain), Dr Jesús Maria Hernández (Servicio de Hematología, Hospital Universitario, Salamanca, Spain), Dr Pedro Martínez (Laboratorio de Genética, Hospital de Toledo, Toledo, Spain), Dr Isabel Granada (Servei d'Hematologia, Hospital Germans Trias i Pujol, Badalona, Spain), Dr Ana Carrió (Servei d'Hematopatologia, Hospital Clínic i Provincial, Barcelona, Spain), Dr Dolors Costa (Servei d'Hematopatologia, Hospital Clínic i Provincial, Barcelona, Spain), Dr Verónica Fernández (Servei d'Hematopatologia, Hospital Clínic i Provincial, Barcelona, Spain), Dr Elias Campo (Servei d'Hematopatologia, Hospital Clínic i Provincial, Barcelona, Spain), Dr Alicia Domingo (Laboratori de Citologia, Hospital de Bellvitge, Barcelona, Spain), and Dr Soledad Woessner (Escola de Citologia Hematológica Soledad Woessner-IMAS, Barcelona, Spain) for providing cases as well as Sergi Mojal for assisting us in the statistical analysis (Departament Assessorament Bioestadistic, Institut Municipal d'Assistència Sanitària, Barcelona, Spain).
This work was supported in part by Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Spain, Red Temática de Investigación Cooperativa en Cáncer (grants RD06/0020/FEDER and RD07/0020/2004).
Authorship
Contribution: M.S. and F.S. designed and performed research, performed cytogenetic analysis, revised, collected, and analyzed clinical data, wrote the paper, and gave final approval; C.B. performed research, collected all cytogenetic data, and gave final approval; D.O. designed and performed research, collected clinical data, and gave final approval; K.S. performed molecular studies, collected clinical data, drafted the IGVH results and discussion sections, assisted in the corrections of the paper, and gave final approval; J.D., S.G., A.A., A.G., M.J.C., and E.L. performed cytogenetic analysis, collected clinical data, and gave final approval; E.M. performed cytologic and immunophenotypic studies, assisted in the corrections of the paper, and gave final approval; A.T.-G. performed morphologic and immunophenotypic studies and IGHV mutational analysis, collected clinical data, and gave final approval; F.B. and P.F. performed morphologic and immunophenotypic studies; C.T. performed cytologic, histologic, and immunophenotypic studies and gave final approval; Z.D. performed molecular studies, collected clinical data, and gave final approval; F.M., G.V., and A.S. collected clinical data and gave final approval; A.F. performed immunophenotypic studies, collected clinical data, and gave final approval; M.M. and T.P. performed histologic studies, collected clinical data, and gave final approval; L.F. performed cytologic and immunophenotypic studies, collected clinical data, and gave final approval; B.E. and I.W. performed cytogenetic analysis and gave final approval; M.G.-G. performed statistical analysis and gave final approval; S.S. performed histologic studies and gave final approval; and M.A.P. designed and performed research and histologic studies and gave final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesc Solé, Servei de Patologia, Laboratori de Citogenètica Molecular, Hospital del Mar, Passeig Marítim, 25-29, 08003 Barcelona, Spain; e-mail: fsole@parcdesalutmar.cat.