Abstract
The cancer testis antigen (CTA) preferentially expressed antigen of melanoma (PRAME) is overexpressed by many hematologic malignancies, but is absent on normal tissues, including hematopoietic progenitor cells, and may therefore be an appropriate candidate for T cell–mediated immunotherapy. Because it is likely that an effective antitumor response will require high-avidity, PRAME-specific cytotoxic T lymphocytes (CTLs), we attempted to generate such CTLs using professional and artificial antigen-presenting cells loaded with a peptide library spanning the entire PRAME protein and consisting of 125 synthetic pentadecapeptides overlapping by 11 amino acids. We successfully generated polyclonal, PRAME-specific CTL lines and elicited high-avidity CTLs, with a high proportion of cells recognizing a previously uninvestigated HLA-A*02–restricted epitope, P435-9mer (NLTHVLYPV). These PRAME-CTLs could be generated both from normal donors and from subjects with PRAME+ hematologic malignancies. The cytotoxic activity of our PRAME-specific CTLs was directed not only against leukemic blasts, but also against leukemic progenitor cells as assessed by colony-forming–inhibition assays, which have been implicated in leukemia relapse. These PRAME-directed CTLs did not affect normal hematopoietic progenitors, indicating that this approach may be of value for immunotherapy of PRAME+ hematologic malignancies.
Introduction
Cytotoxic T lymphocytes (CTLs) directed to tumor-associated antigens (TAAs) have the potential to eradicate malignant diseases.1-4 These CTLs may be generated in vivo by peptide-based vaccination5-7 or ex vivo for subsequent adoptive transfer.2,3 Irrespective of the methodology used for generation, the therapeutic effectiveness of CTLs relies on both the nature of the antigen targeted and the potency and avidity of the specific CTLs elicited. Ideally, the target antigen should be uniquely or highly expressed by tumor cells compared with normal tissues to minimize the occurrence of autoimmunity and to be directly involved in maintaining the tumor phenotype to limit the emergence of tumor escape mutants.
The cancer testis antigen (CTA) preferentially expressed antigen of melanoma (PRAME)8 is a potential target antigen for use in the treatment of tumors. First, PRAME is overexpressed by many hematologic malignancies, such as chronic myelogenous leukemia (CML), acute myeloid leukemia (AML),9-12 and Hodgkin lymphoma (HL),13 as well as by solid tumors,8 but its expression is low or absent in normal tissues, including hematopoietic progenitor cells.10 Secondly, PRAME may significantly contribute to maintaining the tumor phenotype, because its expression can strongly inhibit cell differentiation induced by the retinoic acid receptor-α ligand all-trans retinoic acid,14 a crucial pathway for the proliferation and differentiation of both normal and malignant hematopoietic cells.15 Indeed, it has recently been demonstrated that PRAME overexpression contributes to leukemogenesis by inhibiting myeloid differentiation through blockage of the retinoic acid receptor-α–signaling pathway.14,16
We17 and others18 have generated CTLs targeting PRAME-derived peptides from healthy donors and leukemic patients using antigen-presenting cells (APCs) loaded with specific peptides17 selected by in vitro digestion of long peptides8 or by mass spectrometry of acid elutes obtained from tumor cells.19 Unfortunately, these approaches have produced PRAME epitopes that have preferentially expanded low-avidity CTLs, whose modest functional activity would likely be suboptimal for clinical benefit. Therefore, to exploit PRAME as a potential target antigen in patients with hematologic malignancies and other solid tumors and to induce effective antitumor activity, we sought a means of generating high-avidity, PRAME-specific CTLs.
We now describe natural and artificial APCs loaded directly with a peptide library consisting of 125 synthetic pentadecapeptides, overlapping by 11 amino acids, which span the entire PRAME protein and generate polyclonal, PRAME-specific CTL lines with high affinity for tumor targets. We also describe an immunodominant peptide within PRAME and demonstrate selective killing of putative leukemia-progenitor cells with sparing of normal hemopoietic precursor cells. Because we could generate these PRAME-specific CTLs from normal donors and from subjects with PRAME+ disease, our approach may be of value for immunotherapy of PRAME+ malignancies.
Methods
Cell lines and samples from healthy donors and leukemic patients
The following tumor cell lines were used: KT1 (CML) kindly provided by Dr Fujita (First Department of Internal Medicine, School of Medicine, Ehime University, Japan); BV173 (CML) and L428 (HL) from the German Collection of Cell Cultures (DSMZ, Braunschweig, Germany); and U266B1 and ARH77 (multiple myeloma), K562 (erythroleukemia), and MRC-5 (normal human fetal lung fibroblasts) from the ATCC. Cells were maintained in culture with RPMI 1640 medium (HyClone) containing 10% fetal bovine serum (HyClone), 2mM l-glutamine (GIBCO-BRL), 25 IU/mL of penicillin, and 25 mg/mL of streptomycin (BioWhittaker) in a humidified atmosphere containing 5% CO2 at 37°C. Peripheral blood and bone marrow samples were collected according to the local institutional review board–approved protocol (University of Naples Federico II, Naples, Italy, and Baylor College of Medicine, Houston, TX).
