Abstract
Activated platelets shed surface proteins, potentially modifying platelet function as well as providing a source of bioactive fragments. Previous studies have identified several constituents of the platelet sheddome, but the full extent of shedding is unknown. Here we have taken a global approach, analyzing protein fragments in the supernate of activated platelets using mass spectroscopy and looking for proteins originating from platelet membranes. After removing plasma proteins and microparticles, 1048 proteins were identified, including 69 membrane proteins. Nearly all of the membrane proteins had been detected previously, but only 10 had been shown to be shed in platelets. The remaining 59 are candidates subject to confirmation. Based on spectral counts, protein representation in the sheddome varies considerably. As proof of principle, we validated one of the less frequently detected proteins, semaphorin 7A, which had not previously been identified in platelets. Surface expression, cleavage, and shedding of semaphorin 7A were demonstrated, as was its association with α-granules. Finally, cleavage of semaphorin 7A and 12 other proteins was substantially reduced by an inhibitor of ADAM17, a known sheddase. These results define a subset of membrane proteins as sheddome candidates, forming the basis for further studies examining the impact of ectodomain shedding on platelet function.
Introduction
Platelets are anucleate blood cells that play a critical role in hemostasis, helping to stop bleeding after vascular injury, particularly in the arterial circulation. Proteins on the platelet surface participate in this process, supporting platelet rolling, adhesion, and activation at sites of vascular injury and making possible the cohesive interactions that underlie the growth of the hemostatic mass.1-3 The repertoire of proteins present on the platelet surface is not immutable. Platelet activation recruits new proteins to the plasma membrane, including proteins such as P-selectin that were originally located in the membrane of platelet α-granules. Conversely, some membrane proteins are internalized4,5 or cast off in membrane-derived microparticles6,7 when platelets are activated, reducing their expression on the cell surface. Previous studies have shown that proteins can also be proteolytically shed from the platelet surface.8 Typically, this occurs after platelet activation, although it can also happen in resting platelets.9 Shedding is distinct from secretion, which is a process that results in the release of proteins from within platelet storage granules.
In theory, proteolytic shedding of platelet membrane proteins can serve several roles, including modulating adhesive and cohesive interactions, limiting responses to agonists, and enabling cryptic functions of the cleaved proteins.8 Shedding can also produce bioactive fragments that bind to receptors on other cells, modifying their behavior and contributing to processes as diverse as inflammation and wound healing. Two well-characterized examples of proteins shed from activated platelets are the membrane glycoproteins, GPIbα10 and GPVI.11,12 GPIbα is part of the GPIb/IX/V complex, which allows moving platelets to engage von Willebrand factor.13 GPVI is a signaling receptor for collagen. The short list of other proteins shed from activated platelets includes CD40L (CD154)14 and the semaphorin (sema) family member, sema4D,15 both of which produce bioactive fragments. In cases where the proteinase responsible for shedding has been identified, it has proved to be a member of the ADAM (a disintegrin and metalloproteinase) family. Cleavage of GPIbα10 and sema4D15 is mediated by ADAM17, whereas shedding of GPVI is mediated by ADAM10.11 Both may be involved in cleaving JAM-A.16
Although previous studies have established that ectodomain shedding occurs in platelets,8,17,18 attention has primarily been focused on a limited number of membrane proteins that were being studied for their role in platelet biology and only incidentally found to be shed during platelet activation. Here we have undertaken a different approach, with our primary goal to identify as many as possible of the global set of membrane proteins that contribute to the platelet sheddome. For this purpose, proteins or protein fragments that appeared in the supernate of activated platelets were detected using mass spectroscopy and identified computationally. A bioinformatics approach was used to determine which of the proteins are probably membrane proteins. Internal controls included the 10 proteins whose shedding from platelets had been reported previously. Challenges that had to be overcome included the large number of secreted proteins in platelet storage granules, contamination by unincorporated plasma proteins, leakage of cytosolic proteins, and the possible presence of microparticles carrying membrane proteins that have neither been cleaved nor shed.
In total, we have identified 1048 proteins in the supernate of activated platelets. Among these, the sheddome candidates are 69 membrane proteins with one or more transmembrane domains or a glycosylphosphatidylinositol (GPI) anchor. This number is small compared with the much larger number of membrane proteins (626) detected in a recent survey of the platelet membrane proteome,19 which suggests that not every platelet membrane protein is shed. In support of the general approach, nearly all of the membrane proteins that were identified in the platelet supernate had been detected in more general surveys of the platelet proteome or transcriptome, and all 10 of the proteins that had previously been shown to be shed by platelets were also detected here. The other 59 proteins on the sheddome list have to be viewed as candidates subject to confirmation using other methodologies. As a proof of principle for this form of discovery, we validated one of the less frequently detected proteins, the semaphorin family member, sema7A, which had not previously been shown to be expressed by platelets. Semaphorins are of particular interest because of their role in communication between cells. Surface expression, cleavage, and shedding of sema7A were confirmed, as was its association with platelet α-granules and sensitivity to inhibitors of ADAM17.
