Abstract
Mantle cell lymphoma (MCL) is an aggressive B-cell malignancy with a median survival of 3 years despite chemoimmunotherapy. Rituximab, a chimeric anti–CD20 monoclonal antibody (mAb), has shown only modest activity as single agent in MCL. The humanized mAb milatuzumab targets CD74, an integral membrane protein linked with promotion of B-cell growth and survival, and has shown preclinical activity against B-cell malignancies. Because rituximab and milatuzumab target distinct antigens and potentially signal through different pathways, we explored a preclinical combination strategy in MCL. Treatment of MCL cell lines and primary tumor cells with immobilized milatuzumab and rituximab resulted in rapid cell death, radical oxygen species generation, and loss of mitochondrial membrane potential. Cytoskeletal distrupting agents significantly reduced formation of CD20/CD74 aggregates, cell adhesion, and cell death, highlighting the importance of actin microfilaments in rituximab/milatuzumab–mediated cell death. Cell death was independent of caspase activation, Bcl-2 family proteins or modulation of autophagy. Maximal inhibition of p65 nuclear translocation was observed with combination treatment, indicating disruption of the NF-κB pathway. Significant in vivo therapeutic activity of combination rituximab and milatuzumab was demonstrated in a preclinical model of MCL. These data support clinical evaluation of combination milatuzumab and rituximab therapy in MCL.
Introduction
Mantle cell lymphoma (MCL) is a B-cell malignancy with a variable histology and clinical course, distinguished by the characteristic translocation t(11;14)(q13, q32) that results in overexpression of cyclin D1 and consequent dysregulation of cell-cycle control.1 In addition, MCL exhibits alterations in cell survival pathways, including constitutive activation of phosphatidylinositol 3-kinase (PI3K)/Akt signaling2 and nuclear factor-κB (NF-κB).3 Despite the hallmark genetic translocation in MCL, the clinical course of MCL is variable with some patients experiencing indolent disease,4 whereas others exhibit rapid progression.5 MCL patients have a median overall survival (OS) of approximately 3 years, and no consensus exists for standard first-line therapy.6-9 Although aggressive therapies including chemoimmunotherapy10,11 or stem cell transplantation12,13 have been shown to improve outcomes, no therapy offers the potential for cure. Given the absence of curative therapy and the limited number of options for patients with relapsed/refractory MCL, novel treatment approaches are essential.
Rituximab (Genentech), a chimeric anti–human CD20 monoclonal antibody (mAb), has been used in multiple strategies to treat patients with MCL.14 As a single agent, rituximab has been tested in patients with newly diagnosed and relapsed/refractory MCL with response rates (RR) of 27% to 38% and a median response duration of 6 to 12 months.15,16 Interestingly, the RR obtained in untreated patients was not higher than in relapsed/refractory patients, relegating this antibody to the group of modestly active agents in MCL. However, in combination with anthracycline-based regimens, RR and time to progression, but not OS, of treatment-naive MCL patients was significantly increased compared with patients treated with chemotherapy alone.17
Milatuzumab (hLL1, IMMU-115; Immunomedics) is a fully humanized immunoglobulin-G1κ mAb specific for CD74, a type II transmembrane glycoprotein associated with major histocompatibility complex (MHC) class II α- and β-chain. CD74 was originally thought to function as an MHC class II chaperone; however, was recently found to also play an important role as an accessory signaling molecule and survival receptor in the maturation and proliferation of B cells by activating the PI3K/Akt and the NF-κB pathways.18-20 CD74, which is quickly internalized on binding with its physiologic ligand, the macrophage migration-inhibitory factor21 is expressed on the majority of B-cell malignancies, making it an attractive therapeutic target. CD74 is also expressed on normal B cells, monocytes, macrophages and dendritic cells (DCs).22 However, it has been recently shown that milatuzumab has minimal effects on the viability of normal B cells and DCs.23 Furthermore, it has been shown that milatuzumab has no effect on DC maturation and DC-mediated T-cell function.23 Milatuzumab demonstrated antiproliferative activity in transformed B-cell lines, improved survival in preclinical models,18,22 and is presently being evaluated for the treatment of several hematologic malignancies under clinical trials registered at www.clinicaltrials.gov as NCT00421525, NCT00868478, NCT00603668, and NCT00504972. Unlike rituximab, milatuzumab does not cause cell death via antibody-dependent cell-mediated cytotoxicity or complement-dependent cytotoxicity.22,24
Rituximab and milatuzumab target distinct antigens lacking known association and, as single agents, have demonstrated substantial antitumor activity in B-cell non-Hodgkin lymphoma (NHL) cells,22,25 providing the rationale for exploring this combination treatment strategy in MCL. From a translational standpoint, dual antibody therapy offers several advantages including: favorable toxicity profiles that may permit frequent dosing or maintenance treatment; additional treatment options for heavily pretreated patients or patients with significant comorbidities; potentially increased efficacy compared with single agent regimens because of alternative mechanisms of action; and the ability to overcome resistance mechanisms that may evolve in the setting of single agent mAb therapy. We report in this study the preclinical in vitro and in vivo activity of combination milatuzumab and rituximab, and explore the mechanism by which this combination treatment induces cell death in MCL.
Methods
Primary tumor cells and cell lines
Primary tumor cells were isolated from the peripheral blood of patients with MCL after obtaining informed consent in accordance with the Declaration of Helsinki detailed in a protocol approved by The Ohio State University Institutional Review Board. All patients studied (with clinical characteristics summarized in Figure 2B) were diagnosed with MCL according to the World Health Organization classification of tumors.26 Characteristics of the MCL cell lines, summarized in Table 1, have been previously described. Jeko, Mino, and SP-53 were generously contributed by Dr Raymond Lai (University of Alberta, Edmonton, AB). Rec-1, Granta-519, and HBL-2 were generously contributed by O.A.O.27
Reagents
Rituximab (Genentech) and trastuzumab (Genentech) were obtained commercially. Milatuzumab and veltuzumab were provided by Immunomedics. Ofatumumab was supplied by Genmab. Q-VD-OPH pan-caspase inhibitor was purchased from MP Biomedicals.
Immunophenotypic studies
Immunophenotyping was performed to determine the percentage and mean fluorescence intensity (MFI) of CD20 and CD74 on MCL cell lines and primary tumor cells relative to a specific isotype control. Further details are available in supplemental Data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Synthesis of fluorescence-labeled antibodies
Rituximab and the goat anti–human immunoglobulin-G antibody (Fcγ fragment–specific, anti-Fc; Jackson ImmunoResearch Laboratories) were fluorescently conjugated with N-hydroxysuccinimide ester rhodamine (Pierce). Milatuzumab and total p65 were fluorescently conjugated with Alexa Fluor 488 5-sulfodichlorophenol ester (Invitrogen) as described.28 Further details are available in supplemental Data.
