Abstract
To determine the influence of KIR and HLA class I polymorphism on human NK cell repertoires, 32 different clonotypes representing all possible combinations of 4 inhibitory KIR and NKG2A were analyzed by multicolor flow cytometry. In donors homozygous for the common group A KIR haplotype, a significant influence of HLA-C ligands was seen: KIR repertoires were dominated by clonotypes expressing a single KIR for the respective cognate ligand, either the C1-specific KIR2DL3 or C2-specific KIR2DL1. In contrast, in donors possessing the polymorphic group B haplotypes, a similar adaptation to cognate HLA-C was lacking. We suggest that this discrepancy is largely the result of a suppressive effect of the group B–specific KIR2DL2 on the frequency of KIR2DL1+ NK cells. In functional assays, KIR2DL2 not only recognized C1 but also C2 ligands, showing overlapping specificity with KIR2DL1. Moreover, using an NK cell differentiation assay we show sequential acquisition of KIR2DL2 before KIR2DL1 on developing NK cells. Together, these observations are compatible with a ligand-instructed model of NK cell education, in which recognition of HLA class I by an inhibitory receptor (KIR2DL2) suppresses subsequent expression of a second receptor (KIR2DL1) of related specificity. Importantly, the ligand-instructed model fits to the observed KIR repertoires in both broad KIR haplotype groups.
Introduction
A hallmark of natural killer (NK) cells is the expression of major histocompatibility complex (MHC) class I–specific inhibitory receptors, either encoded by the Ly-49 family in rodents1 or the killer cell immunoglobulin-like receptor (KIR) family in humans.2 According to the missing self-model, lack of MHC class I expression leads to elimination of target cells by NK cells expressing a cognate inhibitory receptor for the respective MHC class I molecule.3 The concept implicates an educational system that shapes the repertoire of inhibitory receptors to ensure NK cell self-tolerance. In this regard, it was shown that expression of MHC class I leads to specific adjustment of NK cell frequency expressing cognate Ly-49 in the mouse.4,5
In human subjects, analysis of NK cell clones demonstrated that functional NK cell repertoires have a high propensity to express at least 1 inhibitory receptor for self-MHC class I.6 However, the influence of the polymorphic human histocompatibility leukocyte antigen (HLA) class I genes on NK cell education is not obvious: firstly, NK cells express inhibitory KIR in the absence of cognate HLA class I ligands at a considerable frequency; secondly, self-tolerant NK cells without any HLA class I–specific inhibitory receptor exist in the periphery.7 Moreover, several studies failed to correlate the frequency of any particular KIR to the presence of cognate HLA class I ligands.8,9 Clear evidence for a direct influence of HLA class I on the KIR repertoire of NK cells came from studies of a Japanese cohort, characterized by very limited KIR gene polymorphism.10 The influence of HLA class I was again challenged by a recent study in which KIR expression frequencies were found to be independent of class I ligands.11
The influence of MHC class I ligands on NK cell function is another actual matter of debate. In this regard, it was shown in mice and humans that those NK cells that express inhibitory receptors for host-encoded MHC class I ligands exhibit an increased responsiveness against MHC-deficient target cells.7,12,13 The model implicates that NK cells lacking inhibitory receptors for self-MHC class I are hyporesponsive or anergic. However, it was recently shown that these so-called ‘unlicensed’ NK cells are readily able to control viral infection.14
Human NK cell repertoires are also influenced by the extensive polymorphism of KIR genes.15 KIR genes are organized in different haplotypes: an abundant group of haplotypes that generally consist of a defined set of 7 functional KIR genes was designated group A haplotypes. Group B haplotypes are more variable and are characterized by presence/absence polymorphisms of 5 additional different stimulatory KIR and the inhibitory KIR2DL5.16,17 Because of the higher complexity of group B haplotypes, detailed information on KIR repertoire formation is so far mostly restricted to donors of the A/A haplotype group as these can be unambiguously monitored by multicolor flow cytometry.10 However, in many ethnic groups, except East Asian populations, group A/B or B/B configurations dominate, and it is so far not known if the rules found in cohorts with A/A haplotypes do similarly apply to other populations and ethnic groups with much higher frequencies of group B haplotypes.
Although the rules governing the formation of NK cell repertoires are a matter of debate, it is already evident that the underlying KIR and KIR ligand genetic polymorphisms are of predictive value in different clinical settings such as chronic hepatitis C infection,18 control of HIV infection,19 resistance to cervical neoplasia,20 and hematopoietic stem cell transplantation (HSCT) for leukemia.21,22 Moreover, during pregnancy, certain combinations of maternal KIR genes and fetal KIR ligands are associated with pre-eclampsia, a condition characterized by a placentation defect that is hazardous for mother and baby.23
In an attempt to shed light on the formation of KIR repertoires across the full range of KIR haplotype diversity, a comparative analysis of donors with group A and group B haplotypes was performed in a genetically diverse cohort of white subjects. We demonstrate that the rules applying to formation of HLA-C–specific KIR repertoires found in group A haplotypes cannot simply be conferred to group B haplotypes. Some of the most evident functional and structural differences could be linked to presence of KIR2DL2 and KIR2DS2 on group B haplotypes, which has an unexpected and strong epistatic influence on the expression of KIR2DL1. The observations are compatible with a ligand-instructed model of receptor acquisition in which initially stochastic KIR repertoires are modified by cognate HLA class I ligands in ways that depend on the genetic background of KIR gene polymorphism.
Methods
KIR and HLA-C genotyping
Blood was obtained from healthy white donors after giving informed consent in accordance with the Declaration of Helsinki. The study was approved by the University of Düsseldorf ethics committee. Genomic DNA was isolated from whole blood via QiaAmp DNA Blood Mini kit (QIAGEN). KIR genotyping was performed by polymerase chain reaction–sequence specific primer (PCR-SSP) as reported previously.24 As quality control, several samples were independently KIR-genotyped with a different PCR-SSP protocol.25 Group A and B haplotypes were defined as previously described leading to the following genotype classification: genotype A/A: KIR2DL1, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2, KIR2DS4; genotype A/B: all group A genes plus at least 1 additional KIR gene; genotype B/B: at least 1 group A gene missing. Notably, some of the donors with genotype A/B might in fact be B/B donors that coincidentally possess all group A genes on a group B background.
HLA-C group 1 (C1) and HLA-C group 2 (C2) genotyping was performed by PCR-SSP as described.9 HLA-C subtyping was performed using Luminex technology according to the manufacturers instructions (One Lambda).
Flow cytometry
For flow cytometric analyses, the following mouse anti–human monoclonal antibodies (mAbs) were used: PC5-conjugated CD56 (clone N901), phycoerythrin (PE)–conjugated CD159a (NKG2A, clone Z199), allophycocyanin (APC)–conjugated CD158a,h (KIR2DL1/S1, clone EB6), ECD-conjugated CD3 (clone UCHT1), PE- or ECD-conjugated CD158e (KIR3DL1/S1, clone Z27.3.7), PE- or APC-cyanin 7– (Cy7) or APC-Alexa Fluor 750–conjugated CD158b,j (KIR2DL2/3/S2, clone GL183) all from Beckman Coulter. Fluorescein isothiocyanate (FITC)–conjugated KIR2DL1 (clone 143211; R&D Systems), and Pacific Blue–conjugated CD3 (clone UCHT1; BD Biosciences) were also used. The CD158k-specific mAb Q66 (KIR3DL2), kindly provided by A. Moretta (Universita di Genova, Genova, Italy), was used in combination with anti–mouse immunoglobulin M–FITC (Beckman Coulter). A 7-color flow cytometric analyses was performed on FACSCanto II (BD Biosciences) using FACSDiva 5.0.1 software.