Methods
Additional methods and details are in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Chemicals and reagents
All chemicals were obtained from Sigma-Aldrich. GM6001 was purchased from Calbiochem, and BMS-561392 was obtained from Bristol-Myers Squibb. Phorbol 12-myristate 13-acetate (PMA) was obtained from Calbiochem. Mouse monoclonal anti–human GPVI (clone 6B12) was a kind gift from Dr Mark L. Kahn (University of Pennsylvania), whereas mouse monoclonal anti–human GPIbα and goat anti–human semaphorin 7A were purchased from R&D Systems. Mouse monoclonal anti–human VWF was purchased from Thermo Scientific. All secondary antibodies were obtained from Southern Biotechnology or Invitrogen. Primary antibodies, goat IgG (Jackson ImmunoResearch Laboratories), and mouse IgG1 (Bethyl Laboratories) whole antibody used in flow cytometry were fluorescently labeled with Alexa Fluor-488 monoclonal antibody labeling kit from Invitrogen. The PAR1 agonist peptide, SFLLRN, was purchased from Bachem.
Blood collection and platelet preparation
Whole blood was obtained from healthy adult volunteers in accordance with a protocol approved by the University of Pennsylvania Institutional Review Board. Blood was collected into ACD (65mM Na3 citrate, 70mM citric acid, 100mM dextrose, pH 4.4) and centrifuged at 129g for 20 minutes to collect platelet-rich plasma. Only the upper third of the platelet-rich plasma was used, to avoid contamination with other blood cells. Platelets were pelleted at 341g for 15 minutes and washed in HEN buffer (150mM NaCl, 1mM Na2-ethylenediaminetetraacetic acid, 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 6.5) containing 1μM PGE1 and 1 U/mL ayprase before resuspending in Tyrode buffer (137mM NaCl, 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 5.6mM dextrose, 1mM MgCl2, 2.7mM KCl, 3.3mM Na2H2PO4, pH 7.4) at a final concentration of 1 × 109 platelets/mL. Platelets were activated with 10μM PMA or 10μM SFLLRN at 37°C. A total of 100μM GM6001 or 1μM BMS-561392 was added to the platelet suspension for 7 minutes at room temperature before platelet activation. Ethylenediaminetetraacetic acid was added to all tubes at a final concentration of 0.01M to stop platelet activation.
Removal of microparticles and common plasma proteins
Microparticles were removed from platelet supernates by centrifugation at 100 000g for 1 hour at 4°C. Common plasma proteins were removed by immunodepletion through the ProteoPrep 20 Jumbo Plasma Immunodepletion Kit (Sigma-Aldrich) or the ProteomeLab IgY Proteome Partitioning Kit (Beckman Coulter).
In-gel trypsin digestion and peptide extraction
Proteins (10μg) were separated by gel electrophoresis and stained with colloidal blue (Invitrogen). Gel slices (20 per lane) were excised and the protein digested with trypsin, before being subjected to reverse-phase liquid chromatography and tandem mass spectrometry (MS).20 Detailed methodology is given in supplemental data. Proteins from platelet supernates after immunodepletion with the ProteomeLab IgY column were subjected to in-solution trypsin digest as described in supplemental data.
Fractionation of peptides by strong cation exchange chromatography
Strong cation exchange chromatography was performed on a PolySulfethyl A column (100 mm × 2.3 mm, 5 μm 300 Å, PolyLC, The Nest Group) attached to a 1100 Series HPLC (Agilent). The collected fractions were lyophilized and stored at −80°C until further analysis.
Reversed-phase capillary liquid chromatography tandem MS analyses
Samples depleted using the ProteoPrep 20 Jumbo Plasma Immunodepletion Kit and subjected to in-gel trypsin digest were subjected to reverse phase separation of the tryptic peptides and tandem MS analysis, as described in supplemental data. Samples following strong cation exchange chromatography were reconstituted with 0.1% formic acid, 1% acetonitrile for reversed phase separation (Dionex Ultimate 3000 nanoflow LC System) on-line to the Thermo Finnigan LTQ-FT linear ion trap mass spectrometer (Thermo Fisher) using electrospray ionization (ESI).
Platelet membrane preparation
Resting or activated platelets were lysed by nitrogen cavitation, and all membranes were sedimented by ultracentrifugation at 100 000g for 1 hour at 4°C. Membrane proteins were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer with β-mercaptoethanol and analyzed by Western blot.