Generation and expansion of PRAME-CTLs
CTL lines were generated from peripheral blood mononuclear cells (PBMCs) as described previously.17 Briefly, we selected CD8+ cells using “magnetic antibodies,” and primed them with autologous APCs (dendritic cells or CD40-activated B lymphocytes generated from autologous PBMCs at a ratio of 1:20 APCs:CD8+ cells that we loaded with HLA-A*02–restricted peptides for 2 hours and then washed twice). The cells were cultured in complete medium (45% RPMI 1640, 45% Click medium [Irvine Scientific] that we supplemented with 5% human AB serum and 2 mmoL of L-glutamine) and IL-7 (10 ng/mL), IL-12 (1 ng/mL), and IL-15 (2 ng/mL; all from R&D Systems). We loaded APCs with: (1) a pool of 4 previously identified PRAME peptides (referred as P4, a peptide pool composed of P100 VLDGLDVLL, P142 SLYSFPEPEA, P300 ALYVDSLFFL, and P425 SLLQHLIGL at 10μM each8,17 ); (2) a newly identified PRAME-derived nonamer (referred to as P435, NLTHVLYPV at 5 μM); (3) a pool of 5 PRAME-peptides (referred as P5, consisting of a P4 pool supplemented with P435 peptide at 10μM each); or (4) a peptide library, PRAME-PepMix (0.6 nmol of each peptide; JPT Technologies) composed of 125 pentadecapeptides spanning the entire PRAME protein and also containing the 4 nonadecamers (P100, P142, P300, and P425) previously identified. To exclude the possibility of antigenic competition between peptides mixed together in the P4 pool, we included control experiments using single peptides. Other controls used APCs loaded with the HLA-A*02–restricted, pp65-derived peptide NLVPMVATV or the HLA-A*02–restricted, MART-1-derived peptide ELAGIGILTV and an irrelevant PepMix. All peptides were obtained from Genemed Synthesis. After priming with APCs, T cells were collected and stimulated weekly with artificial APCs (aAPCs) derived from the K562 cell line17 (PBMC:K562/aAPC ratio, 4:1) loaded with the respective peptides or PepMix. IL-7, IL-12, and IL-15 cytokines were also included in this second stimulation. Subsequently, cells were expanded in complete medium with fetal bovine serum (HyClone) using IL-2 alone (50 IU/mL).17 In selected experiments, these CTL lines were generated from CD8+CD45RO+ or CD8+CD45RA+ T cells prepared by negative immunomagnetic sorting (Miltenyi Biotec).17
Immunophenotyping
We stained T cells with phycoerythrin (PE)–conjugated, fluorescein isothiocyanate (FITC)–conjugated, allophycocyanin and peridinin chlorophyll protein (PerCP)–conjugated CD3, CD4, CD8, CD56, CD45RA, CD45RO, CCR7, CD28, CD27, CD62L, and CD57 monoclonal antibodies (all Becton Dickinson). We also stained cells using the KLRG antibody (Santa Cruz Biotechnology) and PE-conjugated goat anti–rabbit antibody (Jackson ImmunoResearch). Finally the T-cell receptor-Vβ (TCR-Vβ) repertoire was analyzed by fluorescence-activated cell sorting (FACS; IOTest βMark kit; Immunotech). Control samples labeled with an appropriate isotype-matched antibody were included in each experiment. We analyzed these cells using a FACScan (Becton Dickinson) equipped with the filter set for 4 fluorescence signals. T-cell lines were also analyzed for binding of specific tetramers, prepared by the Baylor College of Medicine core facility, as described previously.20 For each sample, a minimum of 100 000 cells were analyzed using a FACSCalibur with CellQuest software (BD Biosciences).
ELISpot assay
We used an IFNγ ELISpot assay, as described previously.21 T cells were plated in triplicate, serially diluted from 1 × 105 to 1 × 104 cells/well, and then peptides (5μM) were added. In all experiments, T cells were also incubated with an irrelevant peptide to show the specificity of IFNγ release. Alternatively, either PRAME-PepMix or an irrelevant PepMix (pp65; 0.06 nmol) was added instead of the single peptide. As a positive control, T cells were stimulated with 25 ng/mL of phorbol myristate acetate and 1 μg/mL of ionomycin (Sigma-Aldrich). The IFNγ+ spot-forming cells (SFCs) were enumerated (ZellNet).
Chromium-release assay
The cytotoxic specificity of T cells was evaluated using a standard 4-hour 51Cr-release assay, as described previously.20 We incubated target cells in medium alone or in 1% Triton X-100 (Sigma-Aldrich) to determine spontaneous and maximum 51Cr release, respectively. The mean percentage of specific lysis of triplicate wells was calculated as follows: [(test counts − spontaneous counts)/(maximum counts − spontaneous counts)] × 100%.
Q-RT-PCR
Colony-forming inhibition assay of leukemic and normal hematopoietic progenitors
We purified mononuclear cells (MNCs) from the bone morrow of 3 healthy HLA-A*02+ donors and from 5 samples (3 bone marrow, 2 peripheral blood) of HLA-A*02+ patients with CML and from bone marrow samples from 2 HLA-A*02− subjects with CML. MNCs (0.5-2 × 104 cells) were then coincubated with PRAME-CTLs (P435-specific, PRAME-PepMix–specific, or P4-specific: P100, 142, 300, and 425) or CTLs specific for an irrelevant peptide (ELA) at an effector:target (E:T) ratio of 10:1 for 6 hours in 0.5 mL of complete medium at 37°C. After coincubation, cells were plated in triplicate in methylcellulose medium supplemented with recombinant cytokines (MethoCult; StemCell Technologies), and incubated at 37°C. Granulocyte-macrophage colony-forming units and erythrocyte colony-forming units (CFU) were scored using a high-quality inverted microscope after 2 weeks of culture. In selected experiments, colonies were collected to assess PRAME and p210 expression by Q-RT-PCR.23
HLA peptide–binding assay
The binding affinity of peptides for the HLA-A*02 molecule was assessed by an HLA stabilization assay, as described previously.24 Briefly, the HLA-A*0201–positive cell line CEM-T2 was plated in 96-well plates at 105 cells per well and incubated overnight with the candidate peptides at concentrations between 0 and 100mM in serum-free RPMI 1640 medium. The CEM-T2 cells were washed twice with 1× phosphate-buffered saline (PBS), and then incubated with anti–HLA-A*02-FITC monoclonal antibody (Becton Dickinson) at 4°C for 20 minutes. The cells were washed and fixed with 0.5% paraformaldehyde before FACS analysis. The fluorescence index (FI) was calculated as FI = (MFI with the given peptide − MFI without peptide)/MFI without peptide). The FI at 6mM was determined as high if > 1.5, intermediate if 1-1.5, and low if < 1.24 Hepatitis B virus core antigen (FLPSDFPSV residues 18-27; referred as the FL peptide) was used as the positive standard for high HLA-A*0201–binding affinity.24
Immunofluorescence
PRAME expression was evaluated at the subcellular level with an immunofluorescence assay, as described previously25 with some modifications. Briefly, 5 × 104 MNCs derived from 3 bone marrow and 2 peripheral blood samples from patients with CML were suspended in PBS and centrifuged on a glass slide using a Shandon CytoSpin 2 (ThermoFisher Scientific). In addition, CD34+ cells obtained after immunomagnetic selection (Miltenyi Biotec) from bone marrow aspirates of 2 healthy donors were centrifuged on a glass slide using a Shandon CytoSpin 2. Cells were then fixed for 10 minutes at room temperature using 1% paraformaldehyde, permeabilized for 2.5 minutes with 0.1% Triton-X (Sigma-Aldrich), and incubated for 30 minutes with an Image-iT FX signal enhancer (Invitrogen) at room temperature. Finally, slides were incubated for 2 hours at room temperature in 0.5% bovine serum albumin with 5 μg/mL of rabbit anti–PRAME antibody (Abcam) and stained using an Alexa Fluor 555–conjugated anti–rabbit immunoglobulin G antibody (Invitrogen) for 45 minutes at room temperature. After counterstaining with 4′,6-diamidino-2-phenylindole, dihydrochloride (Boehringer), fluorescence was visualized using an Olympus BX61 microscope (Zeiss). After the acquisition of fluorescence images using CytoVision 6.5 software, slides were extensively washed in PBS and stained by Diff-Quik solution (Siemens Healthcare Diagnostics) before reexamination using a Genetics GSL10 microscope station.