Assessment of antibody-binding and antigen surface density
Quantitative analysis of CD74 and CD20 surface density was done using the Quantum Simply Cellular kit (Bangs Laboratories), according to the manufacturer's instructions. Further details are available in supplemental Data.
Analysis of cell death by flow cytometry
Cell viability was measured by dual staining with annexin V–fluorescein isothiocyanate and propidium iodide (PI; BD Pharmingen) as described.29 Reactive oxygen species (ROS) generation, mitochondrial membrane potential (ΔΨm) changes research, and actin reorganization experiments were performed as described.30,31 Further details are available in supplemental Data.
Immunoblot analysis
Immunoblot assays were performed as described.29 Antibodies to the following proteins were used: caspase 3, caspase 2, poly(adenosine 5′-diphosphate) ribose polymerase (PARP), and total p65/NF-κB (Cell Signaling Technology); actin, α-tubulin, Brg-1, Mcl-1, Bax, Bcl-2, Bcl-xL, and p53 (Santa Cruz Biotechnology); protein light chain 3 (LC3; Sigma-Aldrich); and p62/SQSTM1 (Medical & Biologic Laboratories).
Microscopy methods
Total p65 localization as well as binding, internalization, and capping of milatuzumab and rituximab were examined by laser scanning confocal microscopy as previously described.28 Cell-cell adhesion was evaluated by visualizing the cells with an inverted microscope (TS100; Nikon) using ×20 PH lens. Further details are available in supplemental Data.
Evaluation of the in vivo therapeutic activity of rituximab in combination with milatuzumab
To examine the in vivo activity of rituximab and/or milatuzumab, our previously described preclinical model of human MCL using the severe combined immunodeficiency (SCID) mouse was implemented.30 Further details are available in supplemental Data.
Statistical analysis
Details of our statistical analysis are available in supplemental Data.
Results
Given the variable biology and clinical behavior of MCL, the activity of milatuzumab, rituximab, and their combination was tested in 6 MCL cell lines to represent the spectrum of this heterogeneous disease. Jeko, Mino, and SP-53 cell lines were used for the mechanistic experiments because of their different histopathologic origins (Jeko, blastoid variant; Mino and SP-53, classic MCL) and P53 status. Jeko and SP-53 cells have no detectable mutations, Mino cells have a single mutation at the level of exon 527,32,33 ; p53 protein is present in Mino and SP-53, with the Mino cell line expressing the highest level, whereas Jeko cells have no detectable p53 level (supplemental Figure 1).
CD20 and CD74 expression on MCL cell lines and patient primary tumor cells
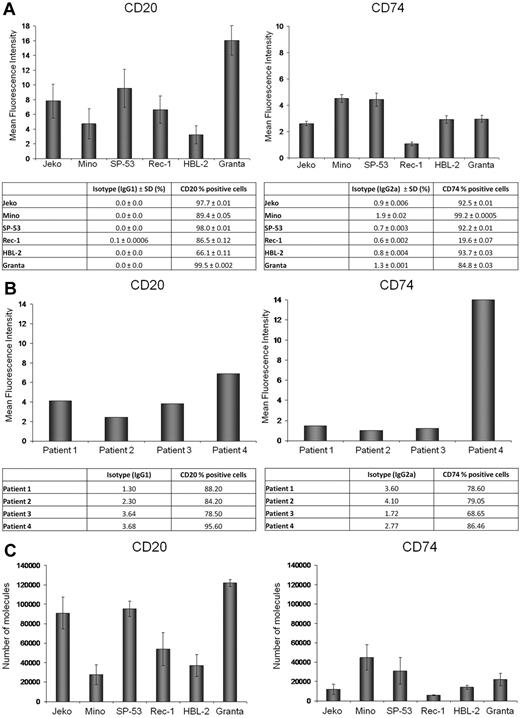
CD20 and CD74 antigen expression and MFI were determined by flow cytometry in 6 MCL cell lines (Figure 1A) and in 4 primary patient samples (Figure 1B). Expression of CD20 and CD74 surface antigens was observed in all cell lines, although with variable intensity. To evaluate the differential expression of CD20 and CD74 between the 6 MCL cell lines, we determined the number of CD74 and CD20 surface molecules using a flow cytometric–based assay. As shown in Figure 1C, Granta, Jeko, and SP-53 cell lines expressed the greatest number of CD20 molecules and Mino and HBL-2 the lowest; Mino and SP-53 expressed the highest number of CD74 molecules and Jeko and Rec-1 the lowest. The number of CD74 molecules was lower in 5 of 6 MCL cell lines compared with expression of CD20.
CD20 and CD74 expression on MCL cell lines and primary MCL tumor cells. (A-B) CD20 and CD74 MFI and the percentage of positive cells based on comparison to isotype control in 6 MCL cell lines (A) and primary cells from 4 different MCL patients (B). (C) The number of CD20 and CD74 molecules per cell in 6 MCL cell lines.
CD20 and CD74 expression on MCL cell lines and primary MCL tumor cells. (A-B) CD20 and CD74 MFI and the percentage of positive cells based on comparison to isotype control in 6 MCL cell lines (A) and primary cells from 4 different MCL patients (B). (C) The number of CD20 and CD74 molecules per cell in 6 MCL cell lines.