Data analysis
Raw data from both liquid chromatography-MS/MS runs was searched against a protein sequence database consisting of the Human International Protein Index (IPI) v3.51 protein sequences (74 049 protein entries; 31 194,560 amino acids) and the reversed human IPI sequences as a decoy component. Two independent algorithms, SEQUEST 3.1 (Thermo Finnigan) and MASCOT 2.1.04 (Matrix Sciences), were used. Results from both algorithms and replicate analyses were combined in a single statistical analysis using the Empirical Bayes Protein Identifier 1.0.21
Protein separation and Western blotting
Platelet proteins were reduced with β-mercaptoethanol (Bio-Rad) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis on 4% to 12% precast BisTris gels (NuPAGE, Invitrogen). Separated proteins were transferred onto a nitrocellulose membrane (Bio-Rad) using the Novex X-Blot cell (Invitrogen) and detected using the enhanced chemiluminescence Western blot detection kit (GE Healthcare).
Flow cytometry
Fluorometric analysis using fluorescently tagged antibodies to various platelet receptors was performed to detect cell surface expression of platelet receptors and to monitor its time-dependent shedding in response to agonist. Washed platelets (5 × 107) were incubated with 1mM CaCl2 and 10μM PMA or 10μM SFLLRN for 15, 30, and 60 minutes, at 37°C. Alexa Fluor-488–conjugated anti-sema7A or the corresponding isotype antibody control was added at a final concentration of 10 μg/mL to the platelet samples for 10 minutes at room temperature. All platelets were fixed with an equal volume of 2% paraformaldehyde for 10 minutes at room temperature and spun down briefly at 2655g for 30 seconds to pellet all platelets. The platelets were washed with 100 μL phosphate-buffered saline (PBS) before resuspending in a final volume of 1 mL PBS and immediately analyzed by flow cytometry (BD Biosciences) with Cellquest Version 5.1.1 software.
Confocal microscopy
Glass coverslips were coated with 10 μg/mL fibrinogen for 3 hours at room temperature and then washed with PBS and blocked with 5% bovine serum albumin in PBS for 30 minutes. Washed human platelets (1 × 108) were added and then activated with 10μM SFLLRN for 15 minutes at 37°C. The platelets were fixed with 3.7% formaldehyde for 5 minutes at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 10 minutes, and blocked with 5% bovine serum albumin for 30 minutes. Platelets were incubated with goat anti–human sema7A antibody (R&D Systems) or isotype-matched goat IgG (Jackson ImmunoResearch Laboratories) at a final concentration of 20 μg/mL for 1 hour at room temperature. A total of 40 μg/mL Alexa Fluor-488–conjugated donkey anti–goat secondary antibodies (Invitrogen) was added to all platelets for a further 45 minutes at room temperature. To determine whether sema7A is localized in the α-granules of platelets, the sema7A-stained activated platelets were counterstained with 20 μg/mL mouse anti–human VWF (Thermo Scientific) and detected with Alexa Fluor-546-conjugated goat anti–mouse secondary antibodies (Invitrogen). Isotype-matched mouse IgG1 (20 μg/mL final concentration; Bethyl Laboratories) was used as a control. The platelets were mounted on Fluoromount-G mounting medium (Southern Biotech) and visualized on a Zeiss LSM510 META NLO laser scanning system on an inverted microscope (Zeiss Axiovert 200M) using a Plan-Apo 63×/1.4 oil objective. Images were analyzed using LSM510 Version 4.0 software.
Results
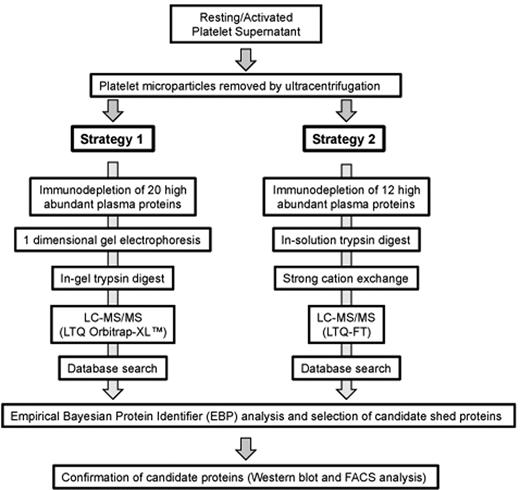
Among the challenges in identifying protein fragments shed from the platelet surface is that they may be present at low levels and therefore obscured by other proteins, including residual plasma proteins, secreted proteins, and any cytosolic proteins that are present because of cell damage during platelet isolation or activation. Microparticles carrying intact membrane proteins can also complicate an analysis of shed proteins. In our initial studies, platelets were activated with PMA, which is a strong stimulus for protein shedding in platelets. Microparticles were removed by ultracentrifugation, after which some of the most abundant plasma proteins were removed by immunodepletion. Remaining proteins were separated and identified by the 2 complementary strategies outlined in Figure 1.