Statistical analysis
All data are presented as means ± SD. Student t test was used to determine the statistical significant differences between samples, and P < .05 was accepted as indicating a significant difference.
Results
Generation of functional PRAME-CTLs from HLA-A*02+ healthy donors using PRAME-PepMix
Using optimized culture conditions and 4 previously described HLA-A*02–restricted PRAME peptides (P100/VLD, P142/SLY, P300/ALY, and P425/SLL), we consistently generated PRAME-CTLs from 8 of 9 healthy donors and from 5 of 6 CML patients.17 However, these T-cell lines were generally characterized by low TCR affinity based on titration of the specific peptides and tetramer staining. To evaluate whether this observation reflected the elimination of PRAME-CTLs with higher TCR affinity (and associated superior antitumor activity) from the circulation, with only low-avidity T cells remaining in the peripheral blood, we loaded APCs with a library of 125 pentadecapeptides spanning the entire PRAME protein (PRAME-PepMix).
We determined whether the immune responses obtained using our approach would merely recapitulate those obtained with peptide-loaded APCs or if new T-cell specificities with enhanced TCR affinity would be generated. After priming and expansion for 2 weeks using PRAME PepMix–loaded APCs and aAPCs, T-cell lines were characterized for their PRAME specificity using the IFNγ ELISpot assay. As shown in Figure 1A, 21 of 23 (91%) of the resulting T-cell lines specifically responded to PRAME-PepMix (IFNγ 416 ± 86 SFCs/105 cells), whereas T-cell lines generated using APCs loaded with an irrelevant PepMix did not (IFNγ 8 ± 2 SFCs/105 cells; P < .001). Surprisingly, CTLs generated using APCs loaded with PRAME-PepMix did not significantly respond to the P4 peptide pool containing the 4 previously identified immunodominant peptides8,17 (IFNγ 6 ± 2 SFCs/105 cells). To evaluate if the response to the P4 peptide pool could be restored using different concentrations of the PepMix, we performed titration experiments. As shown in Figure 1B, similar frequencies of IFNγ spots were obtained when concentrations of 1 or 0.1 μg of PRAME-PepMix were used, whereas no IFNγ spots could be detected when the concentration was reduced to 0.01 μg. No response to the P4 peptide pool was observed (data not shown), suggesting that priming with PRAME-PepMix may determine the generation of CTLs skewed to different PRAME epitopes.
Generation of PRAME-specific CTLs using PepMix. We used a peptide library of 125 pentadecapeptides spanning the entire PRAME protein to load APCs for the generation of PRAME-specific CTLs. (A) Specificity of the generated CTL lines using PRAME-PepMix as assessed by INFγ ELISpot assay against control PepMix, PRAME PepMix, and a pool of the 4 previously identified HLA-A*02–restricted peptides (P4).8,18 Medium was used to evaluate background production of IFNγ by nonstimulated CTL. Each symbol represents 1 of the 23 individual PRAME-CTLs; horizontal lines represent the mean group value. (B) IFNγ production of CTLs generated from 4 donors using the PRAME-PepMix at a concentration of 1, 0.1, or 0.01 μg (corresponding to 6, 0.6, and 0.06 nmol each of the 125 15-mer composing PRAME-PepMix, respectively) against medium, irrelevant PepMix, or PRAME-PepMix. (C) The fold expansion of PRAME-PepMix CTLs primed with autologous APCs and stimulated with aAPCs for 3 weeks. Each symbol represents one of the 15 individual PRAME-CTLs; horizontal lines represent the mean group value. (D) PRAME-CTLs expanded from healthy donors were evaluated for their cytotoxic activity using a standard 4-hour 51Cr-release assay against autologous PHA blasts loaded with irrelevant (gray bar) or PRAME-PepMix (black bars). Data represent the means ± SD of PRAME-CTLs from 9 healthy donors. Shown is the killing at a 20:1 E:T ratio by PRAME-CTLs, which was significantly higher against PRAME PepMix–loaded targets compared with control PepMix targets. In addition, killing of PRAME PepMix–loaded autologous PHA blasts by CTLs was significantly inhibited by preincubation of the targets with an anti–HLA class I antibody, but not by an isotype control, indicating HLA-restricted killing.
Generation of PRAME-specific CTLs using PepMix. We used a peptide library of 125 pentadecapeptides spanning the entire PRAME protein to load APCs for the generation of PRAME-specific CTLs. (A) Specificity of the generated CTL lines using PRAME-PepMix as assessed by INFγ ELISpot assay against control PepMix, PRAME PepMix, and a pool of the 4 previously identified HLA-A*02–restricted peptides (P4).8,18 Medium was used to evaluate background production of IFNγ by nonstimulated CTL. Each symbol represents 1 of the 23 individual PRAME-CTLs; horizontal lines represent the mean group value. (B) IFNγ production of CTLs generated from 4 donors using the PRAME-PepMix at a concentration of 1, 0.1, or 0.01 μg (corresponding to 6, 0.6, and 0.06 nmol each of the 125 15-mer composing PRAME-PepMix, respectively) against medium, irrelevant PepMix, or PRAME-PepMix. (C) The fold expansion of PRAME-PepMix CTLs primed with autologous APCs and stimulated with aAPCs for 3 weeks. Each symbol represents one of the 15 individual PRAME-CTLs; horizontal lines represent the mean group value. (D) PRAME-CTLs expanded from healthy donors were evaluated for their cytotoxic activity using a standard 4-hour 51Cr-release assay against autologous PHA blasts loaded with irrelevant (gray bar) or PRAME-PepMix (black bars). Data represent the means ± SD of PRAME-CTLs from 9 healthy donors. Shown is the killing at a 20:1 E:T ratio by PRAME-CTLs, which was significantly higher against PRAME PepMix–loaded targets compared with control PepMix targets. In addition, killing of PRAME PepMix–loaded autologous PHA blasts by CTLs was significantly inhibited by preincubation of the targets with an anti–HLA class I antibody, but not by an isotype control, indicating HLA-restricted killing.