Combination treatment with milatuzumab and rituximab induces enhanced cell death in MCL cell lines and primary patient tumor cells
Dose titration experiments with Jeko, Mino, and SP-53 cells were performed to determine the optimal dose of milatuzumab. No significant differences in terms of cell death were noticed between 5 and 10 μg/mL at 8, 24, and 48 hours (data not shown); therefore, we chose a dose of 5 μg/mL milatuzumab for all our in vitro experiments. The in vitro survival of 6 MCL cell lines after incubation with milatuzumab, rituximab (10 μg/mL), and the combination, in the presence of a cross-linking antibody (in 5 times excess of binding antibody), was determined at 8, 24, and 48 hours by annexin V–PI staining and flow cytometry. As shown in Figure 2A, incubation of 6 MCL cell lines with cross-linked milatuzumab and rituximab resulted in a statistically significant decrease (P < .01) in cell viability compared with either single agent alone for each of the 6 cell lines averaged across the 3 time points examined. Mean cell survival averaged across 6 cell lines and 3 time points for milatuzumab plus rituximab was 17.8% (± 0.9%); milatuzumab alone 41.5% (± 0.9%); and rituximab alone 23.7% (± 1.1%). Cell death was rapid, with a significant decrease in viability observed as early as 8 hours. SP-53 and Granta cell lines were the most sensitive to combination treatment. Notably, the Jeko cell line, which represents the blastoid variant of MCL and most resistant phenotype to chemotherapeutic and biologic agents,29,30 was the only cell line in which combination treatment resulted in synergistic killing, although both milatuzumab and rituximab as single agents showed only modest activity. To determine whether the anti–CD20 component was not limited to a rituximab-specific effect, we tested combination therapy with ofatumumab and veltuzumab, 2 fully humanized anti–CD20 mAbs that recognize different epitopes on the CD20 protein.34,35 As shown in supplemental Figure 2, similar cytotoxic profiles were seen when all 3 mAbs were used in combination with milatuzumab, suggesting that the combination treatment-induced MCL death is not limited to a drug-specific effect of rituximab, but rather generalized anti-CD20 signaling. As previously shown in non-MCL cell lines,22 we confirmed that milatuzumab did not induce cell death in absence of a cross-linking antibody, and noncross-linked rituximab has only minimal effect on MCL cell viability (data not shown).
Immobilized rituximab and milatuzumab treatment-induced cell death in MCL cells. The 6 MCL cell lines (A), patient characteristics (B), and primary cells from 7 patients (C-D) were treated with rituximab (10 μg/mL) and/or milatuzumab (5 μg/mL), in the presence of a cross-linking antibody. Cell death was determined by annexin V–PI staining and flow cytometry at 8, 24, and 48 hours for the 6 cell lines and at 24 hours for MCL primary cells. Data are shown as the percentage of annexin V−PI cells (live cells) and are normalized to untreated control. Individual patient responses (C) and representative histograms summarizing patient responses (D) are shown. Combination treatment resulted in statistically significant enhanced induction of MCL cell death compared with either agent alone (P < .01).
Immobilized rituximab and milatuzumab treatment-induced cell death in MCL cells. The 6 MCL cell lines (A), patient characteristics (B), and primary cells from 7 patients (C-D) were treated with rituximab (10 μg/mL) and/or milatuzumab (5 μg/mL), in the presence of a cross-linking antibody. Cell death was determined by annexin V–PI staining and flow cytometry at 8, 24, and 48 hours for the 6 cell lines and at 24 hours for MCL primary cells. Data are shown as the percentage of annexin V−PI cells (live cells) and are normalized to untreated control. Individual patient responses (C) and representative histograms summarizing patient responses (D) are shown. Combination treatment resulted in statistically significant enhanced induction of MCL cell death compared with either agent alone (P < .01).
It was previously reported that milatuzumab-mediated cell death of non-MCL cells did not correlate with antigen density.22 However, in MCL cells, statistical analysis showed a significant correlation between CD20/CD74 MFI and cell viability. We observed a statistically significant association between viability of rituximab-treated cells and CD20 MFI across all 6 cell lines at 8, 24, and 48 hours (R2 = 0.74, P = .028; R2 = 0.68, P = .044; R2 = 0.66, P = .049, respectively; supplemental Figure 3). There was also a statistically significant association between viability of milatuzumab-treated cells and CD74 MFI at 24 and 48 hours (R2 = 0.79, P = .017 and R2 = 0.93, P = .002, respectively; supplemental Figure 2). In cells treated with combination rituximab and milatuzumab, CD20 MFI was significantly associated with the percentage of live cells at 8 hours, but not at 24 and 48 hours (R2 = 0.70, P = .039; R2 = 0.45, P = .15; R2 = 0.32, P = .32, respectively). In cells treated with the combination of rituximab and milatuzumab, CD74 MFI was not significantly associated with cell viability at any time point. These data suggest that antitumor responses mediated by single agent rituximab and milatuzumab are related to CD20 MFI and CD74 MFI. In cells treated with the combination of the 2 antibodies, responses are not significantly associated with CD74 MFI (R2 = 0.12, P = .49; R2 = 0.30, P = .26; R2 = 0.51, P = .11, respectively).
We next compared the effects of milatuzumab and rituximab in primary tumor cells from 7 MCL patients (clinical characteristics of patients summarized in Figure 2B) after 24 hours of incubation with miltauzumab (5 μg/mL), rituximab (10 μg/mL), or the combination in the presence of a cross-linking antibody. The combination treatment induced an average 79.6% cell death compared with 60% (P = .002) of the milatuzumab-treated cells and 52% (P = .0001) of the rituximab-treated cells (individual patient responses are shown in Figure 2C, summarization of patient responses are shown in Figure 2D). These data demonstrate combination treatment markedly enhances cell death in MCL cell lines and primary cells regardless of the morphologic variant of MCL compared with single agent treatment.