Proteomic strategies to identify and validate shed proteins of the platelet sheddome. Washed human platelets were activated with 10μM PMA at 37°C for 1 hour and then analyzed using the 2 strategies shown. Details are given in “Methods” and supplemental data.
Proteomic strategies to identify and validate shed proteins of the platelet sheddome. Washed human platelets were activated with 10μM PMA at 37°C for 1 hour and then analyzed using the 2 strategies shown. Details are given in “Methods” and supplemental data.
In the first strategy, proteins immunodepleted using the ProteoPrep 20 immunodepletion column were separated by gel electrophoresis, digested in the gel with trypsin, and then subjected to liquid chromatography-MS/MS on the LTQ-Orbitrap XL. In the second strategy, proteins not retained by the ProteomeLab IgY column were digested in-solution with trypsin without prior electrophoresis and then analyzed by cation exchange chromatography followed by MS on the LTQ-FT. Raw data files from the LTQ-Orbitrap XL and the LTQ-FT were combined and compared against a database containing the human IPI protein sequences with the reversed human IPI sequences serving as a decoy component. Each raw file was analyzed with both Sequest and Mascot search algorithms. Proteins with a false identification rate of less than or equal to 5% were selected, using an integrated approach that has been described previously.21
Based on this analysis, we assigned a semiquantitative value to each protein using spectral counts (the frequency with which all peptides identifying the protein were encountered) as a surrogate for abundance, keeping in mind that spectral counts also reflect factors such as the size of the shed ectodomain, susceptibility to proteolysis by trypsin, and the sensitivity of the fragments produced to the methods used to detect them. Peptides from 1048 proteins were identified in the supernate of the activated platelet (supplemental Table 1). Each was compared with entries in the Swiss-Prot and TrEMBL databases and classified according to its predicted site of origin within the platelet (Figure 2A). Approximately half were from the cytoplasm, 4% were cytoskeletal components, and 22% were from organelles such as the Golgi, endoplasmic reticulum, or lysosomes. Another one-fourth of the proteins were either secreted from platelet storage granules, plasma proteins, or both, because megakaryocytes and platelets can take up plasma proteins as well as synthesize them.22 Only 69 (7%) of the proteins identified were membrane proteins with one or more transmembrane domains or a GPI anchor. Two proteins with more than one transmembrane domain were tetraspanins. Note, however, that the method used to concentrate the proteins would be expected to remove protein fragments whose size was less than 3 kDa, and this may have kept additional proteins from being identified.
Data mining of the platelet sheddome. (A) Subcellular localization of the 1048 proteins identified by MS. Cellular locations were assigned using the Swiss-Prot and TrEMBL databases. (B) Spectral counts for the 69 membrane proteins that were identified. Sema7A is highlighted in green.
Data mining of the platelet sheddome. (A) Subcellular localization of the 1048 proteins identified by MS. Cellular locations were assigned using the Swiss-Prot and TrEMBL databases. (B) Spectral counts for the 69 membrane proteins that were identified. Sema7A is highlighted in green.
Transmembrane and GPI-anchored proteins identified in activated platelet supernates are listed in Table 1 in the order of their spectral counts. Among the single-pass transmembrane proteins, spectral counts varied by 3 orders of magnitude (Figure 2B). The most frequently detected proteins were GPV and amyloid βA4 protein, both of which have previously been reported to be shed from activated platelets. These along with the other 8 proteins that have previously been reported to be shed by platelets are marked in red in Figure 2B. They tend to be among those with the highest spectral counts, but not exclusively.
The least frequently encountered proteins included another member of the GPIb/IX/V complex, GPIX, and transmembrane protein 56, neither of which has previously been shown to be shed. Sema4D, which we have shown to be expressed and shed by human and mouse platelets,15 was also detected, serving as another internal control, as does TREM-like transcript-1 (TLT-1). Among the proteins that have previously been detected in platelets, but not previously shown to be shed from platelets, are the SLAM family member CD84 and 3 members of the CTX (cortical thymus Xenopus) family of adhesion molecules, ESAM, JAM-A, and CD226. There were 4 GPI-anchored proteins. The most frequently encountered, CD109, has been detected previously in platelets. The GPI-anchored semaphorin family member, sema7A (marked in green in Figure 2B), had not previously been detected on platelets. Additional characterization of sema7A is included below in “Sema7A.”