Cell isolation and cell culture
NK cells were isolated from fresh peripheral blood using the RosetteSep method (NK cell enrichment kit; StemCell Technologies). The HLA class I–deficient target cell line K562 and 721.221 as well as the transfected 721.221 (Cw04, Cw07) were cultured in RPMI 1640 (Lonza) with 1% Penicillin/Streptomycin (Invitrogen) and 10% FCS (Biochrom). Cell line 721.221 and the HLA class I–transfected sublines were kindly provided by Salim Khakoo (Imperial College London, London, United Kingdom).
Interferon-γ production and degranulation assay
Freshly isolated NK cells were cultured overnight in the presence of 1000 U/mL recombinant interleukin-2 (Chiron). The following day, NK cells were cocultured with HLA class I–deficient K562 or 721.221 cells or with HLA-C–transfected 721.221 at a 1:1 ratio. Negative controls were performed with NK cells alone. In the positive control, phorbol myristate acetate and ionomycine (both Sigma-Aldrich) were added to NK cell cultures. To measure degranulation, CD107a-FITC (clone H4A3; BD Biosciences) was added. After 1 hour of incubation, monensin (GolgiStop; BD Biosciences) or alternatively brefeldin A (Sigma-Aldrich) was added. The cells were incubated for another 5 hours. The cells were washed and stained with antibodies against CD56, KIR2DL1, KIR2DL2/3, KIR3DL1, and NKG2A. For the interferon-γ (IFN-γ) assay, cells were fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences) and intracellular IFN-γ production was measured using anti–IFN-γ FITC mAb (clone B27; Invitrogen). HLA class I–mediated inhibition of NK cells was assessed by comparison of transfected versus untransfected 721.221 target cells.
Differentiation of NK cells from hematopoietic progenitors
Human CD34+CD38−Lin− hematopoietic progenitor cells were enriched from cord blood using the CD34 Progenitor Cell Isolation kit (Miltenyi Biotec) and subsequent flow cytometric cell sorting. CD34+CD38−Lin− cells were plated in 24-well plates in direct contact with irradiated AFT024 stromal cells and differentiated into mature NK cells as described.26 KIR expression was analyzed by reverse transcription-polymerase chain reaction (RT-PCR) and real-time polymerase chain reaction (PCR) as described.16,22,27
Statistics
Statistical analyses were performed with SPSS 15 software. Significance was determined with ANOVA or 2-tailed t test. Values for P less than .05 were significant.
Results
Differential influence of cognate HLA-C ligands on KIR repertoires in donors with group A and B haplotypes
Repertoires of HLA class I–specific inhibitory receptors were determined in CD56+ NK cells in a cohort of 150 white donors (Table 1) by multicolor flow cytometry, simultaneously using antibodies with specificity for all 4 HLA class I–specific inhibitory KIR (including cross-reactive stimulatory KIR), namely KIR2DL1/KIR2DS1 (C2 group), KIR2DL2/KIR2DL3/KIR2DS2 (C1 group), KIR3DL1/KIR3DS1 (Bw4), and KIR3DL2 (HLA-A3/A11), as well as the HLA-E–specific NKG2A. To simplify designation of the 32 different receptor combinations (clonotypes) that can be distinguished by this approach, an abbreviated nomenclature is used omitting cross-reactive stimulatory KIR, such as clonotype KIR2DL1_2DL2/3 refers to a combination of the antibody specificities KIR2DL1/KIR2DS1 and KIR2DL2/KIR2DL3/KIR2DS2. In concordance with previous studies,11,28 major deviations from the product rule were noted (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Firstly, NKG2A in combination with 1 or more KIR was less frequently observed than expected (P < .05 in 9 of 15 possible combinations) and clonotypes with 2 or more KIR in the absence of NKG2A were more frequently observed (P < .05 in 7 of 11 possible combinations).
Distribution of KIR genotypes in the cohort*
| Haplotype . | Genotype . | KIR Group . | Repertoire analysis . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2DL1 . | 2DL2 . | 2DL3 . | 2DL4 . | 2DL5 . | 3DL1 . | 3DL2 . | 3DL3 . | 2DS1 . | 2DS2 . | 2DS3 . | 2DS4 . | 2DS5 . | 3DS1 . | |||
| AA | 1 |  |  |  |  |  |  |  | 68 | |||||||
| AB | 2 |  |  |  |  |  |  |  |  |  | 23 | |||||
| AB | 3 |  |  |  |  |  |  |  |  |  |  |  | 4 | |||
| AB | 4 |  |  |  |  |  |  |  |  |  |  |  | 6 | |||
| BB | 5 |  |  |  |  |  |  |  |  |  |  | 2 | ||||
| AB | 7 |  |  |  |  |  |  |  |  |  |  |  |  |  | 10 | |
| AB | 8 |  |  |  |  |  |  |  |  |  |  | 2 | ||||
| BB | 9 |  |  |  |  |  |  |  |  |  |  |  |  |  | 1 | |
| BB | 10 |  |  |  |  |  |  |  | 3 | |||||||
| AB | 11 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| AB | 12 |  |  |  |  |  |  |  |  |  |  |  |  |  | 6 | |
| AB | 13 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 14 |  |  |  |  |  |  |  |  |  |  | 7 | ||||
| AB | 15 |  |  |  |  |  |  |  |  | 1 | ||||||
| AB | 16 |  |  |  |  |  |  |  |  | 1 | ||||||
| BB | 17 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| BB | 18 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 19 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 22 |  |  |  |  |  |  |  |  |  |  |  | 3 | |||
| BB | 23 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 25 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 28 |  |  |  |  |  |  |  |  |  | 1 | |||||
| BB | 30 |  |  |  |  |  |  |  |  |  |  |  |  | 2 | ||
| BB | 35 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 42 |  |  |  |  |  |  |  |  |  |  | 1 | ||||
| BB | 44 |  |  |  |  |  |  |  |  |  | 1 | |||||
| Total | 150 | |||||||||||||||
| Haplotype . | Genotype . | KIR Group . | Repertoire analysis . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2DL1 . | 2DL2 . | 2DL3 . | 2DL4 . | 2DL5 . | 3DL1 . | 3DL2 . | 3DL3 . | 2DS1 . | 2DS2 . | 2DS3 . | 2DS4 . | 2DS5 . | 3DS1 . | |||
| AA | 1 |  |  |  |  |  |  |  | 68 | |||||||
| AB | 2 |  |  |  |  |  |  |  |  |  | 23 | |||||
| AB | 3 |  |  |  |  |  |  |  |  |  |  |  | 4 | |||
| AB | 4 |  |  |  |  |  |  |  |  |  |  |  | 6 | |||
| BB | 5 |  |  |  |  |  |  |  |  |  |  | 2 | ||||
| AB | 7 |  |  |  |  |  |  |  |  |  |  |  |  |  | 10 | |
| AB | 8 |  |  |  |  |  |  |  |  |  |  | 2 | ||||
| BB | 9 |  |  |  |  |  |  |  |  |  |  |  |  |  | 1 | |
| BB | 10 |  |  |  |  |  |  |  | 3 | |||||||
| AB | 11 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| AB | 12 |  |  |  |  |  |  |  |  |  |  |  |  |  | 6 | |
| AB | 13 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 14 |  |  |  |  |  |  |  |  |  |  | 7 | ||||
| AB | 15 |  |  |  |  |  |  |  |  | 1 | ||||||
| AB | 16 |  |  |  |  |  |  |  |  | 1 | ||||||
| BB | 17 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| BB | 18 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 19 |  |  |  |  |  |  |  |  |  |  |  |  | 1 | ||
| AB | 22 |  |  |  |  |  |  |  |  |  |  |  | 3 | |||
| BB | 23 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 25 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 28 |  |  |  |  |  |  |  |  |  | 1 | |||||
| BB | 30 |  |  |  |  |  |  |  |  |  |  |  |  | 2 | ||
| BB | 35 |  |  |  |  |  |  |  |  |  |  |  | 1 | |||
| BB | 42 |  |  |  |  |  |  |  |  |  |  | 1 | ||||
| BB | 44 |  |  |  |  |  |  |  |  |  | 1 | |||||
| Total | 150 | |||||||||||||||
Nomenclature of KIR genotypes according to Uhrberg et al24 except genotype 42 and 44, which represent novel genotypes. Presence of a KIR gene is represented by a shaded box.