CD74/CD20 aggregation and cytoskeletal integrity are required for milatuzumab- and rituximab-induced cell death
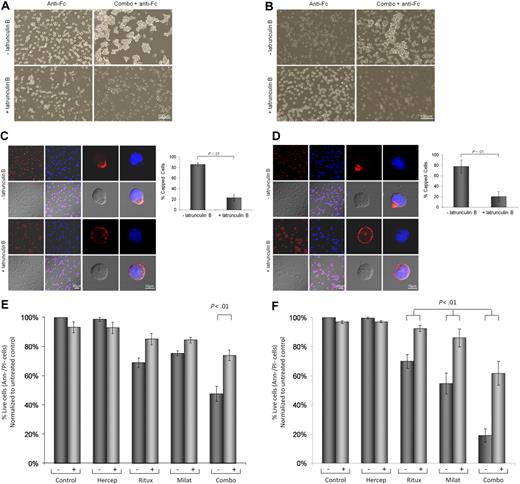
Previous work in non-MCL cell lines showed that cross-linked rituximab induces a redistribution of CD20 molecules on specialized microdomains at the plasma membrane known as the lipid rafts.36 It has also been suggested that membrane rafts may be important in propagating the rituximab-mediated prodeath effects.37 Therefore, we sought to determine whether cross-linking of CD20 and CD74 with rituximab and milatuzumab, respectively, formed aggregates within the plasma membrane. Nonimmobilized rhodamine-conjugated rituximab treatment (10 μg/mL) of Jeko (Figure 3A), and Mino and SP-53 cells (data not shown) induced no change in the distribution of CD20. As expected, incubation of Jeko (Figure 3A), Mino and SP-53 cells (data not shown) with Alexa Fluor 488–conjugated milatuzumab (5 μg/mL), in absence of a cross-linking antibody-induced rapid internalization of CD74. In contrast, cross-linking of fluorescent rituximab and milatuzumab caused clustering of CD74 and coclustering with CD20 on Jeko (Figure 3B) and on Mino and SP-53 (data not shown) cell surfaces. Representative histograms summarizing the percentage of capped cells in the presence or absence of a cross-linking antibody are shown in Figure 3C (Jeko cells left panel and Mino cells right panel). These data clearly show that cross-linking of milatuzumab and rituximab leads to formation and colocalization of large cellular aggregates and capping of CD20 and CD74 proteins on the cell surface and may interrupt the survival signal that has previously been documented with CD74 internalization.21
CD20 and CD74 labeling by specific fluorescent antibodies. Binding and internalization of CD20 and CD74 in Jeko cells were examined by laser scanning confocal microscopy. Jeko cells were incubated with rhodamine-conjugated rituximab (red) and Alexa Fluor 488-conjugated milatuzumab (green) in the absence (A) or presence (B) of a cross-linking antibody for 2 hours at 37°C. DRAQ5 was used for nuclear staining (blue). Cross-linked milatuzumab forms large aggregates and colocalizes with cross-linked rituximab on the surface of MCL cells. (C) Representative histograms summarizing the percentage of capped cells in the presence or absence of a cross-linking antibody are shown (Jeko cells left and Mino cells right). At least 100 cell/conditions were counted for each of the 3 independent experiments. Anti–Fc indicates anti–Fc cross-linking antibody.
CD20 and CD74 labeling by specific fluorescent antibodies. Binding and internalization of CD20 and CD74 in Jeko cells were examined by laser scanning confocal microscopy. Jeko cells were incubated with rhodamine-conjugated rituximab (red) and Alexa Fluor 488-conjugated milatuzumab (green) in the absence (A) or presence (B) of a cross-linking antibody for 2 hours at 37°C. DRAQ5 was used for nuclear staining (blue). Cross-linked milatuzumab forms large aggregates and colocalizes with cross-linked rituximab on the surface of MCL cells. (C) Representative histograms summarizing the percentage of capped cells in the presence or absence of a cross-linking antibody are shown (Jeko cells left and Mino cells right). At least 100 cell/conditions were counted for each of the 3 independent experiments. Anti–Fc indicates anti–Fc cross-linking antibody.
Cytoskeletal reorganization has been demonstrated to be necessary for CD47,38 HLA-DR,39 and CD5240 cross-linked mAb-mediated cell death in chronic lymphocytic leukemia (CLL) cells. We therefore sought to determine whether rituximab- and milatuzumab-mediated cell death required CD20/CD74 aggregation and concurrent cytoskeletal association. Jeko cells (Figure 4A), Mino cells (Figure 4B), and SP-53 (data not shown) demonstrate that pretreatment of MCL cells with 10μM latrunculin B (Figure 4B) and cytochalasin D (data not shown), an agent that prevents actin polymerization, significantly reduced cell aggregation, as assessed by light microscopy 4 hours later. Furthermore, as shown in Figure 4C (Jeko), Figure 4D (Mino), and data not shown (SP-53), pretreatment with latrunculin B significantly reduced the capping and colocalization of CD74 and CD20 antigens (P < .01), as assessed by confocal microscopy 4 hours after the addition of the antibodies.
Cell-cell interactions and cytoskeleton organization are involved in MCL cell death evoked by immobilized rituximab and milatuzumab. Jeko (A) and Mino (B) cells were treated with dimethylsulfoxide (DMSO) or the actin polymerization inhibitor, latrunculin B (10μM), for 45 minutes before the addition of rituximab and milatuzumab, in the presence of a cross-linking antibody. Cell aggregation was assessed 4 hours later by light microscopy. Pretreatment with latrunculin B significantly reduced cell aggregation. Jeko (C) and Mino (D) cells were treated with DMSO or an actin polymerization inhibitor (latrunculin B), for 45 minutes before the addition of rituximab and milatuzumab in the presence of a rhodamine-conjugated cross-linking antibody. The formation of aggregates and caps are shown on Jeko in panel C (left) and Mino in panel D (left) cell surface was evaluated 4 hours later by confocal microscopy. Representative histograms summarizing the percentage of capped cells in the presence or absence of latrunculin B are shown in panel C (Jeko right) and panel D (Mino right). At least 100 cell/conditions were counted for each of the 3 independent experiments. Pretreatment with latrunculin B significantly (P < .01) reduced the capping and colocalization of CD74 and CD20 antigens. Cell death evaluation of Jeko (E) and Mino (F) cells treated with DMSO or latrunculin B, for 45 minutes before the addition of rituximab and milatuzumab in the presence of a cross-linking antibody. Cell death was determined 4 hours later by AnnexinV–PI and flow cytometry. Pretreatment with lactrunculin B resulted in statistically significant reduced cell death induced by the combination treatment (P < .01).
Cell-cell interactions and cytoskeleton organization are involved in MCL cell death evoked by immobilized rituximab and milatuzumab. Jeko (A) and Mino (B) cells were treated with dimethylsulfoxide (DMSO) or the actin polymerization inhibitor, latrunculin B (10μM), for 45 minutes before the addition of rituximab and milatuzumab, in the presence of a cross-linking antibody. Cell aggregation was assessed 4 hours later by light microscopy. Pretreatment with latrunculin B significantly reduced cell aggregation. Jeko (C) and Mino (D) cells were treated with DMSO or an actin polymerization inhibitor (latrunculin B), for 45 minutes before the addition of rituximab and milatuzumab in the presence of a rhodamine-conjugated cross-linking antibody. The formation of aggregates and caps are shown on Jeko in panel C (left) and Mino in panel D (left) cell surface was evaluated 4 hours later by confocal microscopy. Representative histograms summarizing the percentage of capped cells in the presence or absence of latrunculin B are shown in panel C (Jeko right) and panel D (Mino right). At least 100 cell/conditions were counted for each of the 3 independent experiments. Pretreatment with latrunculin B significantly (P < .01) reduced the capping and colocalization of CD74 and CD20 antigens. Cell death evaluation of Jeko (E) and Mino (F) cells treated with DMSO or latrunculin B, for 45 minutes before the addition of rituximab and milatuzumab in the presence of a cross-linking antibody. Cell death was determined 4 hours later by AnnexinV–PI and flow cytometry. Pretreatment with lactrunculin B resulted in statistically significant reduced cell death induced by the combination treatment (P < .01).