As shown in Figure 1C, T-cell lines expanded in culture by repeated stimulations with PRAME-PepMix–loaded APCs and aAPCs (median expansion, 23-fold; range, 5- to 124-fold after 4 stimulations). T-cell lines also had specific cytotoxic activity against PRAME-PepMix–loaded autologous phytohemagglutinin (PHA) blasts (specific lysis of 60% ± 5% at a 20:1 E:T ratio) compared with PHA blasts loaded with an irrelevant PepMix (18% ± 3%; P < .01), as assessed by a standard 4-hour 51Cr-release assay. Cytotoxic activity was major histocompatibility complex (MHC) class I restricted because it was significantly inhibited by preincubation of target cells with anti–HLA class I antibody (30% ± 5% at a 20:1 E:T ratio), but not by preincubation with the appropriate isotype control antibody (50% ± 6%; P < .01; Figure 1D).
PRAME-CTLs generated using the PRAME-PepMix are polyclonal and preferentially recognize distinct HLA-A*02–restricted PRAME epitopes
Because the PRAME-CTLs we generated using APCs and aAPCs loaded with PRAME-PepMix failed to recognize HLA-A*02–restricted PRAME peptides previously identified as immunodominant,8,17 we further characterized their epitope specificity against an array consisting of 13 subpools of overlapping peptides spanning the entire PRAME protein (Table 1). PRAME-CTLs generated from 14 HLA-A*02+ healthy donors were incubated with each subpool, and IFNγ release was measured using ELISpot assays. Half of the T-cell lines (57%) were polyclonal, because they produced IFNγ+ SFCs in response to at least 2 subpools (Table 2 and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The polyclonality of these PRAME PepMix-CTL lines was also confirmed by TCR-Vβ analysis (supplemental Table 1).
The majority (77% ± 21%) of the ex vivo–expanded CD8+ T cells had an effector phenotype lacking CD62L, CD45RA, and CCR7, and some lines also had a small proportion (11% ± 13%) of CD45RO+ and CD62L+ effector-memory cells (supplemental Figure 2). A variable proportion of cells expressed CD27 (28% ± 26%) and/or CD28 (15% ± 21%). Our T cells lacked KLRG-1 (2.6% ± 1.2%), which identifies senescent effector cells with diminished replicative capacity. No natural killer cells (CD3−CD56+; < 0.1%) and few natural killer T cells (CD3+CD57+; 3% ± 2.2%) were observed.
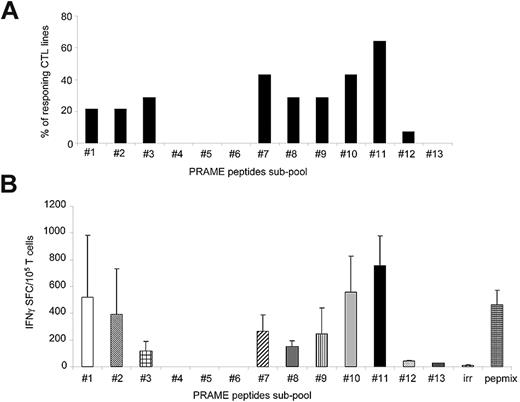
Although the majority of CTLs responded to subpools 11 (57%; 755 ± 224 IFNγ+ SFCs/105 cells) and 7 (43%; 265 ± 23 IFNγ+ SFCs/105 cells for subpool 7; Figure 2A), the most robust IFNγ response was against subpool 11, which spans the C-terminal region of PRAME protein between amino acids 401 and 451 (Figure 2B). None of the CTL lines responded to subpool 4, which contains the epitope P142/SLY.8 Although 3 of 14 CTL lines (21%) responded to subpools 3 and 8, which contain the epitopes P100/VLD and P300/ALY,8 respectively, the production of IFNγ+ spots in response to these subpools was significantly lower (116 ± 71 and 150 ± 44 IFNγ+ SFCs/105 cells, respectively) compared with subpool 11 (P = .03; Figure 2B). Because subpool 11 contains the immunodominant epitope P425/SLL,8 which failed to be recognized when it was tested as part of the P4 pool (Figure 1A), we further mapped this region by generating ten 15-mer overlapping peptides (amino acids 401-451) and found that the reactivity was predominantly directed against 2 peptides, P429 and P433 (651 ± 479 and 429 ± 319 IFNγ+ SFCs/105 cells, respectively; Figure 3A). Two computational algorithms (SYFPEITHI and BIMAS) were used to predict possible HLA-A*02–binding peptide motifs in this region. Five minimal putative peptides with the highest predicted binding scores (Table 3), referred as P429-9mer, P429-10mer, P432-9mer, P432-10mer, and P435-9mer (Figure 3B), were then tested in vitro for their ability to elicit IFNγ production by PRAME-CTL lines using an ELISpot assay. Although the P429-9mer peptide (HLIGLSNLT) led to some IFNγ production (42 ± 8 SFCs/105 cells), the most significant response was observed for P435-9mer (NLTHVLYPV; amino acids 435-443; 677 ± 22 SFCs/105 cells; Figure 3C). We evaluated the binding affinity of the new epitope P435/NLT for the HLA-A*02 molecule using a T2-binding assay, and found that this epitope greatly enhanced the stabilization of the HLA-A*02 molecule (FI > 1.5) on T2 cells when used at 6mM compared with the previously described P300/ALY epitope8 (FI > 1.5) when used at a 12mM peptide concentration (Figure 3D), indicating that HLA molecules recognizing P435/NLT have higher affinity compared with the P300/ALY epitope for which we previously detected the most robust CTL generation.17
PRAME-CTLs generated using PRAME-PepMix are polyclonal. We evaluated the polyclonality of the expanded lines by assessing their reactivity against an array consisting of 13 subpools of overlapping 15-mers spanning the entire PRAME protein. (A) Percentage of CTL lines (generated from 14 donors) responding to each of the 13 subpools. The majority of the CTL lines reacted against subpools 7 and 11. (B) Frequency of these CTLs responding to the PRAME-PepMix and each subpool as assessed by IFNγ ELISpot assay. Shown are the means ± SD of the responding lines. The highest reactivity of PRAME-PepMix CTLs was observed for subpool 11.