Identifying ADAM17 substrates within the platelet sheddome
As noted in the Introduction, GPIbα, GPV, and sema4D have been reported to be shed from activated platelets by the metalloproteinase, ADAM17, based on either pharmacologic or genetic evidence, or both.10,15,23 As a general screen of the relationship of ADAM17 to the larger sheddome, we repeated the sheddome analysis, preincubating the platelets with BMS-561392, a potent and selective inhibitor of ADAM17.32 Of the membrane proteins identified, shedding of 13 was inhibited by half or more (Table 2). This analysis is only semiquantitative because it is based on spectral counts rather than a more accurate measure of abundance. Nonetheless, shedding of sema4D, which is known to require ADAM17, was inhibited by 85%. Conversely, shedding of GPVI, which is known to be mediated by ADAM10, was inhibited by only 2%.
Analysis of GPIbα and GPVI
As an internal control, Western blotting and flow cytometry were used to confirm the shedding of 2 well-characterized proteins, GPIbα and GPVI, both of which have previously been shown to be shed by activated platelets. Platelets were incubated with PMA for up to 60 minutes (Figure 3). Western blots show bands at approximately 120 and 55 kDa, which correspond to the ectodomain fragments of GPIbα and GPVI, respectively. The GPVI fragment was not present in the supernate of resting platelets but accumulated when the platelets were activated. The GPIbα fragment also accumulated with time but, in contrast to GPVI, could also be detected in the supernate of resting platelets. Constitutive shedding of GPIbα has been reported previously, and soluble GPIbα has been shown to circulate in plasma.9
Shedding of GPIbα and GPVI during platelet activation. Platelets were activated with 10μM PMA at 37°C for the times indicated. Representative Western blots of platelet supernatant for (A) GPIbα and (B) GPVI. Summary plots from 3 experiments (mean ± SEM).
Shedding of GPIbα and GPVI during platelet activation. Platelets were activated with 10μM PMA at 37°C for the times indicated. Representative Western blots of platelet supernatant for (A) GPIbα and (B) GPVI. Summary plots from 3 experiments (mean ± SEM).
Sema7A
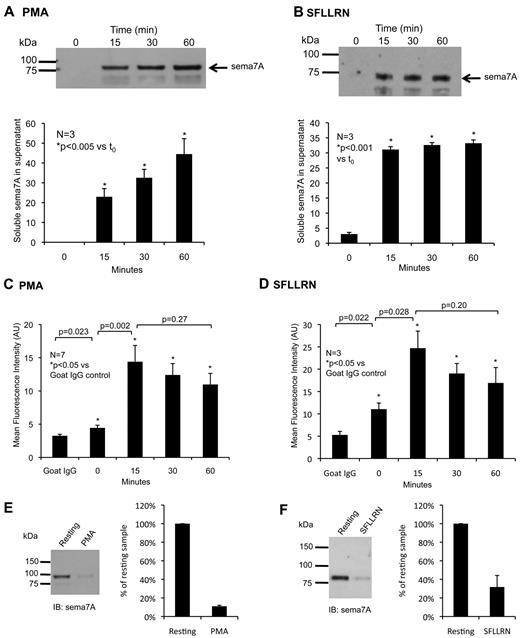
The 69 membrane proteins that we detected vary greatly in the frequency with which their fragments were detected. Because the 10 that had been shown previously to be shed tended to have higher spectral counts (red bars in Figure 2B), we chose one with low spectral counts that had not been shown to be expressed by platelets and sought confirmation that it really is shed during platelet activation. Sema7A is a GPI-anchored member of the semaphorin family that is also expressed by erythrocytes, where it carries the JMH antigen. Full-length sema7A migrates at approximately 80 kDa on Western blots of platelet and brain lysates (Figure 4A). Confocal microscopy was applied to human platelets that had been activated with the PAR1 (thrombin receptor) agonist peptide, SFLLRN, and allowed to spread on fibrinogen. Sema7A was detectable on the plasma membrane and in intracellular compartments (Figure 4B). Based on colocalization with von Willebrand factor, the latter are probably α-granules. Activating platelets with either PMA or SFLLRN caused an accumulation of sema7A in the platelet supernate (Figure 5A-B). The size of the soluble sema7A is similar to the intact protein, suggesting that cleavage occurs very close to the GPI anchor. Flow cytometry performed on intact, nonpermeabilized platelets shows that there is a limited amount of sema7A on the surface of resting platelets. This increases when platelets are activated for 15 minutes, after which surface expression declines (Figure 5C-D). Western blots show that most of the intact sema7A detectable in membranes prepared from resting platelets disappears when the platelets are incubated with an agonist for 60 minutes. Only 11% plus or minus 1% (mean ± SEM, N = 3) remains in the PMA-treated samples and 32% plus or minus 12% in the SFLLRN-treated samples (Figure 5E-F). These results suggest that most sema7A translocates to the platelet surface and is cleaved when platelets are activated, but that some is present on the surface before hand and some remains afterward.