To further evaluate the link between glycolipid-enriched membrane domain aggregation, alteration of MCL cells cytoskeletal elements, and the transmission of a death signal, we examined the effect of latrunculin B on MCL cells viability. Cells were treated with latrunculin B prior of the addition of immobilized milatuzumab and/or rituximab. After 4 hours of incubation, cell viability was determined by annexin V–PI and flow cytometry. As shown in Figure 4E (Jeko), Figure 4F (Mino), and data not shown (SP-53), pretreatment with lactrunculin B (and cytochalasin D, data not shown) resulted in statistically significant reduced cell death induced by the combination treatment (P < .01 after Holm adjustment for multiple comparisons). Our data suggest that colocalization of CD74 and CD20 and cytoskeletal association are necessary events in triggering the milatuzumab/rituxmab-mediated cell death signal in MCL cells.
Milatuzumab- and rituximab-mediated cytotoxicity is partially dependent on generation of ROS and loss of mitochondrial transmembrane potential
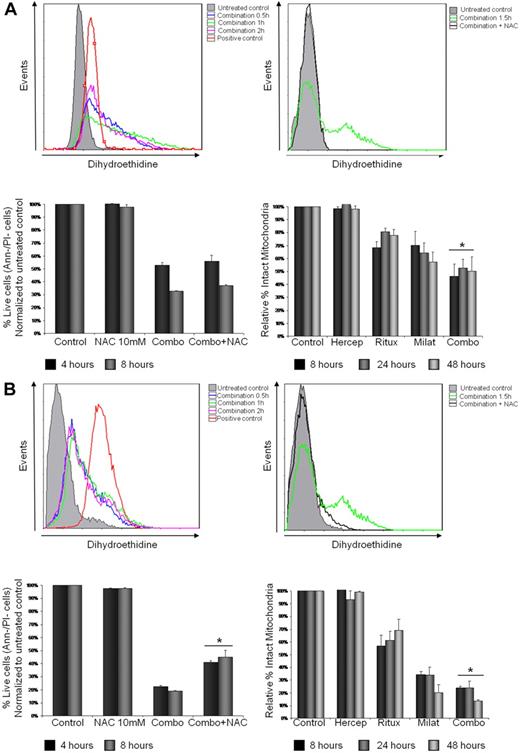
To directly determine whether milatuzumab- and rituximab-induced cytotoxicity was mediated by generation of ROS, Jeko (Figure 5A), Mino (Figure 5B), and SP-53 (data not shown) cells were treated with combination rituximab (10 μg/mL) and milatuzumab (5 μg/mL), in the presence of a cross-linking antibody for 0.5, 1, 1.5 and 2 hours, and cells examined for changes in ROS generation using the specific fluorescence probe, dihydroethidine. The combination treatment induced an increase of dihydroethidine fluorescence indicating an increase in ROS generation as early as 0.5 hours, peaking at 1 to 1.5 hours and reducing at 2 hours, in all the 3 MCL cell lines examined (Figure 5A-B top left panel and data not shown). The levels of ROS in untreated control cells remained unchanged over all incubation time periods. To further explore the implication of ROS in milatuzumab- and rituximab-mediated cell death, Jeko, Mino, and SP-53 cells were incubated with N-acetylcysteine (NAC, 10mM), a nonspecific ROS scavenger, for 1.5 hours, in the absence or presence of the combination treatment. NAC was added 15 minutes before the addition of the mAbs and, as shown in Figure 5A-B top right panel and data not shown, NAC was able to block ROS generation in treated cells. We next determined cell viability by annexin V–PI staining and flow cytometry. As shown in Figure 5B bottom left panel, the addition of NAC led to a statistically significant increase (P < .01) in the viability of Mino cells compared with milatuzumab- and rituximab-treated cells averaged across 4 and 8 hours incubation. Similar results were observed in SP-53 cells (data not shown). In contrast, NAC pretreatment failed to rescue milatuzumab- and rituximab-treated Jeko cells (P = .064 and P = .054) at 4 and 8 hours, respectively (Figure 5A bottom left panel).
Rituximab and milatuzumab cytotoxicity is partially dependent on ROS generation and loss of ΔΨm. (A-B) Jeko in panel A (top left) and Mino in panel B (top left) cells were treated with rituximab and milatuzumab in the presence of a cross-linking antibody for the indicated time points. ROS generation was determined by flow cytometric analysis using dihydroethidine dye. ROS generation is indicated by right shift of the dihydroethidine curves. Hydrogen peroxide was used as positive control. Top right panel shows pretreatment of MCL cells with the nonspecific ROS scavenger NAC (10mM) and inhibition of ROS generation induced by antibody treatment. For rescue experiments, Jeko (bottom left panel) and Mino (bottom left panel) cells were incubated with NAC for 4 and 8 hours in the absence or presence of the antibodies, and cell viability was determined by annexin V–PI staining and flow cytometry. Bottom right panel shows ΔΨm changes in Jeko and Mino cells treated with rituxmab, milatuzumab, or the combination of both in the presence of a cross-linking antibody, for 4, 8 and 24 hours. The ΔΨm changes were quantified by flow cytometry determination using JC-1. Data are shown as the percentage of treated cells with intact mitochondria relative to untreated cells at the same time points.
Rituximab and milatuzumab cytotoxicity is partially dependent on ROS generation and loss of ΔΨm. (A-B) Jeko in panel A (top left) and Mino in panel B (top left) cells were treated with rituximab and milatuzumab in the presence of a cross-linking antibody for the indicated time points. ROS generation was determined by flow cytometric analysis using dihydroethidine dye. ROS generation is indicated by right shift of the dihydroethidine curves. Hydrogen peroxide was used as positive control. Top right panel shows pretreatment of MCL cells with the nonspecific ROS scavenger NAC (10mM) and inhibition of ROS generation induced by antibody treatment. For rescue experiments, Jeko (bottom left panel) and Mino (bottom left panel) cells were incubated with NAC for 4 and 8 hours in the absence or presence of the antibodies, and cell viability was determined by annexin V–PI staining and flow cytometry. Bottom right panel shows ΔΨm changes in Jeko and Mino cells treated with rituxmab, milatuzumab, or the combination of both in the presence of a cross-linking antibody, for 4, 8 and 24 hours. The ΔΨm changes were quantified by flow cytometry determination using JC-1. Data are shown as the percentage of treated cells with intact mitochondria relative to untreated cells at the same time points.