PRAME-CTLs generated using PRAME-PepMix are polyclonal. We evaluated the polyclonality of the expanded lines by assessing their reactivity against an array consisting of 13 subpools of overlapping 15-mers spanning the entire PRAME protein. (A) Percentage of CTL lines (generated from 14 donors) responding to each of the 13 subpools. The majority of the CTL lines reacted against subpools 7 and 11. (B) Frequency of these CTLs responding to the PRAME-PepMix and each subpool as assessed by IFNγ ELISpot assay. Shown are the means ± SD of the responding lines. The highest reactivity of PRAME-PepMix CTLs was observed for subpool 11.
Identification of an immunodominant, HLA-A*02–restricted PRAME epitope. When we minimized subpool 11 in ten 15-mer overlapping peptides, IFNγ production by PRAME-CTL was directed against the P435/NLT peptide. (A) Reactivity of PRAME-CTLs was predominantly directed against two 15-mers, P429 and P433. (B) Five predicted HLA-A*02–binding peptide motifs in this region using 2 computational algorithms (SYFPEITHI and BIMAS; Table 3). (C) IFNγ production by 8 PRAME-CTL lines against the 5 minimized epitopes. Although some IFNγ production was observed against the P429-9mer (HLI), the most significant response was observed for the P435-9mer (NLT). (D) Stabilization assay of the new epitope P435/NLT for the HLA-A*02 molecule using a T2-binding assay. An HLA-A11–restricted peptide was used as a negative control. FL, a hepatitis B virus core antigen with high HLA-A*0201–binding affinity, was used as a positive control. The affinity of P435/NLT was superior to that of the previously described P300/ALY.
Identification of an immunodominant, HLA-A*02–restricted PRAME epitope. When we minimized subpool 11 in ten 15-mer overlapping peptides, IFNγ production by PRAME-CTL was directed against the P435/NLT peptide. (A) Reactivity of PRAME-CTLs was predominantly directed against two 15-mers, P429 and P433. (B) Five predicted HLA-A*02–binding peptide motifs in this region using 2 computational algorithms (SYFPEITHI and BIMAS; Table 3). (C) IFNγ production by 8 PRAME-CTL lines against the 5 minimized epitopes. Although some IFNγ production was observed against the P429-9mer (HLI), the most significant response was observed for the P435-9mer (NLT). (D) Stabilization assay of the new epitope P435/NLT for the HLA-A*02 molecule using a T2-binding assay. An HLA-A11–restricted peptide was used as a negative control. FL, a hepatitis B virus core antigen with high HLA-A*0201–binding affinity, was used as a positive control. The affinity of P435/NLT was superior to that of the previously described P300/ALY.
Characterization of P435/NLT–specific CTLs generated from HLA-A*02+ healthy donors
Having identified this highly immunogenic PRAME epitope (P435/NLT) within the PepMix, we then investigated whether it could be used directly to generate PRAME-CTLs in vitro.17 We stimulated CD8+ T cells obtained from 10 healthy HLA-A*02+ donors with APCs and aAPCs loaded with either P435/NLT or the P4 pool. As shown in Figure 4A, APCs loaded with the P435/NLT peptide successfully expanded specific CTLs in 9 of 10 subjects. In particular, this peptide induced the generation of specific CTLs in samples obtained from 3 subjects in whom the P4 pool failed to elicit specific CTL responses. Furthermore, even in the P4-responder patients, the frequency of IFNγ+ spots in response to P435/NLT was higher (521 ± 100 SFCs/105 cells) than for P4 (248 ± 101 SFCs/105 cells). Supplemental Figure 3 shows data confirming our previous observation17 that there is a lower frequency of PRAME-specific CTLs to the P4 pool, and that this observation cannot simply be attributed to antigenic competition between peptides. The degree of expansion of P435/NLT–specific CTL lines after 3 rounds of stimulation with loaded aAPCs was similar to that observed for CTL lines obtained using stimulation with the P4 pool or PRAME-PepMix (range, 4- to 142-fold; Figure 4B). P435/NLT–specific CTLs also specifically bound a specific tetramer in 9 of 10 CTL lines tested (range, 0.4%-42%; Figure 4C).
Functional P435/NLT–specific CTLs can be expanded from HLA-A*02+ healthy donors. P435/NLT could be used to generate PRAME-CTLs. (A) IFNγ response of P435/NLT–specific or P4-specific CTLs generated from 10 healthy HLA-A*02+ donors. P435/NLT–specific CTLs could be generated from 9 of 10 samples tested. (B) Expansion of P435/NLT–specific or P4-specific CTL lines after 3 rounds of stimulation. (C) Staining with the P435/NLT–specific tetramer of CTL lines generated from 2 representative healthy donors. (D) IFNγ+ SFCs in response to P435/NLT-peptide or NLV-pp65 (from the pp65 protein of CMV) peptide of P435/NLT–specific CTLs expanded from sorted naive (CD45RA+) and memory (CD45RO+) T-cell subsets. T cells producing IFNγ in response to the P435/NLT peptide were significantly higher in CTL lines that originated from the naive (CD45RA+) T-cell subset. In contrast, CTLs generated against the viral peptide NVL originated predominantly from the memory (CD45RO+) T-cell subset. Shown is 1 of 4 representative donors.
Functional P435/NLT–specific CTLs can be expanded from HLA-A*02+ healthy donors. P435/NLT could be used to generate PRAME-CTLs. (A) IFNγ response of P435/NLT–specific or P4-specific CTLs generated from 10 healthy HLA-A*02+ donors. P435/NLT–specific CTLs could be generated from 9 of 10 samples tested. (B) Expansion of P435/NLT–specific or P4-specific CTL lines after 3 rounds of stimulation. (C) Staining with the P435/NLT–specific tetramer of CTL lines generated from 2 representative healthy donors. (D) IFNγ+ SFCs in response to P435/NLT-peptide or NLV-pp65 (from the pp65 protein of CMV) peptide of P435/NLT–specific CTLs expanded from sorted naive (CD45RA+) and memory (CD45RO+) T-cell subsets. T cells producing IFNγ in response to the P435/NLT peptide were significantly higher in CTL lines that originated from the naive (CD45RA+) T-cell subset. In contrast, CTLs generated against the viral peptide NVL originated predominantly from the memory (CD45RO+) T-cell subset. Shown is 1 of 4 representative donors.
As further evidence that P435/NLT–specific cells were consistently selected and expanded during PRAME-CTL generation, we expanded PRAME-specific T cells from 3 healthy donors using a mixture of P100/VLD, P142/SLY, P300/ALY, P425/SLL, and P435/NLT peptides (designated as the P5 pool), and analyzed the reactivity of the CTLs using the ELISpot assay. We found that these T-cell lines were mainly reactive against the P435/NTL peptide (439 ± 215 SFCs/105 cells), further suggesting that this peptide is indeed more immunogenic (supplemental Figure 4).