Sema7A is present in human platelets. (A) Western blot of human platelet and brain lysates. (B) Confocal microscopy of activated human platelets allowed spreading on a fibrinogen-coated surface showing sema7A on the surface and colocalizing with VWF in α-granules.
Sema7A is present in human platelets. (A) Western blot of human platelet and brain lysates. (B) Confocal microscopy of activated human platelets allowed spreading on a fibrinogen-coated surface showing sema7A on the surface and colocalizing with VWF in α-granules.
Sema7A is cleaved and shed during platelet activation. (A-B) Shed sema7A in platelet supernatants with increasing time after (A) 10μM PMA or (B) 10μM SFLLRN activation. (C-D) Surface expression of sema7A determined by flow cytometry after platelet activation with (C) PMA or (D) SFLLRN activation. Alexa Fluor-488–conjugated goat IgG was used as the isotype control. (E-F) Representative Western blots of sema7A on platelet membranes.
Sema7A is cleaved and shed during platelet activation. (A-B) Shed sema7A in platelet supernatants with increasing time after (A) 10μM PMA or (B) 10μM SFLLRN activation. (C-D) Surface expression of sema7A determined by flow cytometry after platelet activation with (C) PMA or (D) SFLLRN activation. Alexa Fluor-488–conjugated goat IgG was used as the isotype control. (E-F) Representative Western blots of sema7A on platelet membranes.
Because the analysis of the proteomics results suggested that sema7A might be one of the proteins cleaved by ADAM17, Western blots were also performed on the supernate of activated platelets prepared in the presence of either BMS-561392, to inhibit ADAM17, or GM6001, a general inhibitor of metalloproteinases (Figure 6). The results show that GM6001 inhibits sema7A cleavage by 68% when platelets are activated by PMA and 77% when platelets are activated by SFLLRN. The corresponding numbers for BMS-561392 are 66% and 59%, respectively. Thus, the results of the immunofluorescence microscopy, flow cytometry, and Western blotting studies validate the detection of sema7A in the sheddome analysis and suggest that ADAM17 is probably the sheddase.
Shedding of sema7A is inhibited by metalloproteinase inhibitors. Western blots of sema7A in the supernate of platelets activated with (A) PMA or (B) SFLLRN in the presence of the metalloprotease inhibitor GM6001 or the ADAM17-selective inhibitor BMS-561392 (mean ± SEM, N = 3).
Shedding of sema7A is inhibited by metalloproteinase inhibitors. Western blots of sema7A in the supernate of platelets activated with (A) PMA or (B) SFLLRN in the presence of the metalloprotease inhibitor GM6001 or the ADAM17-selective inhibitor BMS-561392 (mean ± SEM, N = 3).
Discussion
Programmed autologous cleavage of platelet cell surface molecules has emerged as a potential mechanism for regulating platelet function.8 In cells other than platelets, shedding can release bioactive fragments and is implicated in controlling normal and pathologic processes, including inflammation and cancer.33,34 In platelets, ectodomain shedding has been well documented, but its role is less well understood, as is the full repertoire of proteins that are shed. Here we have taken a global approach, attempting to identify as many as possible of the membrane proteins that give rise to fragments in the supernate of activated platelets. Technical challenges that had to be overcome included the presence of residual plasma proteins in the platelet preparation and the large number of proteins that are secreted from platelet storage granules and leaked from damaged platelets. These proteins are a challenge because they hamper detection of shed protein fragments, especially those that are present in low abundance. Another challenge is presented by the platelet microparticles that bud from the surface of activated platelets, carrying intact membrane proteins with them. To the greatest extent possible, abundant plasma proteins were removed by immunodepletion. Microparticles were removed by ultracentrifugation.
Sensitivity of MS protein detection was enhanced by applying both gel electrophoresis- and chromatography-based approaches to protein separation before high resolution MS. Specificity of protein identification was augmented by analyzing the peptide mass spectra with 2 search algorithms (Sequest and Mascot) that are based on distinct, orthogonal scoring methods for evaluation of the similarity between observed and theoretical masses of peptide fragments. Sensitivity and error rate were controlled by statistical modeling in an integrated analysis of all of these data and decoy database searching.21 The statistical model penalized nonconsensual identifications from the 2 search algorithms and technical replicates but allowed nonconsensual identification from both protein separation methods. This identified a large number of proteins (1048) with high confidence (false identification rate < 5%). Of these, only 69 are membrane proteins as defined by the presence of one or more transmembrane domains or a GPI anchor. Using spectral counts as an indicator, the 69 proteins varied 1000-fold in the frequency with which they were detected in the supernate. Although this is not the same as saying that the shed protein fragments varied 1000-fold in abundance, it is probably not coincidental that the 5 proteins with the highest counts have been shown previously by other methodologies to be cleaved and shed from the platelet surface (red bars in Figure 2B). Conversely, there could be proteins that we did not detect and the sheddome could be even larger than the 69 proteins listed in Table 1.