The cytotoxic effect of milatuzumab in combination with rituximab was associated with a gradual and significant loss of ΔΨm as early as 8 hours, as evidenced by disaggregation of JC-1 in Jeko, Mino (Figure 5A-B bottom right panel), and SP-53 (data not shown) cell lines. This effect was concurrent with an increase in Annexin binding, indicative of early stages of cell death. The loss in ΔΨm caused by the combination treatment was greater than either drug alone with Mino and SP-53 cells and was statistically significant (P < .01) at each of the 3 time points examined. In Jeko cells, the combination treatment resulted in a statistically significant loss of ΔΨm compared with the rituximab-treated cells (P < .01) but not compared with milatuzumab-treated cells (P = .079). These results indicate that ROS generation and mitochondrial membrane dysfunction are involved in the combination treatment-mediated cell death in Mino and SP-53 cells but only partially in Jeko cells. Interestingly, Jeko is the only cell line in which the combination treatment induced a synergistic cell death.
Milatuzumab and rituximab induce cell death via a nonclassic apoptotic mechanism
Rituximab-induced cell death in B-cell lymphoproliferative disorders has been shown to be associated with the generation of ROS and loss of ΔΨm in a caspase-independent fashion.31 It has recently been shown that milatuzumab induced caspase 2 cleavage in CLL-treated cells.24 We therefore sought to determine whether caspase 2 and caspase 3 activation and PARP cleavage were involved in MCL cell death induced by milatuzumab and rituximab. As shown in supplemental Figure 4, 24-hour incubation of Jeko and Mino cells with milatuzumab (5 μg/mL), rituximab (10 μg/mL), or the combination in the presence of a cross-linking antibody failed to induce caspase 2 and caspase 3 activation and PARP cleavage. To confirm these findings, we also showed that pretreatment of MCL cell lines with the pan-caspase inhibitor, Q-VD-OPH, was able to block cleavage of caspase 2, caspase 3, and PARP (caused by bortezomib), but it had no significant effect on milatuzumab- and rituximab-induced cell death (data not shown).
To determine whether the milatuzumab- and rituximab-mediated cytotoxicity was because of down-regulation of antiapoptotic proteins, we tested the effect of this combination treatment on the expression of Bcl-2, Bcl-xL, Mcl-1, and Mcl-3 critical antiapoptotic proteins in MCL. As shown in supplemental Figure 5, the expression of Bax, Bcl-2, Bcl-xL, and Mcl-1 was not altered in response to treatment in Jeko and Mino cells.
Given the absence of an activated classic apoptotic death program in MCL cells treated with milatuzumab and rituximab as evidenced by the absence of caspase 2 and caspase 3 activation and changes in expression in Bcl-2 family members, we explored alternative mechanisms of cell death. Autophagy describes a physiologic mechanism that may serve as a means of temporary survival as is triggered by starvation and metabolic stress. However, if the cellular stress continues, autophagy-mediated cell death may occur.41 Several studies have shown that autophagy plays a major role in the induction of tumor cell death triggered by antitumor agents such as tamoxifen42 and arsenic trioxide.43 To establish whether the autophagic machinery was involved in milatuzumab- and rituximab-mediated cell death, we examined the effect of this combination treatment on LC3-II and p62 levels. During autophagy, LC3-I is converted to LC3-II through lipidation,44 Protein p62 directly interacts with LC3 and is degradated by the autophagic-lysosome pathway.44 Jeko and Mino cells were both incubated for 4 and 24 hours with milatuzumab and rituximab in the presence or absence of chloroquine, which is known to inhibit lysosomal degradation but not autophagosome formation. If milatuzumab and rituximab treatment led to cell death dependent on autophagic mechanisms, then treatment with this combination in presence of chloroquine should result in a significant increase of LC3-II. Treatment with milatuzumab and rituximab in combination with chloroquine did not cause further increase of LC3-II or p62 levels compared with cells treated with chloroquine alone (supplemental Figure 6 and data not shown). Importantly, MCL cells appear to be relatively resistant to enhanced induction of autophagy caused by rapamycin, a well-established reagent that accelerates the autophagic process (supplemental Figure 6). Taken together, these results suggest that milatuzumab and rituximab treatment of MCL cells is not associated with caspase cleavage, Bcl-2 family member dysregulation, or induction of autophagy.
NF-κB inhibition with immobilized milatuzumab and rituximab in MCL cell lines
Previous studies have demonstrated that constitutive activation of NF-κB is an important contributing factor to the pathogenesis of MCL.3,45 Furthermore, it has already been shown that ligation of CD74 induces CD74 cleavage and internalization and initiates a signaling cascade leading to B-cell proliferation and survival via the NF-κB pathway.21 Similarly, it has been shown that rituximab is able to inhibit the NF-κB signaling pathway in B-cell NHL cell lines.46 We therefore investigated the effects of milatuzumab and rituximab on the NF-κB pathway in MCL cells. We isolated nuclear and cytoplasmic protein fractions from Jeko, Mino, and SP-53 cells after 4-hour incubation with cross-linked milatuzumab, rituximab, or the combination of both. Fractions were analyzed by immunoblot for nuclear p65, α-tubulin (cytoplasmic control), and Brg-1 (nuclear control). As shown in Figure 6, nuclear p65 levels were not significantly reduced in Jeko, Mino, and SP-53 cells (data not shown) treated with either single agent milatuzumab or rituximab 4 hours after exposure to the respective drug. However, combined exposure to milatuzumab and rituximab resulted in near elimination of nuclear p65 in Mino (Figure 6B) and SP-53 cells (data not shown), whereas the effect on Jeko cells was less clear (Figure 6A).
Rituximab and milatuzumab combination treatment inhibited nuclear total p65 translocation in MCL cell lines. Top panels show nuclear and cytosolic protein fractions collected at 4 hours prepared from Jeko (A) and Mino (B) cell lines and subjected to immunoblot analysis for p65, α-tubulin (cytoplasmic control), and Brg-1 (nuclear control). Bottom panels show confocal microscopic analysis of intracellular localization of total p65 (green) in Jeko (A) and Mino (B) cells. MCL cells were treated with cross-linking antibody, trastuzumab, rituximab, milatuzumab, and the combination of rituximab and milatuzumab in the presence of a cross-linking antibody. After 4 hours, cells were collected and prepared for confocal microscopy analysis. DRAQ5 was used for nuclear staining.