We next determined whether the P435/NLT–specific T-cell lines were of higher affinity because the responding cells were derived from the memory rather than the naive T-cell compartment. Using the same culture conditions, we generated P435/NLT–specific CTLs from the CD45RO+ or CD45RA+ T-cell fractions. In 4 healthy subjects, we detected P435/NLT-peptide–responsive T cells only in CTL lines originating from the naive (CD45RA+) T-cell subset (Figure 4D). In contrast, virus-specific CTLs recognizing the NLVPMVATV peptide derived from the pp65 protein of cytomegalovirus were obtained exclusively from the CD45RO+ memory T-cell fraction. The immunophenotype of the P435-specific CTL line was similar to that of the PepMix-specific CTLs (data not shown).
We then evaluated the cytotoxic activity of P435/NLT–specific CTLs against autologous PHA blasts loaded with the P435/NLT peptide using a standard 4-hour 51Cr-release assay. As shown in Figure 5A, P435/NLT–specific CTLs specifically lysed P435/NLT–loaded PHA blasts (67% ± 11% at a 20:1 E:T ratio), but not PHA blasts loaded with an irrelevant peptide (P < .001). The cytotoxic effect was MHC class I restricted, because it was significantly inhibited by preincubation of target cells with an anti–HLA class I antibody (30% ± 8% at a 20:1 E:T ratio), but not by preincubation with the appropriate isotype control antibody (50% ± 11%; P = .004; Figure 5B). We next assessed the capacity of P435/NLT–specific CTLs to recognize target cells naturally expressing the PRAME protein by measuring the cytotoxic activity of P435/NLT–specific CTLs against the PRAME+ HLA-A*02+ tumor cell lines U266, ARH77, KT1, and BV173.17 As shown in Figure 5C, killing of U266, ARH77, KT1, and BV173 tumor cells by P435-specific CTL lines was superior (55% ± 17%, 43% ± 14%, 61% ± 7%, and 58% ± 14%, respectively) to that of L428 (12% ± 8%), which is PRAME+ but HLA-A*02−. We confirmed that killing was mediated through HLA class I by blocking cytotoxicity after preincubation of target cells with an anti–HLA class I antibody (range of killing at the 20:1 E:T ratio, 28%-44%), but not with the appropriate isotype control antibody (range of killing at the 20:1 E:T ratio, 40%-64%). Similarly, IFNγ production against the PRAME+ HLA-A*02+ tumor cell lines U266, HDLM-2, and ARH77 was increased compared with control CTLs or the PRAME− HLA-A*02+ cell line (Figure 5D)
P435/NLT–specific CTLs are cytotoxic to PRAME+ tumor cell lines. (A) Cytotoxic activity of P435/NLT–specific CTLs toward autologous PHA blasts loaded with NLV-irrelevant peptide or loaded with the NLT peptide. Data represent the means ± SD of PRAME-CTLs from 7 healthy donors. (B) Data at a 20:1 E:T ratio of P435/NLT–specific CTLs against autologous PHA blasts loaded with NLV-irrelevant peptide or loaded with the NLT peptide preincubated with isotype control or an anti–HLA class I antibody to confirm MHC-restricted killing. (C) Cytotoxic activity of P435/NLT–specific CTLs toward PRAME+ HLA-A*02+ tumor cell lines evaluated using a standard 51Cr-release assay. As negative controls, we used the L428 tumor cell line (PRAME+ but HLA-A*02−). Killing of PRAME+HLA-A*02+ cell lines was significantly higher than that of the PRAME+HLA-A*02− L428, and was inhibited by preincubation with an anti–HLA class I antibody but not by an isotype control. (D) IFNγ ELISpot release of P435/NLT–specific CTLs or control/irrelevant CTLs against the indicated PRAME+ HLA-A*02+ cell lines or the control PRAME− HLA-A*02+ cell line.
P435/NLT–specific CTLs are cytotoxic to PRAME+ tumor cell lines. (A) Cytotoxic activity of P435/NLT–specific CTLs toward autologous PHA blasts loaded with NLV-irrelevant peptide or loaded with the NLT peptide. Data represent the means ± SD of PRAME-CTLs from 7 healthy donors. (B) Data at a 20:1 E:T ratio of P435/NLT–specific CTLs against autologous PHA blasts loaded with NLV-irrelevant peptide or loaded with the NLT peptide preincubated with isotype control or an anti–HLA class I antibody to confirm MHC-restricted killing. (C) Cytotoxic activity of P435/NLT–specific CTLs toward PRAME+ HLA-A*02+ tumor cell lines evaluated using a standard 51Cr-release assay. As negative controls, we used the L428 tumor cell line (PRAME+ but HLA-A*02−). Killing of PRAME+HLA-A*02+ cell lines was significantly higher than that of the PRAME+HLA-A*02− L428, and was inhibited by preincubation with an anti–HLA class I antibody but not by an isotype control. (D) IFNγ ELISpot release of P435/NLT–specific CTLs or control/irrelevant CTLs against the indicated PRAME+ HLA-A*02+ cell lines or the control PRAME− HLA-A*02+ cell line.
P435/NLT–specific CTLs selectively inhibit the growth of leukemic precursor cells
To measure the cytotoxic activity of P435/NLT–specific CTLs against leukemic precursor cells, we used clonogenic assays of 3 bone marrow and 2 peripheral blood samples from 5 HLA-A*02+ patients with untreated CML. All 5 patients had PRAME+ disease (as assessed by Q-RT-PCR; data not shown) and a high expression of PRAME protein in leukemic cells derived from both the bone marrow (supplemental Figure 5A) and the peripheral blood (supplemental Figure 5B). In contrast, patient T and B lymphocytes lacked discernible PRAME protein expression (data not shown). Similarly, CD34+ cells obtained from the bone marrow of healthy donors lacked both PRAME transcripts and protein (supplemental Figure 6), confirming that PRAME expression was restricted to leukemic cells.