We next subdivided the cohort on the basis of KIR haplotype groups and HLA-C ligands (Table 2). In the A/A haplotype subgroup (Figure 1A), the repertoires of C1 and C2 donors were largely similar except for 3 clonotypes: firstly, a pronounced increase of single-positive KIR2DL1 (single-KIR2DL1+) NK cells was observed in presence of cognate C2/C2 ligands; secondly, single-KIR2DL3+ cells were more prevalent in donors with cognate C1/C1 ligands; in both cases the respective self-specific clonotype represented the most frequent KIR-expressing clonotype in that cohort; thirdly, the KIR2DL1_2DL2/3 clonotype was more abundant in C2 donors. In contrast, no such increase of single-KIR+ clonotypes for cognate HLA-C ligands was seen in the B/B haplotype subgroup (Figure 1B). A minor increase in presence of cognate C2 was seen for 3 clonotypes involving KIR2DL1 (KIR2DL1_3DL2, KIR2DL1_NKG2A, and KIR2DL1_2DL2/3_NKG2A). It should be mentioned that in group B donors, it was not possible to differentiate between KIR2DL2, KIR2DL3, and KIR2DS2 because of cross-reactivity of the GL183 antibody.
Distribution of KIR haplotypes*
| HLA-C group . | Haplotype group . | |||
|---|---|---|---|---|
| A/A . | A/B . | B/B . | Total . | |
| C1/C1 | 28 | 30 | 6 | 65 |
| C1/C2 | 30 | 22 | 7 | 58 |
| C2/C2 | 10 | 13 | 4 | 27 |
| Total | 68 | 65 | 17 | 150 |
| HLA-C group . | Haplotype group . | |||
|---|---|---|---|---|
| A/A . | A/B . | B/B . | Total . | |
| C1/C1 | 28 | 30 | 6 | 65 |
| C1/C2 | 30 | 22 | 7 | 58 |
| C2/C2 | 10 | 13 | 4 | 27 |
| Total | 68 | 65 | 17 | 150 |
Distribution depends on the subgroups of HLA-C ligands.
KIR repertoires are shaped by HLA class I–dependent mechanisms. The repertoire of donors with group A/A haplotypes (A) and group B/B haplotypes (B) is shown for C1/C1 (□) versus C2/C2 (■) donors. HLA class I ligand and haplotype group distribution of samples is shown in Table 2. Designation of clonotypes does not include cross-reactive stimulatory KIR. KIR−NKG2A− refers to NK cells expressing no inhibitory receptors. Statistical significance was determined by 2-tailed t test at *P < .05, **P < .01, ***P < .001. SD is indicated by error bar.
KIR repertoires are shaped by HLA class I–dependent mechanisms. The repertoire of donors with group A/A haplotypes (A) and group B/B haplotypes (B) is shown for C1/C1 (□) versus C2/C2 (■) donors. HLA class I ligand and haplotype group distribution of samples is shown in Table 2. Designation of clonotypes does not include cross-reactive stimulatory KIR. KIR−NKG2A− refers to NK cells expressing no inhibitory receptors. Statistical significance was determined by 2-tailed t test at *P < .05, **P < .01, ***P < .001. SD is indicated by error bar.
Separate analysis for the 3 subgroups of HLA-C ligands revealed that in donors homozygous for group A haplotypes (A/A), the overall frequency of NK cells expressing KIR2DL1 (sum of all 16 clonotypes expressing KIR2DL1) as well as single-KIR2DL1+ NK cells increased in presence of cognate ligands in a gene dose-dependent way (Figure 2A). A similar, but less pronounced increase in response to cognate ligand was seen for the frequency of KIR2DL3-expressing NK cells. Remarkably, in group A/A donors the ratio of single-KIR2DL1+/single-KIR2DL3+ frequencies is reversed from 0.6 in C1/C1 donors to 4.3 in the presence of cognate C2/C2 ligands (Table 3). Increased NK cell frequency went along with decreased cell surface expression levels for KIR2DL1 but not KIR2DL3 (supplemental Figure 2), as previously described.10 In contrast to the group A haplotype cohort, no significant changes in this ratio and no bias of KIR2DL1 expression toward cognate ligands were seen for group B haplotypes (A/B and B/B; Table 3 and Figure 2B).
HLA-C ligands differentially affect frequencies but not expression levels of cognate KIR in donors with group A and group B haplotypes. (A) The cohort was divided into 4 genetic subgroups: donors with group A (A/A) haplotypes (n = 68), donors with group B (A/B and B/B) haplotypes (n = 82), donors with genotype 2 (KIR2DL2+; n = 23), and donors with group B haplotypes lacking KIR2DL2 (n = 13). In panels A through D, the frequency of NK cells expressing KIR2DL1 or KIR2DL2/3 is separately shown for the 3 subgroups of HLA-C ligands. Results are shown as stapled bars with overall frequency of NK cells expressing a given KIR ( ) and frequency of single-positive NK cells, that is NK cells expressing a given KIR without coexpression of any other detectable KIR or NKG2A (
) and frequency of single-positive NK cells, that is NK cells expressing a given KIR without coexpression of any other detectable KIR or NKG2A ( ). Results represent mean and an error bar represents SD. Statistical significance was determined by ANOVA at *P < .05, **P < .01, ***P < .001) and is shown for single-positive KIR frequencies.
). Results represent mean and an error bar represents SD. Statistical significance was determined by ANOVA at *P < .05, **P < .01, ***P < .001) and is shown for single-positive KIR frequencies.