Rituximab and milatuzumab combination treatment inhibited nuclear total p65 translocation in MCL cell lines. Top panels show nuclear and cytosolic protein fractions collected at 4 hours prepared from Jeko (A) and Mino (B) cell lines and subjected to immunoblot analysis for p65, α-tubulin (cytoplasmic control), and Brg-1 (nuclear control). Bottom panels show confocal microscopic analysis of intracellular localization of total p65 (green) in Jeko (A) and Mino (B) cells. MCL cells were treated with cross-linking antibody, trastuzumab, rituximab, milatuzumab, and the combination of rituximab and milatuzumab in the presence of a cross-linking antibody. After 4 hours, cells were collected and prepared for confocal microscopy analysis. DRAQ5 was used for nuclear staining.
To verify these results, we performed the same experiment, and evaluated cytosolic and nuclear level of total p65 in Jeko, Mino, and SP-53 cells using confocal microscopy. As shown in Figure 6A-B, combination treatment with immobilized milatuzumab and rituximab led to a significant reduction in nuclear p65 compared with single agent milatuzumab and rituximab in Mino cells and SP-53 cells (data not shown) but, again, the effect was less clear in Jeko cells. These results indicate that disruption of the NF-κB pathway and subsequent loss of the survival signal is induced by the combination treatment in Mino and SP-53 cells and, perhaps, to a lesser degree in Jeko cells.
In vivo therapeutic activity of combination treatment with milatuzumab and rituximab
We next evaluated the in vivo effect of milatuzumab in combination with rituximab in a preclinical model of human MCL. Female SCID mice 6 to 8 weeks of age (cb17 scid/scid), natural killer cells depleted with intraperitoneal injections of anti–mouse interleukin-2 receptorβ mAb, were engrafted with 40 × 106 Jeko cells via tail vein injection. Animals in groups of 10, starting at day 15 after engraftment, received trastuzumab (15 mg/kg), as a control, rituximab (15 mg/kg), milatuzumab (15 mg/kg), or the combination of rituximab and milatuzumab every 3 days via intraperitoneal injection. The primary end point of the study was OS defined as the time to develop symptoms indicating lethal tumor burden after initiation of treatment. As shown in Figure 7, the mean survival for the combination treated group was 44.5 days (95% confidence interval [CI], 39%-51%), compared with 28 days for trastuzumab-treated mice (95% CI, 24%-30%), 33.5 days for the milatuzumab-treated mice (95% CI, 28%-36%), and 38 days for the rituximab-treated mice (95% CI, 36%-42%). The combination treatment significantly prolonged survival of this group compared with trastuzumab control (P < .0001), milatuzumab (P = .001), and rituximab (P = .03).
Evaluation of in vivo therapeutic activity of rituximab and milatuzumab in the preclinical MCL model. SCID mice (in groups of 10) were injected intravenously with 40 × 106 Jeko cells and observed daily for signs of tumor burden. The mAbs (trastuzumab 15 mg/kg, rituximab 15 mg/kg, milatuzumab 15 mg/kg, or the combination of rituximab and milatuzumab) were given every 3 days via intraperitoneal.injection, starting at day 15 after engraftment. The mean survival for rituximab- and milatuzumab-treated mice was 44.5 days (95% CI, 39%-51%), compared with 33.5 days for the milatuzumab-treated mice (95% CI, 28%-36%), and 38 days for the rituximab-treated mice (95% CI, 36%-42%).
Evaluation of in vivo therapeutic activity of rituximab and milatuzumab in the preclinical MCL model. SCID mice (in groups of 10) were injected intravenously with 40 × 106 Jeko cells and observed daily for signs of tumor burden. The mAbs (trastuzumab 15 mg/kg, rituximab 15 mg/kg, milatuzumab 15 mg/kg, or the combination of rituximab and milatuzumab) were given every 3 days via intraperitoneal.injection, starting at day 15 after engraftment. The mean survival for rituximab- and milatuzumab-treated mice was 44.5 days (95% CI, 39%-51%), compared with 33.5 days for the milatuzumab-treated mice (95% CI, 28%-36%), and 38 days for the rituximab-treated mice (95% CI, 36%-42%).
Discussion
The natural history of MCL is a course of progressive relapses that are increasingly short-lived as the disease becomes more resistant to therapy. Therefore, the development of new therapeutic options is crucial to improve outcomes for patients with this incurable disease.
In this study, we have shown that the treatment with rituximab and milatuzumab resulted in statistically significant enhanced cell death in all 6 MCL cell lines and all primary tumor cell samples from 7 MCL patients, compared with either agent alone. It has to be noted that the primary cells were obtained from the peripheral blood of patients who have MCL with leukemic tumor burden. Although peripheral blood involvement is present in only a subset of patients who have MCL with cytotoxicity, experiments in such a group may have some limitations. It is important to note that peripheral blood involvement represents an independent negative prognostic factor and is included in the MCL International Prognostic index score.47
We decided on in vitro and in vivo dosing parameters to best reflect concentrations of antibody that were achieved in phase I studies. Because there are no published preclinical data on the pharmacokinetics and pharmacodynamics of milatuzumab, the in vitro and in vivo dosing schedule used in our experiments were based on reported in NHL and multiple myeloma studies.18,22 Notably, a pharmacokinetics study performed in refractory/relapsed multiple myeloma patients treated with milatuzumab (1.5, 4, 8, or 16 mg/kg) twice weekly for 4 weeks showed that at the lowest human dose (1.5 mg/kg), the peak levels were 10 to 20 μg/mL, more than twice the in vitro concentration we used in our study. The dosing in our mice was at 1.2 mg/kg human equivalent dose, approximately the lowest dose used in the clinical study (1.5 mg/kg),48 perhaps making our findings more clinically relevant.
The combination treatment led to rapid cell death through a death pathway that was caspase-independent, nonautophagic and did not involve the Bcl-2 family members. Instead, cell death involved ROS generation, loss of ΔΨm, changes in actin dynamics, and disruption of the NF-κB pathway.
In contrast with the previously published report by Stein et al22 in non-MCL cell lines, single agent milatuzumab-mediated cell death significantly correlated with antigen density; however, this correlation was lost when milatuzumab was combined with rituximab, making this combination an attractive treatment option for a disease with variability of CD74 antigen expression. Notably, the combination treatment induced enhanced cell death in all MCL cell lines and patient samples regardless of the expression of DNA damage sensor genes such as P53 (silenced in Jeko cells and 2 patients samples), ATM (defective in Granta cells), and cell-cycle checkpoint regulators such as P16 (deleted in Rec-1 and Granta cells).