MNCs from these subjects were incubated with P435/NLT–specific CTLs, P4-specific CTLs, or control CTLs, and then placed in standard hematopoietic colony-forming cultures. As shown in Figure 6A, P435/NLT-CTLs had specific cytotoxic effects against PRAME+HLA-A*02+ leukemic precursors, which resulted in a consistent and significant reduction in CFUs when MNCs were coincubated with P435/NLT–specific CTLs (59 ± 11 CFUs/5 × 104 cells) compared with control CTLs (181 ± 48 CFUs/5 × 104 cells; P = .01). P4-specific CTLs showed a trend of inhibition (107 ± 45 CFUs/5 × 104 cells), although this was not statistically significant compared with control CTLs. Growth inhibition by P435/NLT–specific CTLs was highly specific for PRAME+ precursors, because these CTLs had no effect on the growth of bone marrow–derived MNCs obtained from 3 HLA-A*02+PRAME− normal donors or from 2 donors who had HLA-A*02−PRAME+ CML blasts. The CFUs that outgrew in the presence of the P435/NLT CTLs were profoundly depleted of PRAME expression and BCR/ABL p210 transcripts (as detected by Q-RT-PCR in single-cell colonies). Therefore, as shown in Figure 6B, P435/NLT–specific CTLs almost completely eliminated the growth of PRAME+ CFUs and significantly reduced the growth of BCR/ABL+ CFUs. We also collected and analyzed CFUs from 2 healthy donors, and neither PRAME+ nor BCR/ABL+ precursor cells were detected (data not shown), further indicating that PRAME is not expressed by normal progenitor cells.
P435/NLT–specific CTLs target primary tumor cells and tumor progenitors. We performed clonogenic assays using peripheral and bone marrow MNCs from patients with CML to test antitumor activity by PRAME-CTLs against tumor progenitors. (A) CFUs generated from MNCs of PRAME+HLA-A*02+ and PRAME+HLA-A*02− CML patients with active disease and HLA-A*02+CD34+ cells from healthy donors incubated with P435/NLT–specific CTLs, P4-specific CTLs, or control CTLs (targeting the MART1-derived ELA epitope or the pp65-derived NLV peptide). P435/NLT–specific CTLs had a specific cytotoxic activity against PRAME+HLA-A*02+ leukemic precursors, because the number of CFUs was significantly lower when MNCs were coincubated with P435/NLT–specific CTLs compared with control CTLs (P = .01). (B) Expression of PRAME and the oncogenic fusion BCR-ABL (p210) transcript using Q-RT-PCR in colonies collected from 2 independent experiments. P435/NLT–specific CTLs almost completely eliminated the growth of PRAME+ CFUs and significantly reduced the growth of BCR/ABL+ CFUs.
P435/NLT–specific CTLs target primary tumor cells and tumor progenitors. We performed clonogenic assays using peripheral and bone marrow MNCs from patients with CML to test antitumor activity by PRAME-CTLs against tumor progenitors. (A) CFUs generated from MNCs of PRAME+HLA-A*02+ and PRAME+HLA-A*02− CML patients with active disease and HLA-A*02+CD34+ cells from healthy donors incubated with P435/NLT–specific CTLs, P4-specific CTLs, or control CTLs (targeting the MART1-derived ELA epitope or the pp65-derived NLV peptide). P435/NLT–specific CTLs had a specific cytotoxic activity against PRAME+HLA-A*02+ leukemic precursors, because the number of CFUs was significantly lower when MNCs were coincubated with P435/NLT–specific CTLs compared with control CTLs (P = .01). (B) Expression of PRAME and the oncogenic fusion BCR-ABL (p210) transcript using Q-RT-PCR in colonies collected from 2 independent experiments. P435/NLT–specific CTLs almost completely eliminated the growth of PRAME+ CFUs and significantly reduced the growth of BCR/ABL+ CFUs.
Generation of PRAME-CTLs from patients with leukemia
We next obtained peripheral blood from 11 HLA-A*02+ patients affected by PRAME+ AML (3 patients), CML (5 patients), HL (2 patients), or follicular lymphoma (1 patient). PBMCs were primed with autologous dendritic cells loaded with PRAME-derived peptides (P435/NLT, PRAME-PepMix, or the P4 pool) in the presence of IL-7, IL-12, and IL-15, and then stimulated with peptide-loaded aAPCs and IL-2. As shown in Figure 7A, in all 11 patients, we were successful in expanding T cells that responded to the PRAME-PepMix. In 10 of 11 subjects, the CTLs responded to P435/NLT, and 4 of the 9 tested CTL lines responded to the P4 peptides, suggesting that the frequency of precursor specific for the P435 epitope is higher compared with the previously described epitopes. As in healthy donors, responses were more robust against PRAME-PepMix and P435/NLT (709 ± 371 and 306 ± 166 IFNγ+ SFCs, respectively) than against the P4 peptides (200 ± 86 IFNγ+ SFCs).
P435/NLT–specific CTLs can be generated from HLA-A*02+ patients. (A) Frequency of IFNγ+ CTLs specific for irrelevant (white bars), PRAME PepMix (black bars), P435 (gray bars) and P4 (striped bars) against specific peptides in 11 HLA-A*02+ patients with AML, CML, HL, or follicular lymphoma. NT indicates not tested. (B) PRAME-CTLs produced IFNγ+ SFCs in response to autologous CML blasts but not against HLA-A*02− CML blasts, and this effect was HLA restricted because IFNγ+ SFCs were reduced when samples were preincubated with an anti–HLA class I antibody, but not with an isotype control. (C) Staining with the P435/NLT–specific tetramer of CTL lines generated from 3 representative patients.
P435/NLT–specific CTLs can be generated from HLA-A*02+ patients. (A) Frequency of IFNγ+ CTLs specific for irrelevant (white bars), PRAME PepMix (black bars), P435 (gray bars) and P4 (striped bars) against specific peptides in 11 HLA-A*02+ patients with AML, CML, HL, or follicular lymphoma. NT indicates not tested. (B) PRAME-CTLs produced IFNγ+ SFCs in response to autologous CML blasts but not against HLA-A*02− CML blasts, and this effect was HLA restricted because IFNγ+ SFCs were reduced when samples were preincubated with an anti–HLA class I antibody, but not with an isotype control. (C) Staining with the P435/NLT–specific tetramer of CTL lines generated from 3 representative patients.
To confirm antitumor activity, we tested these patient-derived CTLs against autologous tumor blasts in patients with CML. As shown in Figure 7B, we found that P435/NLT–specific CTLs produced IFNγ in an ELISpot assay when incubated with autologous PRAME+ CML blasts (57 ± 10 SFCs/105 T cells). In contrast, CTLs generated from the same patient against control peptide did not produce IFNγ when they were incubated with P435/NLT peptide or with autologous CML blasts (< 5 SFCs/105 T cells; data not shown); once again, T-cell specificity was HLA restricted, because it was inhibited by preincubation with an anti–HLA class I antibody. As observed for healthy donors, in 4 of 6 P435/NLT–specific T-cell lines tested, after the third stimulation, we could detect P435/NLT-tetramer+ cells (range, 2%-88%; Figure 7C), whereas no P435/NLT-tetramer+ T cells were detected in PBMCs derived from patients before ex vivo expansion.