HLA-C ligands differentially affect frequencies but not expression levels of cognate KIR in donors with group A and group B haplotypes. (A) The cohort was divided into 4 genetic subgroups: donors with group A (A/A) haplotypes (n = 68), donors with group B (A/B and B/B) haplotypes (n = 82), donors with genotype 2 (KIR2DL2+; n = 23), and donors with group B haplotypes lacking KIR2DL2 (n = 13). In panels A through D, the frequency of NK cells expressing KIR2DL1 or KIR2DL2/3 is separately shown for the 3 subgroups of HLA-C ligands. Results are shown as stapled bars with overall frequency of NK cells expressing a given KIR ( ) and frequency of single-positive NK cells, that is NK cells expressing a given KIR without coexpression of any other detectable KIR or NKG2A (
) and frequency of single-positive NK cells, that is NK cells expressing a given KIR without coexpression of any other detectable KIR or NKG2A ( ). Results represent mean and an error bar represents SD. Statistical significance was determined by ANOVA at *P < .05, **P < .01, ***P < .001) and is shown for single-positive KIR frequencies.
). Results represent mean and an error bar represents SD. Statistical significance was determined by ANOVA at *P < .05, **P < .01, ***P < .001) and is shown for single-positive KIR frequencies.
Ratio of KIR2DL1- to KIR2DL2/3-expressing NK cells
| KIR Haplotype group . | HLA-C ligand . | Ratio KIR2DL1/KIR2DL2/3 (single/single KIR)* . | P† . |
|---|---|---|---|
| Group A | C1/C1 | 0.69 (0.55) | < .001 (< .001) |
| C1/C2 | 0.87 (1.04) | ||
| C2/C2 | 1.51 (4.39) | ||
| Group B | C1/C1 | 0.40 (0.28) | .330 (.356) |
| C1/C2 | 0.67 (0.81) | ||
| C2/C2 | 0.58 (0.59) | ||
| Group B KIR2DL2+ | C1/C1 | 0.40 (0.27) | .748 (.660) |
| C1/C2 | 0.62 (0.80) | ||
| C2/C2 | 0.42 (0.36) | ||
| Group B KIR2DL2− | C1/C1 | 0.61 (0.49) | .011 (.005) |
| C1/C2 | 0.84 (0.84) | ||
| C2/C2 | 1.42 (1.79) |
| KIR Haplotype group . | HLA-C ligand . | Ratio KIR2DL1/KIR2DL2/3 (single/single KIR)* . | P† . |
|---|---|---|---|
| Group A | C1/C1 | 0.69 (0.55) | < .001 (< .001) |
| C1/C2 | 0.87 (1.04) | ||
| C2/C2 | 1.51 (4.39) | ||
| Group B | C1/C1 | 0.40 (0.28) | .330 (.356) |
| C1/C2 | 0.67 (0.81) | ||
| C2/C2 | 0.58 (0.59) | ||
| Group B KIR2DL2+ | C1/C1 | 0.40 (0.27) | .748 (.660) |
| C1/C2 | 0.62 (0.80) | ||
| C2/C2 | 0.42 (0.36) | ||
| Group B KIR2DL2− | C1/C1 | 0.61 (0.49) | .011 (.005) |
| C1/C2 | 0.84 (0.84) | ||
| C2/C2 | 1.42 (1.79) |
Expression frequency of KIR2DL1 divided by expression frequency of KIR2DL2/3. Parentheses are ratio of single-KIR2DL1+/single-KIR2DL2/3.
P values were calculated by ANOVA. Values for P less than .05 are significant.
The KIR2DL2/3 allelic pair affects frequency and function of the KIR2DL1+ NK cell subpopulation
The next aim was to find out which features of the genetically heterogeneous group B haplotypes might explain the apparent differences in HLA-C–specific NK cell repertoires. When analysis was restricted to the most frequent group B genotype (genotype 2),24 possessing all KIR genes of group A haplotypes plus KIR2DL2 and KIR2DS2, the frequency of KIR2DL1+ NK cells again did not increase in the presence of cognate ligands (Figure 2C). This suggested that the observed difference in the frequency of KIR2DL1+ NK cells between group A and B haplotypes was associated with KIR2DL2 and KIR2DS2. To cross-check this association, we next restricted the analysis to those group B genotypes lacking KIR2DL2 and KIR2DS2 (Figure 2D). This cohort indeed showed changes similar to the A/A haplotype group, such as frequency of KIR2DL1 and KIR2DL3 was increased in the presence of the respective cognate ligand. In the remaining group B genotypes (KIR2DL2+/2DS2+ but not genotype 2) again no significant influence of HLA-C ligands was observed (data not shown). Remarkably, the KIR2DL1/KIR2DL3 frequency ratio was strongly affected by the presence of the KIR2DL2 gene: in group B donors lacking KIR2DL2, ratios were highly responsive to the presence of cognate ligands similar to what was seen in donors with group A/A haplotypes (Table 3). In contrast, in group B donors having KIR2DL2, the ratio remained unaffected.
These study observations already suggested that the frequency of KIR2DL1+ NK cells is influenced by the allelic state of the KIR2DL2/3 gene. Indeed, the frequency of KIR2DL1+ NK cells decreased significantly and in a gene-dose dependent way in presence of 0, 1, or 2 KIR2DL2 alleles (Figure 3A). Consequently, the frequency of cytotoxic KIR2DL1+ NK cells is influenced by the allelic state of the KIR2DL2/3 gene, (Figure 3B): flow cytometric analysis of CD107 expression revealed a significantly reduced fraction of degranulating KIR2DL1-expressing NK cells in KIR2DL2+ donors. In contrast, the frequency of degranulating KIR2DL2/3-expressing NK cells remained unchanged (Figure 3C). Thus, KIR2DL2+ donors possess less NK cells that are exclusively specific for C2 ligands than KIR2DL2− donors.
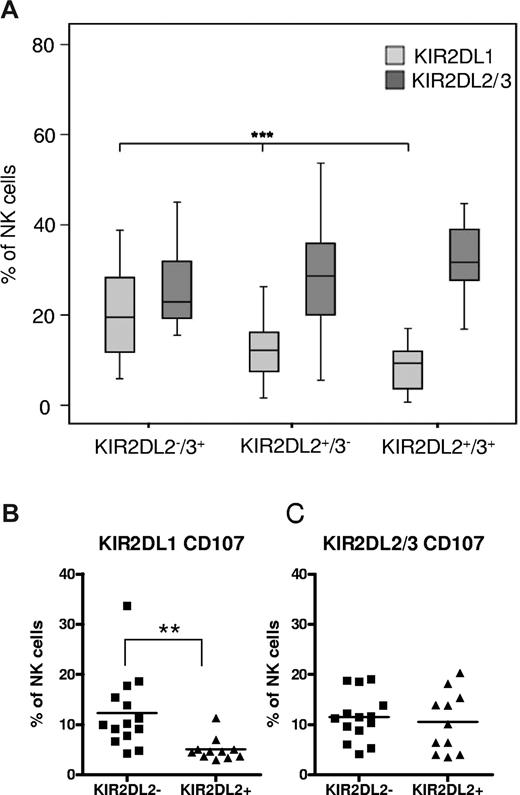
The KIR2DL1+ NK cell compartment is influenced by the allelic state of KIR2DL2/3. (A) The frequency of NK cells expressing KIR2DL1 and KIR2DL2/3 is shown for donors with group B haplotypes (A/B and B/B) who were divided into 3 groups according to the allelic state of the KIR2DL2/3 gene. Data are represented as box plots: that represent the median and 25/75 percentiles, whiskers the lowest and largest non-outlier data points (at most 1.5 times the box width). Statistical significance was determined by ANOVA at ***P < .001. (B-C) Frequency of NK cells expressing KIR2DL1 (B) or KIR2DL2/3 (C) in a CD107 mobilization assay against K562 target cells. Samples from donors without (n = 14) and with (n = 11) KIR2DL2 genes are presented in scatter plots with each dot representing 1 sample. Results are expressed as fraction of all CD56+ NK cells. Statistical significance was calculated by 2-tailed t test as **P < .01.