Most of the proteins that were identified (60 of 69) have one transmembrane domain; 5 have more than one and 4 have GPI anchors. Consistent with the idea of ectodomain cleavage, nearly all of the fragments map to the extracellular or transmembrane domains (supplemental Table 3, complete maps). However, a fragment mapping to the cytoplasmic domain was found for 8 proteins: integrin αIIb, integrin β3, dystroglycan, CLIC4, CD226, PECAM, desmoglein-1, and sodium channel subunit β4 (green asterisks in Figure 2B). Of these proteins, there is prior evidence of shedding for only one, PECAM (Table 1). This could mean that the other 7 are present as intact protein contaminants, perhaps carried by residual microparticles or platelet fragments. However, the absence of cytoplasmic domain fragments from other highly expressed platelet plasma membrane proteins argues against this interpretation. Two of the identified proteins are tetraspanins, CD9 and T-Span9. The fragments that were identified from these proteins map to the extracellular domains. Given the cytoplasmic location of the N- and C-termini of tetraspanins, this may mean that more than one cleavage event has occurred.
Detection of a membrane protein in the supernate of activated platelets means that it is a candidate for inclusion in the sheddome, but not proof that cleavage and shedding actually occur. Confirmation by other methods is required. Of the 69 proteins that we identified, only 10 had been shown previously to be shed from the surface of activated platelets (Table 1; Figure 2B). As far as we know, these are the only proteins that have been shown to be shed by platelets. However, an additional protein, JAM-A, has been reported to be shed from endothelial and epithelial cells,16 and shedding of ICAM2 and VCAM1 has been reported in Jurkat cells28 and endothelial cells,31 respectively. Subtracting these 13 proteins (the 10 shown in platelets plus JAM-A, ICAM2, and VCAM1) leaves 56 others that require validation, including a number that have the lowest spectral counts. However, it should not be assumed that low frequency of detection means either misidentification or an inappropriate assignment to the sheddome. As a proof of principle, we used Western blotting, flow cytometry, and fluorescence microscopy to establish that one of the least abundant proteins, sema7A, is expressed on platelets, cleaved during platelet activation and then shed into the solution phase. Confocal microscopy suggests that sema7A is present on α-granule membranes as well as the plasma membrane and translocates to the platelet surface in response to agonists.
Studies in cells other than platelets have shown that ectodomain shedding is often mediated by the ADAM family of proteinases.33-35 In platelets, ADAM17 mediates shedding of GPIbα,10 GPV,23 and sema4D.15 GPVI is cleaved by ADAM10.11 To determine the involvement of ADAM17 in the shedding of other platelet proteins, we used a selective inhibitor, BMS-561392.36 Using spectral counts as an endpoint, cleavage of 9 proteins was inhibited by more than 80% (Table 2). As noted earlier, the decrease in spectral counts when BMS-561392 was present is not sufficient for precisely quantifying the extent of inhibition but is useful for highlighting proteins for validation by other methods. The subset of affected proteins included sema4D and sema7A, whose cleavage dependence on ADAM17 was independently established. Cleavage of 16 of the proteins, including GPVI, was essentially unaffected.
The impact of shedding
Among the proteins identified in the platelet sheddome, there are a number that fall into distinct families whose role is worth considering in the context of shedding. In addition to well-known glycoproteins that serve as adhesion and cohesion receptors in platelets, the list includes several semaphorins and CTX family members. Semaphorins are of interest because of the evidence that some members of the family, including sema4D, can serve as ligands for cell surface receptors and, in addition, give rise to bioactive fragments when cleaved. We have previously shown this to be the case for sema4D, which amplifies platelet responses to collagen. Sema4D is cleaved and shed from activated platelets, releasing a large fragment that retains biologic activity on its receptors, including plexin-B1 on endothelial cells.37 The sema4D knockout impairs collagen-induced platelet aggregation in vitro and inhibits platelet accumulation after vascular injury in vivo.15,38 Sema7A (CD108) was originally identified in the immune system39 and as the JMH antigen on erythrocytes,40 but it is also expressed by neurons and has a role in axon guidance.41 Known binding partners for sema7A include plexin C142 and integrin α1β1.41,43 Sema7A expressed on activated T cells is able to stimulate cytokine production in monocytes and macrophages through α1β1.