A large body of literature supports the association between CD20, lipid rafts, cytoskeletal elements and initiation of cell death.37,49,50 However, little is known about the cytoskeletal associations that might control the aggregation of CD74. In this study, we showed that immobilized milatuzumab and rituximab led to capping of antibody-antigen complexes on the majority of the MCL cells examined. Unlike rituximab, nonimmobilized milatuzumab induced quick internalization of the receptor leading us to hypothesize that the clustering of CD74 on the cell surface by immobilized milatuzumab may mediate downstream pathways that trigger a death signal. In addition, the colocalization of CD74 and CD20 in cross-linked antibody-treated cells may promote an amplification of the death signal as seen by other studies.37,49 To further characterize the CD74 and CD20 capping and to explain the receptor redistribution through a linkage to the cytoskeleton, we demonstrated that pretreatment with actin polymerization inhibitors such as latrunculin B and cytochalasin D significantly reduced cross-linked antibody-mediated capping, cell-cell adhesion, and cell death in MCL cells treated with the combination of milatuzumab and rituximab and a cross-linking antibody.
Previous work in non-MCL cell lines has determined that rituximab-induced cell death was caspase independent,31 which was confirmed in our analysis. Stein et al22 showed that milatuzumab-mediated cell death in Burkitt lymphoma cell lines was dependent on caspase 3 and caspase 8 activation. More recently, caspase 2 cleavage was observed in CLL cells treated with milatuzumab.24 We established that the engagement of CD20 and CD74 resulted in ROS generation and loss of ΔΨm, but cell death occurred in the absence of caspase activation or alteration of Bcl-2 family member expression.
Prior studies have demonstrated that constitutive activation of NF-κB is a contributing factor to the pathogenesis of MCL.3,45 It has also been previously shown in non-MCL cell lines that rituximab disrupts the NF-κB pathway and sensitizes tumor cells to chemotherapeutic agents, in part by selectively down-modulating Bcl-2 and Bcl-xL.46 Ligation of CD74 by its natural ligand migration-inhibitory factor also resulted in NF-κB activation through the PI3K/Akt survival pathway and via Syk phosphorylation.51,52 In this study, we showed that the combination treatment was able to disrupt the NF-κB pathway by inhibiting the translocation of p65 from the cytosol to the nucleus. However we failed to show down-modulation of NF-κB targets such as Bcl-2 and Bcl-xL.3 Interestingly, Starlets et al21 showed that CD74 stimulation of CLL cells initiates a cascade of events that leads to proliferation and survival through activation of the NF-κB pathway and up-regulation of Bcl-xL; however, only up-regulation of the transcript was shown with no evaluation of protein levels.
Based on these findings, it is plausible that the initial and common event in MCL cell death mediated by rituximab and milatuzumab could be the patching/capping on cell surface that subsequently triggers ROS generation, loss of ΔΨm, and involves the underlying actin cytoskeleton rearrangements. Our results also suggest that disruption of the NF-κB pathway, through inhibition of p65 nuclear translocation, may contribute to milatuzumab- and rituximab-induced cell death.
The intriguing results obtained with the in vitro experiments led us to investigate the combination of milatuzumab and rituximab in our recently described in vivo model of MCL.30 The Jeko model was used because it represents the most aggressive and stringent preclinical model to evaluate potential experimental therapeutic strategies in MCL. Importantly, the combination of milatuzumab and rituximab significantly prolonged survival compared with trastuzumab-treated controls (P < .0001) but also with single agent milatuzumab (P = .001) and single agent rituximab (P = .03).
In summary, milatuzumab and rituximab combination therapy resulted in in vitro enhanced cell death in MCL cell lines and primary tumor samples, regardless of antigen density and p53 status. Furthermore, enhanced survival was observed with combination therapy in an in vivo murine model of MCL. To our knowledge to date, our findings are the first to show that milatuzumab and rituximab could potentially represent an active therapeutic strategy for the treatment of MCL patients. Our data support the clinical development of milatuzumab and rituximab in MCL, and based on these collective findings, a phase I/II study is currently underway at our institution evaluating combination milatuzumab and veltuzumab in B-cell NHL (registered at www.clinicaltrials.gov as NCT00989586).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Lymphoma Research Foundation, Mantle Cell Lymphoma Research Initiative (R.A.B.), the American Cancer Society grants ODSR 2009-2 and IRG-67-003-47 (M.P.-I.), the Leukemia & Lymphoma Society and the D. Warren Brown Foundation (N.M. and J.C.B.), and a National Institutes of Health grant 1PO1-CA103985 (D.M.G.).
National Institutes of Health
Authorship
Contribution: L.A. designed and performed research, analyzed data, wrote and reviewed the paper, and approved the final version of the manuscript; B.Y. performed research, analyzed data, and approved the final version of the manuscript; B.A.C. performed research, analyzed data, reviewed drafts, and approved the final version of the manuscript; F.Y. performed research and approved the final version of the manuscript; J.S. performed research and approved the final version of the manuscript; R.L. performed research and approved the final version of the manuscript; E.H. performed research and approved the final version of the manuscript; M.E.L. performed statistical analysis and approved the final version of the manuscript; X.Z. performed statistical analysis and approved the final version of the manuscript; C.Q. performed research, analyzed data, and approved the final version of the manuscript; G.L. performed research, analyzed data, and approved the final version of the manuscript; N.M. designed research, reviewed drafts, and approved the final version of the manuscript; M.P.-I. performed research, analyzed data, reviewed drafts, and approved the final version of the manuscript; O.A.O. designed research, provided cell lines, reviewed drafts, and approved the final version of the manuscript; D.M.G. provided reagent, reviewed drafts, and approved the final version of the manuscript; J.C.B. designed research, reviewed drafts, and approved the final version of the manuscript; K.A.B. provided samples, reviewed drafts, and approved the final version of the manuscript; and R.A.B. designed and supervised research, obtained funding for the work, reviewed drafts, and approved the final version of the manuscript.
Conflict-of-interest disclosure: D.M.G is an officer and member of the Board of Directors of Immunomedics Inc, which owns milatuzumab. The remaining authors declare no competing financial interests.
Correspondence: Robert Baiocchi MD, PhD, B420 Starling Loving Hall, 320 W 10th Ave, Columbus, OH 43210; e-mail: robert.baiocchi@osumc.edu.