Discussion
Leukemias such as CML and AML and lymphomas can be highly susceptible to control by the immune system, and effector T cells recognizing minor histocompatibility antigens and other TAAs overexpressed in tumor cells play an important role in this process.26-31 Recently, the CTA PRAME, which is expressed by leukemic blasts9,11,32 and HLs,13 has been investigated as a potential target for immunotherapy of these malignancies.17,18,33 Although we and others have previously been able to generate ex vivo PRAME-specific CTLs from both healthy donors and leukemic patients,17 the low affinity of these cells for the target antigen has limited their suitability for clinical application.17 We now show that loading professional and artificial APCs with a PRAME peptide library can generate high-avidity CTLs, and have identified a new immunodominant PRAME epitope. These high-affinity CTLs can be generated from both healthy donors and patients with CML, AML, and lymphomas, and have cytotoxic activity against both leukemic blasts and leukemic precursor cells while sparing normal hematopoietic precursors.
To effectively eliminate tumor cells, CTLs must recognize specific epitopes from tumor antigens in the context of the appropriate HLA molecules.34 Therefore, for a tumor to be recognized by T cells, its TAAs must be degraded and processed by the tumor cells in vivo. Our understanding of the proteolytic process that leads to MHC peptide presentation remains incomplete, making it difficult to accurately predict which epitopes will be presented with the highest frequency by the tumor cells. Several strategies have been used to identify or predict antigen epitopes, including screening of cDNA libraries with clones derived from tumor-infiltrating lymphocytes and the use of specific algorithms (the so-called “reverse immunology” approach).35,36 Kessler et al have identified HLA-A*02–restricted, PRAME-derived peptides by incorporating an in vitro, proteosome-mediated digestion of 27-mer polypeptides encompassing high-affinity binding peptides in the epitope prediction procedure.8 Although we were able to generate PRAME-reactive CTLs using the 4 peptides described by Kessler et al, in our experience, the majority of T cells generated from both healthy donors and CML patients contain low-affinity TCRs,17 and in only one report was it possible to detect high-avidity T cells—and then only with specificity for one of these peptides and only after allogeneic hemopoietic stem-cell transplantation.37 Moreover, several high-affinity epitopes predicted by the algorithms were excluded from these earlier functional analyses based on the assumption that the correct C-termini would not be generated after the in vitro, proteasome-mediated digestion analysis.8 However, peptidases that modify the C-termini of peptide precursors have subsequently been identified.38 These more recent data suggest that the initial assumption about epitope suitability may have been incorrect, and this possibility is borne out by our observation that APCs loaded with a peptide library consisting of 125 synthetic pentadecapeptides, overlapping by 11 amino acids and spanning the entire PRAME protein, can indeed generate high-avidity CTLs that recognize the “incorrect” P435/NLT epitope. The high-avidity CTLs we made that recognized the P435/NLT epitope had antitumor activity against HLA-A*02+ PRAME+ tumor cell lines and primary leukemic blasts, and this activity was manifest in both cytotoxicity and IFNγ ELISpot assays, indicating that this peptide is efficiently processed and presented by tumor cells. Although the level of cytotoxicity was donor dependent, it was invariably present.
The level of expression of specific antigens in different tumor-cell subsets may influence their susceptibility to CTL recognition.39,40 For many leukemias, including CML, there is evidence for an origin from a phenotypically distinct progenitor-cell compartment. Because these malignant progenitor cells may contribute to leukemia recurrence,41 we evaluated whether they too were susceptible to recognition and elimination by high-avidity CTLs specific for the P435/NLT peptide. Using in vitro CFU assays as a surrogate assay for leukemic progenitors, we indeed found that high-avidity P435/NLT CTLs significantly reduced CFU formation from CML samples without affecting normal hematopoietic precursors.
Our ability to generate high-avidity CTLs recognizing the P435/NLT epitope even from subjects with PRAME+ malignancy is striking, because the isolation and expansion of high-affinity T cells specific for self-antigens is impaired by both the physiologic deletion of these cells during thymic selection and by the tumor-specific induced T-cell apoptosis previously described for other antigens such proteinase 342 and for other PRAME-associated epitopes.17 Because high-avidity P435/NLT-epitope–specific CTLs can be obtained in both healthy donors and leukemic patients, it seems unlikely that prior thymic deletion occurred, and it is possible that not all (normal and malignant) cells may be capable of processing and presenting this particular epitope.
Although we have used our approach only for subjects with an HLA-A*02 polymorphism, there is no reason why the methodology could not be applied to identify high-affinity epitopes for other common HLA phenotypes using the appropriate artificial APCs expressing the relevant HLA polymorphisms.43 Unlike approaches using ex vivo digestion with immune proteasomes, PepMix libraries are readily available and standardized and could be useful for generating CTLs in vivo by vaccination and ex vivo for adoptive transfer to subjects with PRAME+ hematologic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Marco Cosenza, Dr Ida Pisano, Dr Novella Pugliese, and Dr Raffaella Accetta for all of the advice and the data collection assistance in the immunofluorescence analysis. We also thank Linda Chapman of Golden Funds for the support.
This work was supported by National Institutes of Health grants P01CA094237, P50CA126752, and R21CA150109; by a Support of Competitive Research (SCOR) grant from the Leukemia & Lymphoma Society; and by grants from Regione Campania (DGRC 2326/07) and the Italian Ministry of Research (MIUR; PS 35-126/Ind). H.E.H. is supported by a Dan L. Duncan Chair and M.K.B. by a Fayez Sarofim Chair.
National Institutes of Health
Authorship
Contribution: C.Q., G.D., and B.S. designed the research, performed experiments, analyzed the data, and wrote the manuscript; S.T.H., B.D.A., V.H., and S.E. performed some experiments; M.M., L.L., J.S., and F.P. provided patient samples; A.M.L., H.E.H., and C.M.R. provided expertise in T-cell generation and critically reviewed the manuscript; M.K.B. reviewed the experimental design and the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara Savoldo, MD, Center for Cell and Gene Therapy, Baylor College of Medicine, 6621 Fannin St, MC 3-3320, Houston, TX 77030; e-mail: bsavoldo@bcm.tmc.edu.