The KIR2DL1+ NK cell compartment is influenced by the allelic state of KIR2DL2/3. (A) The frequency of NK cells expressing KIR2DL1 and KIR2DL2/3 is shown for donors with group B haplotypes (A/B and B/B) who were divided into 3 groups according to the allelic state of the KIR2DL2/3 gene. Data are represented as box plots: that represent the median and 25/75 percentiles, whiskers the lowest and largest non-outlier data points (at most 1.5 times the box width). Statistical significance was determined by ANOVA at ***P < .001. (B-C) Frequency of NK cells expressing KIR2DL1 (B) or KIR2DL2/3 (C) in a CD107 mobilization assay against K562 target cells. Samples from donors without (n = 14) and with (n = 11) KIR2DL2 genes are presented in scatter plots with each dot representing 1 sample. Results are expressed as fraction of all CD56+ NK cells. Statistical significance was calculated by 2-tailed t test as **P < .01.
The correlation of KIR2DL1 expression with the allelic state of KIR2DL2/3 was observed across all combinations of HLA-C ligands (Figure 4A). We further concentrated our analysis on the subgroup of C2/C2 donors, as their NK cell repertoire is functionally most strongly affected by the observed changes in KIR2DL1 frequency. Among the 8 KIR2DL1+ clonotypes that were most abundant (frequency > 1%) in donors lacking KIR2DL2, all but 1 clonotype exhibited strongly decreased levels in donors possessing the KIR2DL2 gene (Figure 4B). The correlation was gene dose-dependent and most strongly seen in donors with 2 copies of KIR2DL2. Interestingly, no dependency on the presence of KIR2DL2 was seen in case of the KIR2DL1_3DL2 clonotype, suggesting that KIR2DL1 expression is not affected by the presence of the HLA-A3– and HLA-A11–specific inhibitory receptor KIR3DL2.
KIR2DL2-associated changes in KIR2DL1 expression are independent of the presence of cognate HLA-C ligands and expression of KIR2DS1. (A) Frequency of single-KIR2DL1+ NK cells is shown for donors with group B haplotypes (A/B and B/B) who were divided into 9 groups according to the allelic state of the KIR2DL2/3 gene and the HLA-C ligands C1 and C2. (B) All 16 different clonotypes that express KIR2DL1 are shown for C2/C2 donors with zero (□), 1 ( ), or 2 (
), or 2 ( ) copies of the KIR2DL2 gene. (C) Frequency of single-KIR2DL1+, KIR2DL1+KIR2DS1+, and single-KIR2DS1+ NK cells was measured with multicolor flow cytometric analysis with the following antibody combinations: KIR2DL1(143211)–FITC, CD158a,h(EB6)–APC, KIR3DL1/KIR2DL2/3/NKG2A-PE, CD56-PC5 and CD3-ECD. Donors had a KIR2DL1+/KIR2DS1+ genotype (n = 29) and were divided into KIR2DL2−KIR2DL3+, KIR2DL2+KIR2DL3+, and KIR2DL2+KIR2DL3− as indicated on the x-axis. Statistical significance was determined by ANOVA as *P < .05, **P < .01, ***P < .001).
) copies of the KIR2DL2 gene. (C) Frequency of single-KIR2DL1+, KIR2DL1+KIR2DS1+, and single-KIR2DS1+ NK cells was measured with multicolor flow cytometric analysis with the following antibody combinations: KIR2DL1(143211)–FITC, CD158a,h(EB6)–APC, KIR3DL1/KIR2DL2/3/NKG2A-PE, CD56-PC5 and CD3-ECD. Donors had a KIR2DL1+/KIR2DS1+ genotype (n = 29) and were divided into KIR2DL2−KIR2DL3+, KIR2DL2+KIR2DL3+, and KIR2DL2+KIR2DL3− as indicated on the x-axis. Statistical significance was determined by ANOVA as *P < .05, **P < .01, ***P < .001).
KIR2DL2-associated changes in KIR2DL1 expression are independent of the presence of cognate HLA-C ligands and expression of KIR2DS1. (A) Frequency of single-KIR2DL1+ NK cells is shown for donors with group B haplotypes (A/B and B/B) who were divided into 9 groups according to the allelic state of the KIR2DL2/3 gene and the HLA-C ligands C1 and C2. (B) All 16 different clonotypes that express KIR2DL1 are shown for C2/C2 donors with zero (□), 1 ( ), or 2 (
), or 2 ( ) copies of the KIR2DL2 gene. (C) Frequency of single-KIR2DL1+, KIR2DL1+KIR2DS1+, and single-KIR2DS1+ NK cells was measured with multicolor flow cytometric analysis with the following antibody combinations: KIR2DL1(143211)–FITC, CD158a,h(EB6)–APC, KIR3DL1/KIR2DL2/3/NKG2A-PE, CD56-PC5 and CD3-ECD. Donors had a KIR2DL1+/KIR2DS1+ genotype (n = 29) and were divided into KIR2DL2−KIR2DL3+, KIR2DL2+KIR2DL3+, and KIR2DL2+KIR2DL3− as indicated on the x-axis. Statistical significance was determined by ANOVA as *P < .05, **P < .01, ***P < .001).
) copies of the KIR2DL2 gene. (C) Frequency of single-KIR2DL1+, KIR2DL1+KIR2DS1+, and single-KIR2DS1+ NK cells was measured with multicolor flow cytometric analysis with the following antibody combinations: KIR2DL1(143211)–FITC, CD158a,h(EB6)–APC, KIR3DL1/KIR2DL2/3/NKG2A-PE, CD56-PC5 and CD3-ECD. Donors had a KIR2DL1+/KIR2DS1+ genotype (n = 29) and were divided into KIR2DL2−KIR2DL3+, KIR2DL2+KIR2DL3+, and KIR2DL2+KIR2DL3− as indicated on the x-axis. Statistical significance was determined by ANOVA as *P < .05, **P < .01, ***P < .001).