Comparisons between the sheddome and the membrane proteome
Although this is the first concerted effort to identify the platelet sheddome, a number of studies have explored other aspects of the platelet proteome. Some comparisons with the results that we obtained are revealing. Supplemental Table 2 indicates whether each of the 69 membrane proteins identified in the present study was detected in 14 previous surveys, including a global survey of the megakaryocyte transcriptome44 and an analysis of the membrane proteome.19 Overall, 59 (86%) of the 69 membrane proteins reported here were detected in at least one of the other studies; 49 in the transcriptome44 and 42 in the membrane proteome19 (Figure 7). The 10 proteins that were not previously detected tended to be ones with the lowest spectral counts (red asterisks in Figure 2B). Notably, the most inclusive membrane protein study used isolated membranes rather than platelet lysates as its starting material and identified 626 membrane proteins, approximately 10 times as many as we detected in the platelet supernate. This suggests that only a minority of membrane proteins are shed from activated platelets, at least at the level of detection and the conditions used here. Smaller numbers of membrane proteins were reported by Moebius et al45 and Senis et al,46 possibly because of differences in their isolation protocols. In contrast, there is little overlap between the 69 membrane proteins that we identified and the proteins detected in 2 studies of platelet microparticles,6,7 which may reflect distinct specificities between shedding and microparticle formation.
Comparisons with other proteomes. The Venn diagrams in the figure indicate how many of the 69 membrane proteins identified in the present study were detected in (A) a study of proteins in which the starting material was isolated in platelet membranes and (B) 3 studies of either isolated platelet α-granules or platelet releasate. In the releasate studies, platelets were incubated with an agonist for 5 minutes or less, considerably shorter than the 60-minute incubation adopted here to allow time for delayed shedding to proceed to or close to completion.
Comparisons with other proteomes. The Venn diagrams in the figure indicate how many of the 69 membrane proteins identified in the present study were detected in (A) a study of proteins in which the starting material was isolated in platelet membranes and (B) 3 studies of either isolated platelet α-granules or platelet releasate. In the releasate studies, platelets were incubated with an agonist for 5 minutes or less, considerably shorter than the 60-minute incubation adopted here to allow time for delayed shedding to proceed to or close to completion.
Two previous studies have explored the platelet secretome by identifying as many proteins as possible in the releasate of activated platelets.47,48 In theory, there should be a considerable overlap between those studies and ours because in each case the starting material was the solution phase of activated platelets. However, in the releasate studies, the platelets were activated for 5 minutes or less, compared with the 60-minute incubation period that we used to allow time for shedding to reach completion. As a result, only a small number of membrane proteins were found in common: 10 in one case and 3 in the other (supplemental Table 2; Figure 7). In contrast, an α-granule proteome study identified 284 proteins, 24 of which were also identified in the present study (supplemental Table 2; Figure 7).22 These tended to be among the most abundant membrane proteins but also included P-selectin, which is a well-established constituent of α-granule membranes, and sema7A, which appears to be as well (Figure 4).
In conclusion, we have determined that 69 membrane proteins can be detected in the supernate of activated platelets after platelets and microparticles have been removed. Most of these have a single transmembrane domain, but several GPI-anchored proteins and proteins with multiple transmembrane domains were also detected. This study represents the first effort to use an unbiased approach to identify as much as possible of the platelet sheddome. In addition to the 10 proteins that were already known to be shed, a sizeable number of proteins were identified that were either not known to be expressed by platelets or not known to be shed, among them a new semaphorin family member, sema7A. Ectodomain shedding of platelet cell surface proteins offers a novel mechanism to regulate platelet function by modifying adhesive interactions, removing ligand-receptor signaling pairs and creating fragments that can retain biologic activity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Wistar Proteomics Facility, including the technical assistance of Lynn A. Beer and Thomas E. Beer with proteomics Strategy 1, and Tony Chang-Wong for his invaluable assistance with bioinformatics, as well as Bristol-Myers Squibb for the use of BMS-561392.
This work was supported in part by the National Institutes of Health: grant HL038794 (D.W.S.), grant HL081012 (T.G.), and grants HL86358, HL81012, and HL40387 (L.F.B.).
National Institutes of Health
Authorship
Contribution: K.P.F., C.B., T.G., I.A.B., D.W.S., and L.F.B. designed research, K.P.F., C.B., A.N.T., E.A.T., and K.M.W. performed research; K.P.F., T.G., H.-Y.T., and K.D.S. analyzed data; and K.P.F., C.B., H.-Y.T., T.G., and L.F.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lawrence F. Brass, University of Pennsylvania, 915 Biomedical Research Bldg II/III, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: brass@mail.med.upenn.edu.