KIR2DL1 but not KIR2DS1 expression is affected by the presence of KIR2DL2
As a result of serologic cross-reactivity of the EB6 antibody with KIR2DL1 and KIR2DS1, it was unclear in how far both receptors were affected by the presence of KIR2DL2. Generally, single-KIR2DS1+ NK cells should be rare in the presence of C2 ligands as these cells are potentially autoreactive. Indeed, when we analyzed the NK cell repertoire of a donor possessing KIR2DS1 but not KIR2DL1 (genotype 23),24 single-KIR2DS1+ NK cells were rare (< 0.3% of 6% overall KIR2DS1 frequency; supplemental Figure 3), an observation that is in line with previous studies of NK cell clones.6,29,30 To exactly determine how the KIR2DS1+ subset of NK cells is influenced by KIR2DL2, the subset of KIR2DL1+/KIR2DS1+ donors (n = 29) were analyzed with 2 different KIR2DL1-specific antibodies, 1 cross-reacting (mAb EB6) and 1 not cross-reacting (mAb 143211) with KIR2DS1.31 Whereas the frequency of single-KIR2DL1+ as well as double-positive NK cells significantly decreased in the presence of KIR2DL2, the frequency of single-KIR2DS1+ NK cells was not influenced (Figure 4C). These data demonstrate that expression of the inhibitory KIR2DL1, but not the closely related stimulatory KIR2DS1, is affected by KIR2DL2.
KIR2DL2 can functionally replace KIR2DL1 as C2-specific inhibitory receptor
These study data demonstrate that KIR2DL1 expression is suppressed by KIR2DL2 in NK cell repertoires of most donors with group B/B haplotypes. One key question was whether the lack of KIR2DL1-expressing NK cells leads to a general lack of NK cells that are able to recognize C2 ligands in these donors. In this regard, it was recently shown that KIR2DL2 and to a lesser extent KIR2DL3 are not solely specific for HLA-C alleles with C1 epitopes, as originally reported, but can also bind to C2 ligands.32,33 To this end, we compared the effector functions of NK cells from donors lacking KIR2DL2 with those having 1 or 2 copies of KIR2DL2 (Figure 5A). Single-KIR2DL3+ NK cells (KIR2DL2− donors) were strongly inhibited by 721.221 transfectants expressing the cognate C1 group ligand Cw*0701 but not by cells expressing the C2 group ligand Cw*0401. Significant differences in effector functions were seen on the level of CD107 mobilization (Figure 5B) as well as production of interferon-γ (data not shown). In contrast, single-KIR2DL2/3+ NK cells from KIR2DL2+ donors were inhibited by C1 as well as C2 transfectants without showing significant differences in the degree of inhibition (Figure 5B). In control experiments, single-KIR2DL1+ cells were strongly inhibited by C2 but not C1 ligands, as expected (data not shown). These data demonstrate that the decreased frequency of KIR2DL1-expressing NK cells in donors having the KIR2DL2 gene does not lead to a general lack of NK cells that are able to recognize C2 ligands. Instead, it suggests that KIR2DL2+ NK cells can take over the function as C2-specific receptor but without the ability to discriminate between C1 and C2 ligands.
KIR2DL2-expressing NK cells are inhibited by C1 as well as C2 ligands. Flow cytometric analysis of CD107 mobilization and intracellular IFN-γ on NK cells using 721.221 and HLA-transfected sublines Cw*0702 and Cw*0401 as target cell lines. (A) Histograms show CD107 expression of KIR2DL2/3+ cells from a donor with KIR2DL2 (genotype 12) and 1 donor without KIR2DL2 (genotype 1). (B) Functional inhibition was calculated by measuring reduction of CD107 mobilization NK cells in response to Cw*0401 (C2), Cw*0702 (C1)–transfected 721.221, and no targets (negative control). Values are calculated relative to NK cells cultured with untransfected 721.221. Analysis was gated on single-KIR2DL2/3+ NK cells in donors with (n = 6) or without (n = 5) KIR2DL2. Statistical analysis was performed by 2-tailed t test for *P < .05.
KIR2DL2-expressing NK cells are inhibited by C1 as well as C2 ligands. Flow cytometric analysis of CD107 mobilization and intracellular IFN-γ on NK cells using 721.221 and HLA-transfected sublines Cw*0702 and Cw*0401 as target cell lines. (A) Histograms show CD107 expression of KIR2DL2/3+ cells from a donor with KIR2DL2 (genotype 12) and 1 donor without KIR2DL2 (genotype 1). (B) Functional inhibition was calculated by measuring reduction of CD107 mobilization NK cells in response to Cw*0401 (C2), Cw*0702 (C1)–transfected 721.221, and no targets (negative control). Values are calculated relative to NK cells cultured with untransfected 721.221. Analysis was gated on single-KIR2DL2/3+ NK cells in donors with (n = 6) or without (n = 5) KIR2DL2. Statistical analysis was performed by 2-tailed t test for *P < .05.
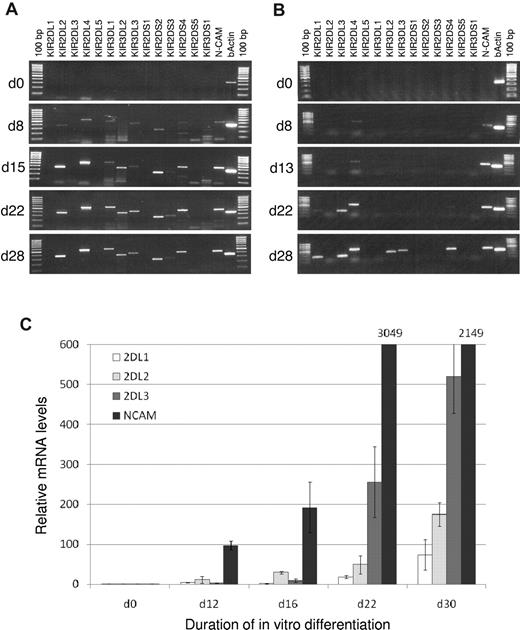
Sequential acquisition of HLA-C-specific KIR during NK cell differentiation in vitro
These study data suggest that KIR2DL3 is mainly C1-specific, whereas KIR2DL1 as well as KIR2DL2 both can serve as C2-specific receptors. The question remained as to why KIR2DL1 is largely replaced by KIR2DL2 in C2/C2 donors: if both KIR types can serve a similar function as C2-specific receptor they should be selected at comparable frequencies as self-specific receptors. Notably, we have previously shown that in the course of NK cell differentiation, KIR2DL3 is expressed before and at a higher frequency than KIR2DL1.22 Here, the same in vitro NK cell differentiation assay, with a hepatic stroma cell line as support, was used to analyze if the nonrandom sequential acquisition of KIR similarly applies to KIR2DL2. To this end, CD34+CD38−Lin− hematopoietic progenitors from cord blood were differentiated into mature NK cells. To distinguish between KIR2DL2 and KIR2DS2, analyses were performed on the mRNA expression level. In a KIR2DL2+KIR2DL3− donor, KIR2DL2 mRNA was detected on day 15 of in vitro culture together with several other KIR (Figure 6A). Remarkably, no signal for KIR2DL1 was detectable until the last time point of analysis (day 28). In a KIR2DL2−KIR2DL3+ donor (group A/A), a similar sequential expression mode (albeit with a somewhat slower kinetics) with detection of KIR2DL3 mRNA (day 22) before KIR2DL1 (day 28) as well as other KIR was seen, (Figure 6B). Finally, in a KIR2DL2+KIR2DL3+ donor, a semi-quantitative evaluation of expression was done by real-time PCR. Again, KIR2DL2 was earlier and stronger expressed than KIR2DL1. Notably, KIR2DL3 was strongest expressed at later time points (Figure 6C). Thus, KIR2DL1 was consistently expressed subsequent to KIR2DL2 (or KIR2DL3).
Sequential acquisition of KIR during NK cell differentiation. In vitro differentiation of NK cells from hematopoietic stem cells was performed as described in “Methods.” RT-PCR results from a donor with KIR genotype 5 (KIR2DL2+KIR2DL3−; A) and a donor with KIR genotype 1 (KIR2DL2−KIR2DL3+; B). Neural cell adhesion molecule (NCAM, CD56) intensity was used as correlate for NK cell development, β-actin as positive control for PCR. (C) Expression levels of KIR2DL1, KIR2DL2, KIR2DL3, and NCAM normalized against β-actin from a donor with KIR genotype 7 (KIR2DL2+KIR2DL3+) were measured with real-time PCR. NCAM levels greater than 600 are indicated.
Sequential acquisition of KIR during NK cell differentiation. In vitro differentiation of NK cells from hematopoietic stem cells was performed as described in “Methods.” RT-PCR results from a donor with KIR genotype 5 (KIR2DL2+KIR2DL3−; A) and a donor with KIR genotype 1 (KIR2DL2−KIR2DL3+; B). Neural cell adhesion molecule (NCAM, CD56) intensity was used as correlate for NK cell development, β-actin as positive control for PCR. (C) Expression levels of KIR2DL1, KIR2DL2, KIR2DL3, and NCAM normalized against β-actin from a donor with KIR genotype 7 (KIR2DL2+KIR2DL3+) were measured with real-time PCR. NCAM levels greater than 600 are indicated.
Discussion
The intrinsic and extrinsic factors that shape human NK cell repertoires are incompletely defined, in particular the contribution of stochastic versus ligand-instructed mechanisms.8,10,11 The 3 major determinants that influence the structure and function of KIR repertoires could be distinguished, namely cognate HLA class I ligands, KIR haplotype diversity, and a hardwired program of sequential receptor acquisition during NK cell development. Together, these factors seem to efficiently adjust a basically stochastic repertoire toward specific recognition of self-HLA class I.
The influence of HLA class I was clearly evident in group A/A haplotype donors. Analysis of individual clonotypes revealed highly specific changes in response to HLA class I ligands: in C1/C1 as well as C2/C2 donors the specific single-KIR+ clonotype that recognizes cognate HLA in a mono-specific way was found at frequencies exceeding any other KIR-expressing clonotype. Moreover, the ratio of KIR2DL1+/KIR2DL3+ NK cells was significantly different between C1/C1 and C2/C2 donors. Especially the ratio of single-KIR2DL1+/single-KIR2DL3+ frequencies provided a highly reliable parameter that was consistently well below 1 in C1/C1 and well above 1 in C2/C2 donors. It thus appears that the relative proportions of C1- and C2-specific NK cells are a parameter that is tightly controlled in the process of HLA class I–dependent KIR repertoire development. It should be noted that also the frequency of KIR3DL1-expressing NK cells significantly increases with the presence of the respective Bw4 ligand (data not shown). Our results are thus in agreement with a previous study in the Japanese population showing adjustment of KIR expression frequency by cognate HLA class I.10
The second major component shaping HLA-C-specific NK cell repertoires is the genetic diversity of KIR, which is characterized by extensive variation in KIR haplotypes. Comparative analysis of clonotype profiles in group A and group B donors revealed pronounced differences: the expansion of single-KIR+ NK cells for cognate HLA-C, a highly significant and consistent finding in group A/A donors, was not detectable in donors with group B haplotypes. Subsequent analyses demonstrate that this difference is not a general property of all group B haplotypes but segregates with the presence of KIR2DL2. Not only the frequency but also the function of KIR2DL1+ NK cells was influenced by the presence of KIR2DL2. There were less KIR2DL1+ NK cells that degranulated in the presence of HLA-deficient target in donors with KIR2DL2 compared with donors with homozygous KIR2DL3 (Figure 3B). Moreover, KIR2DL2 exhibited specific inhibition of IFN-γ production not only by C1 but also by C2 ligands, In fact, inhibition of KIR2DL2 by the C2 allele Cw*0401 was slightly lower but not significantly different to the inhibition of KIR2DL1. Because Cw*0401 does represent a ligand of intermediate affinity in binding assays,32 it is likely that many other C2 alleles will similarly serve as inhibitory ligands for KIR2DL2. Notably, HLA-C subtyping of C2/C2 donors (supplemental Table 2) did not reveal any HLA-C alleles not supporting the KIR2DL2-specific effect on HLA-C–specific KIR repertoires. These findings together with previous studies suggest that the specificity of KIR2DL2 transcends the distinction of HLA-C ligands in C1 and C2.32,34
The presence of KIR2DL2 had a dominant suppressive effect on expression of KIR2DL1. We hypothesize that the impact of KIR2DL2 on the formation of HLA-C–specific KIR repertoires is not only based on its broad specificity but also on the mode of receptor acquisition during NK cell differentiation. In an in vitro NK cell differentiation model it could be shown that KIR2DL2 and KIR2DL3 expression preceded expression of KIR2DL1, both on mRNA and, as shown by us and others, cell surface expression levels.22,26 Similar observations were made during NK cell reconstitution in clinical stem cell transplantation.35,36 The molecular basis for the observed sequential KIR acquisition mode is unclear. Although KIR regulatory regions are highly homologous, distinct differences exist between promoters of KIR2DL1 and KIR2DL2/3 in terms of transcription factor binding sites.37 It is thus possible, that small differences in promoter strength or the recently described antisense activity of KIR promoters38 might be involved in the control of sequential KIR acquisition.
We suggest that the presence of KIR2DL2 on developing NK cells does not favor subsequent expression of KIR2DL1 on the same clones. Because single-KIR2DL2+ NK cells are already self-specific in C1/C1 as well as C2/C2 donors, further acquisition of KIR2DL1 is not required for induction of tolerance in any circumstance. In contrast, single-KIR2DL3+ NK cells are largely specific for C1 ligands and would thus not be selected for in C2/C2 donors. Thus, in these donors KIR2DL1 is required for induction of tolerance to C2 ligands. These considerations readily explain why in C2/C2 donors that possess the KIR2DL2 allele, single-KIR2DL1+ NK cells are exceedingly rare, whereas C2/C2 donors possessing 2 KIR2DL3 alleles exhibit a substantial expansion of single-KIR2DL1+ NK cells. In this context, we could exclude a major role for KIR2DS1, as the presence of KIR2DL2 did not affect expression level or frequency of KIR2DS1. Notably, it was recently shown that KIR2DS1 expression is not influenced by HLA-C either.39,40 In contrast, we could not differentiate between expression of KIR2DL2 and KIR2DS2 by flow cytometry. Because KIR2DL2 and KIR2DS2 are in strong linkage disequilibrium and we did not have any donors lacking 1 but not the other gene in our cohort, we cannot exclude the KIR2DS2 gene as a confounding factor. Nonetheless, as KIR2DS2 has no detectable affinity for HLA-C ligands, KIR2DL2 is likely to be the main factor leading to the shift in HLA-C2–specific NK cell frequencies. Another ambiguity that remains to be resolved as a result of the lack of discriminating antibodies is the effect of KIR2DL2 on KIR2DL3. In fact it is likely that KIR2DL3 frequencies are lower in KIR2DL2/3 heterozygotes than in KIR2DL3 homozygotes as a result of a gene dose effect.
The study strongly suggests that the order in which NK cell receptors are acquired during NK cell development is a crucial factor that shapes HLA class I–specific NK cell repertoires, a notion that pertains to KIR as well as NKG2A. During NK cell development, NKG2A expression is initiated at an earlier stage than KIR expression (K.S., unpublished observations, November 2009).26 Those NK cell that express NKG2A as their only inhibitory receptor, are already self-specific and thus do not require further acquisition of cognate inhibitory KIR. Indeed, the NKG2A+KIR− clonotype is significantly more frequent than expected by the product rule, an effect that is independent of KIR haplotype groups. Vice versa, combinations of NKG2A with cognate KIR are less frequently found than statistically expected, an effect previously described for NK cell clones.6
The observed bias toward single-KIR+ NK cells for cognate ligands, the repression of KIR2DL1 expression in the presence of KIR2DL2 in C2/C2 donors, as well as the preponderance of single-NKG2A+ NK cells all point toward a system that favors NK cells with restricted specificity for 1 major HLA class I ligand (HLA class I or HLA-E, respectively). In line with this observation, human NK cell repertoires are dominated by clonotypes with few HLA class I–specific inhibitory receptors: among the 32 different clonotypes, NK cells expressing combinations of 0 to 2 receptors (representing 16 different clonotypes) comprised more than 90% of the repertoire (Figure 1 and supplemental Figure 1). These observations are compatible with a ligand-instructed model: upon interaction of an inhibitory receptor with a self-ligand, up-regulation of additional receptors ceases or at least becomes less likely. Only in the absence of a cognate ligand, more inhibitory receptors are up-regulated until self-specificity is ensured. Of note, the functional significance of the KIR−NKG2A− population, which is the largest clonotype in all analyzed cohorts, is still unclear. It could be speculated that it provides a reservoir of still immature NK cells that is shaped in the periphery toward expression of cognate HLA class I. This notion is compatible with the observation that the overall frequency of KIR−NKG2A− cells is inversely correlated to the frequency of KIR+NKG2A− NK cells.28 In this context, it has to be noted that we have not assessed expression of LILRB1, an inhibitory receptor with ubiquitous specificity for HLA class I.41 Given the comparably low affinity for HLA class I and the fact that LILRB1 in contrast to KIR and NKG2A does not confer enhanced missing self responses,28 the impact of LILRB1 expression on KIR repertoire acquisition is likely to be limited.
The present results appear to contradict a recent study claiming that KIR repertoires are generated through a sequential but random acquisition mode without any selection.11 The model is based on the observation that combinations of 2 or more KIR are more abundant than expected by the product rule and independent of selection by cognate HLA class I. Indeed, we also observe that NK cells expressing 3 to 5 KIR receptors in the absence of NKG2A are more frequent than expected by the product rule. Although these clonotypes are rare and add up to a cumulative frequency of 2% to 3% of NK cells, the effect is highly significant (supplemental Figure 4). However, this observation does not exclude an influence of HLA class I on the formation of KIR repertoires. The failure of Andersson et al11 to detect HLA class I–mediated adaptation could rather be because the authors did not break down the analysis to single clonotypes and did not consider the major class I ligands separately. In fact, if our data are calculated similarly, that is cumulative consideration of KIR ligands and KIR receptors, the influence of HLA class I ligands is no longer detectable: donors with 1 major HLA class I ligand do not express significantly more inhibitory receptors than donors with 2 or more HLA class I ligands (supplemental Figure 4).
It is important to note that the rules governing formation of KIR repertoires in group A/A donors are different for C1/C1 and C2/C2 subgroups. Because of the early acquisition of KIR2DL2/3, the repertoire of a C1/C1 donor is inherently biased toward expression of self-specific KIR. In contrast, the repertoire of a C2/C2 donor is dependent on selection of KIR2DL1+ NK cells, which are per se less frequent in an unbiased KIR repertoire because of their late appearance. This asymmetry, caused by the nonrandom sequential acquisition mode, has various consequences for the formation of KIR repertoires that are clearly visible in the present study, such as single-KIR2DL1+ NK cells show much stronger adaptation toward cognate class I than single-KIR2DL3+ NK cells. Moreover, the KIR2DL1_2DL3 clonotype is significantly more frequent in C2 than in C1 donors (Figure. 1A). Together, these observations clearly indicate that it is generally crucial to perform the analysis of individual KIR clonotypes in the context of individual HLA class I ligands to provide the necessary resolution to detect the influence of HLA class I on the KIR repertoire.
In the clinical setting of allogeneic HSCT, the observed differences in expression of HLA-C–specific KIR between group A and group B donors might have important consequences for the composition of alloreactive NK cell repertoires. As shown in this study, a gradual depletion of KIR2DL1+ NK cells is seen in donors having 1 and an even greater depletion in donors with 2 copies of KIR2DL2. Notably, only NK cells expressing the C2-specific KIR2DL1+ have the potential to recognize a C2/C1 mismatch in graft-versus-host direction, whereas KIR2DL2+ NK cells would not discriminate between the 2 ligands. Thus, even if 2 donors share the same HLA class I type they still might provide qualitatively and quantitatively divergent alloreactive NK cell repertoires just on the basis of the KIR2DL2/3 polymorphism. It is likely that specificity and activation threshold of NK cells is further modified by coexpression of stimulatory KIR. Interestingly, Pende et al33 have recently shown that after haploidentical HSCT, donor-derived KIR2DL2/3+ NK cells poorly recognize C2 leukemic blasts unless KIR2DS1 was coexpressed. This as well as observations showing that donors with group B haplotypes are associated with less bacterial infections and improved outcome in HSCT suggest that inhibitory KIR repertoires indeed are modified by coexpression of stimulatory KIR.42,43
In summary, the present study shows how HLA-C–specific NK cell repertoires are structurally and functionally affected by both HLA class I and KIR genotype. We suggest that the assessment of KIR and HLA class I type in conjunction with high-dimensional flow cytometry provides a promising approach to predict NK cell function in the clinical setting such as haploidentical HSCT or novel adoptive immunotherapy protocols.21,44
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all donors that volunteered for blood donation. We also thank Dr A. Moretta for the CD158k-specific antibody (Q66, KIR3DL2) and Dr S. Khakoo for the HLA-deficient 721.221 and the HLA class I–transfected sublines.
This study was supported by the Deutsche Forschungsgemeinschaft grants UH 91/2-1 and UH91/5-1 (M.U.).
Authorship
Contribution: K.S. designed the project, performed the experiments, and wrote the manuscript; M.S. performed the NK cell differentiation assays; J.E. performed the HLA-C typing; J.C.F. designed the project and performed statistical analysis; and M.U. designed the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Markus Uhrberg, PhD, University Clinic of Düsseldorf, Institute for Transplantation Diagnostics, and Cell Therapeutics, Bldg 14.80, Moorenstrasse 5, D-40225 Düsseldorf, Germany; e-mail: uhrberg@itz.uni-duesseldorf.de.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal