Abstract
The p53 protein is a pivotal tumor suppressor that is frequently mutated in many human cancers, although precisely how p53 prevents tumors is still unclear. To add to its complexity, several isoforms of human p53 have now been reported. The Δ133p53 isoform is generated from an alternative transcription initiation site in intron 4 of the p53 gene (Tp53) and lacks the N-terminus. Elevated levels of Δ133p53 have been observed in a variety of tumors. To explore the functions of Δ133p53, we created a mouse expressing an N-terminal deletion mutant of p53 (Δ122p53) that corresponds to Δ133p53. Δ122p53 mice show decreased survival and a different and more aggressive tumor spectrum compared with p53 null mice, implying that Δ122p53 is a dominant oncogene. Consistent with this, Δ122p53 also confers a marked proliferative advantage on cells and reduced apoptosis. In addition to tumor development, Δ122p53 mice show a profound proinflammatory phenotype having increased serum concentrations of interleukin-6 and other proinflammatory cytokines and lymphocyte aggregates in the lung and liver as well as other pathologies. Based on these observations, we propose that human Δ133p53 also functions to promote cell proliferation and inflammation, one or both of which contribute to tumor development.
Introduction
p53 is most important for preventing cancers. We know this because mice deleted for the p53 gene (Trp53) are highly tumor prone1 ; in humans, Li-Fraumeni syndrome, characterized by multiple tumor phenotypes, is the result of germline inherited mutations in the p53 gene (Tp53)2 ; and most common human cancers contain mutations in Tp53 (www.p53.iarc.fr), generally rendering the protein functionally impaired. Ten isoforms of human p53 have been reported that are generated by the use of alternative translation initiation sites, splicing, or alternative promoters.3-9 Two p53 isoforms (Δ40p53 and Δ133p53) lack the N-terminus of p53, whereas 4 others (Δ40p53β, Δ40p53γ, Δ133p53β, and Δ133p53γ) also lack part of the C-terminus beyond codon 331. In addition, 3 more isoforms have recently been described (Δ160p53, Δ160p53β, Δ160p53γ) that use an alternative start codon at position 161 in the transcript for the Δ133p53 isoform family.8 The isoforms are generally expressed in a variable and to some extent tissue specific manner, although the Δ133p53 isoform appears to be ubiquitous.5 Aberrant expression of the Δ133p53 isoforms occurs in a variety of tumors, including breast,5 head and neck,10 acute myeloid leukemia,11 melanoma,12 colon cancer,13 and ovarian cancer,14 suggesting that Δ133p53 contributes to tumor formation. In zebrafish, the homolog of Δ133p53 (Δ113p53) attenuates p53-dependent apoptosis by activating the homolog (bcl2l) of the antiapoptotic protein Bcl-xl,15 and knockdown of Δ113p53 using silencing RNA induced p53-dependent apoptosis. In another study, overexpression of Δ133p53 extended the life span of normal human fibroblasts by inhibiting replicative senescence induced by the p53β isoform,13 and a recent report showed that up-regulation of Δ133p53 in cell culture inhibits apoptosis and G1 arrest induction mediated by full-length p53 but does not affect induction of G2 arrest.16 Thus, these reports suggest that Δ133p53 affects full-length p53 functions in a selective manner. To explore the functions of Δ133p53 in vivo, we created a mouse expressing an N-terminal deletion mutant of p53 (Δ122p53). Our results show that Δ122p53 is a dominant oncogene, confers a proliferative advantage on cells, and is a trigger for inflammation. By implication, we argue that human Δ133p53 has similar functions and is therefore also an oncogene.
Methods
Mice
The Δ122p53 mice were constructed as outlined in supplemental Figure 1A (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). At the commencement of the study, all mice were on a predominantly C57/BL6 background (> 80%), which was further enriched by crossing with C57/BL6 p53+/+ mice for at least 3 generations. p53 null mice were also on the C57/BL6 background. All animal research was granted ethical approval by the institutional authorities of all participating institutions.
Retroviruses and transduced cell lines
Retroviruses expressing Δ122p53 and Δ133p53 were used to generate mouse cell lines according to previously described procedures.17 In brief, cells were infected with retroviral constructs, and those cultures showing more than 85% transduction frequency, based on GFP expression, were selected.
Western blotting
From tissue samples.
Tissues were homogenized after dissection, or cells isolated and treated with 1 μg/mL amsacrine and incubated in complete RPMI media for 5 hours from 4- to 6-week-old animals. Protein lysates were prepared in the presence of protease inhibitors and the IgG and albumin content removed using the albumin/IgG removal kit (Pierce Chemical). A total of 50 to 60 μg of protein was separated on 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Blots were probed with the primary antibodies. Horseradish peroxidase-conjugated antibodies were detected using SuperSignal West Femto detection agent (Pierce Chemical).
From retrovirus transduced cell lines.
Cells were treated and protein lysates prepared. A total of 20 to 30 μg of protein was separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting was carried out. Alkaline phosphatase-conjugated antibodies were detected using the Western Breeze Immunodetection kit (Invitrogen). All blots were probed with primary antibodies against the N-terminus of p53 (1C12, Cell Signaling Technology) or the central region of p53 (PAb 240, Calbiochem) and β-actin (AC-15, Abcam) according to the manufacturer's instructions.
Immunofluorescence
Fibroblast cells expressing Δ122p53 were seeded into chamber slide wells at 2 × 104 cells/well (2-chamber polystyrene vessel tissue culture-treated glass slide, BD Biosciences) and then fixed with paraformaldehyde (2% volume/volume). Cells were then permeablized with phosphate-buffered saline/0.5% volume/volume Triton X-100. Fixed cells were stained with PAb240 and detected using AlexaFluor-488 goat antimouse secondary antibody, and nuclei were labeled with 4′-6-diamidino-2-phenylindole (DAPI).
Gene expression analysis
Pooled single-cell suspensions were prepared from spleens of 4 5- to 6-week-old mice of the following genotypes: Δ122p53, p53+/+, and p53−/−. Cells (2 × 106 cells/mL) were cultured in Dulbecco modified Eagle medium supplemented with 20% fetal calf serum, l-glutamine (2mM), and penicillin and streptomycin and left untreated or treated with 1 μg/mL of amsacrine for 5 hours. Total RNA was extracted using TRI reagent (Sigma-Aldrich), and integrity and purity were confirmed by spectrophotometry and visualized by gel electrophoresis. cDNA was synthesized using oligo-dT-primed SuperScript III (Invitrogen) reverse transcriptase. For detection of Δ122p53 and p53 transcripts, polymerase chain reaction (PCR) was performed and amplicons run on 2% agarose gels. For quantification of p53 target genes, quantitative real-time (RT)-PCR was performed using 5-ng RNA equivalents with QuantiTect SYBR Green PCR master mix (QIAGEN) according to the manufacturer's instructions and performed on a Rotor-Gene 6000 Real time PCR machine (QIAGEN). Primer sequences for detecting p21, Bax, Puma, and Mdm2 are available on request. The ΔΔCt method was used for comparative quantification of gene expression. Data from each time point were normalized against GAPDH (see Figure 2) or β2-Microglobulin (see Figure 3) expression.
Microarray analyses
Splenocytes were treated with amsacrine and RNA extracted, then amplified (250 ng) and labeled using the Illumina TotalPrep RNA Amplification Kit according to the manufacturer's instructions (Ambion). A total of 750 ng of biotinylated cRNA was then hybridized to an Illumina BeadChip array (Illumina Mouse reference no. 8 WGGX V1.1) according to the manufacturer's instructions (Illumina), scanned with an Illumina BeadStation scanner, and analyzed with GenomeStudio software (Version 2009.1). For gene ontology analysis and functional classification, expression data were exported from GenomeStudio and imported into BRB-ArrayTools for normalization (lumi R package) and filtering (BRB-ArrayTools, Version 4.1.0 Beta_2 Release developed by Dr Richard Simon and BRB-ArrayTools Development Team). A transcript was excluded if < 20% of the expression data had at least a 2-fold change in either direction from the gene's median value, or missing data exceeded 50%. Normalization and filtering resulted in a list of 498 Illumina IDs representing approximately 460 transcripts that was used as input for gene ontology functional classification using DAVID18,19 and pathway and network analysis using MetaCore (GeneGo). Unsupervised hierarchical clustering of filtered, normalized, and median-centered log2-transformed data were performed using GenePattern20 with a Pearson correlation distance measure and an average linkage clustering method, and viewed using Java TreeView, Version 1.1.1.21 The microarray data have been deposited in GEO as Series GSE27586 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27586).
In vitro apoptosis analyses
Thymocytes were isolated from 5- to 6-week-old mice and cultured at a density of 2 × 106 cells/mL in Dulbecco modified Eagle medium with 20% fetal calf serum. Cells were left untreated or irradiated with a pulse of 4 Gy γ-irradiation. A 30% ethanol solution was added as a vehicle control to untreated cells for amsacrine experiments. For the apoptosis assay, 1 × 105 cells were doubled stained with annexin-V/allophycocyanin and propidium iodide as described by the manufacturer (BD Biosciences), and analyzed by flow cytometry.
Cell proliferation
Transduced p53 null 10–1 fibroblasts were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and l-glutamine (2mM). Cells were then harvested at indicated times, and cell counting was done by the method of trypan blue exclusion.
Colony formation
Bone marrow isolated from 6-week-old mice (6 mice per genotype) was added to Mouse Methylcellulose Complete Media and cultured (3 × 104 total cells per assay) according to the manufacturer's instructions (the Mouse Colony Forming Cell Assay using Methylcellulose-based media, R&D Systems). The total number of granulocyte and macrophage colonies per assay was counted at day 10 days using an inverted microscope and a scoring grid, according to the manufacturer's instructions (R&D Systems).
In vivo cell cycle arrest/proliferation analysis
Bone marrow.
Mice (3 per genotype) were irradiated with a pulse of 4 Gy γ-irradiation at 5 to 6 weeks of age. For detection of cells actively synthesizing DNA, mice were administered 1.5 mg of BrdU (BD Biosciences) by intraperitoneal injection 24 hours after irradiation. Mice were killed 90 minutes later, and bone marrow was removed. Incorporated BrdU was stained with anti-BrdU/allophycocyanin, and total DNA was labeled with 7-amino-actinomycin D and then analyzed by flow cytometry. Specific details for preparing cells for flow cytometry are outlined in the BrdU Flow Kit instruction manual (BD Biosciences).
Other tissues.
Mice (3 per genotype) at 5 to 6 weeks of age were administered 1.0 mg of BrdU (BD Biosciences) by intraperitoneal injection. Mice were killed 100 minutes (thymus and spleen dissected) or 24 hours later (spleen, lymph node, liver, kidney, pancreas, salivary gland, seminal vesicle, heart, skin, and lung dissected). Tissues were fixed in 10% neutral-buffered formalin. Incorporated BrdU was stained with anti-BrdU/horseradish peroxidase according to the manufacturer's instruction manual (BrdU In-Situ Detection Kit, BD Biosciences). BrdU+ cells were visualized using light microscopy, and the percentage cells per 2000 to 5000 cells counted was determined.
Survival, tumor, and premalignant pathologic analyses
Mice were aged for 600 days and killed when visible tumor burden was apparent or at day 601. For assessment of mice before tumor development, mice were killed at 5 to 7 weeks or 7 to 10 months of age. Tissues and tumors were paraffin-embedded, and sections were stained with hemtoxylin and eosin to identify tumor type and other associated pathology. Immunohistochemisty analysis was performed on tumor sections to confirm histopathologic examinations.
Cytokine and chemokine analysis
Measurement of cytokines and chemokines was performed on mouse sera incubated with the Bio-Plex Pro Mouse Cytokine 23-Plex Panel plate and processed according to the manufacturer's instructions (Bio-Rad). Data were collected and analyzed on a Bio-Plex suspension array system (Bio-Rad).
Results
Generation and characterization of Δ122p53
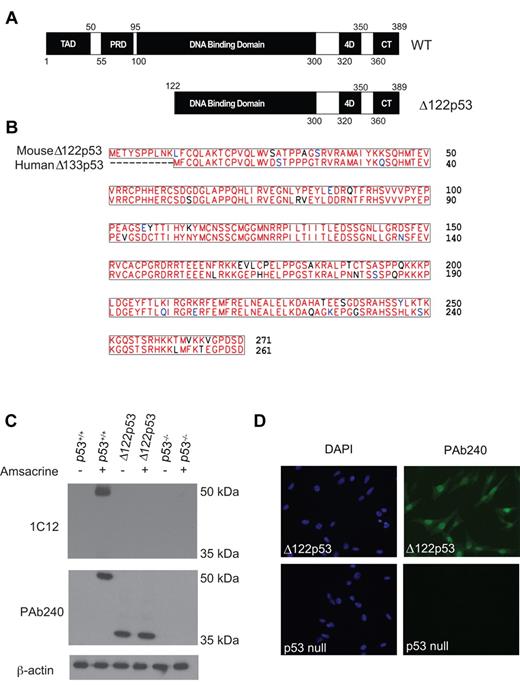
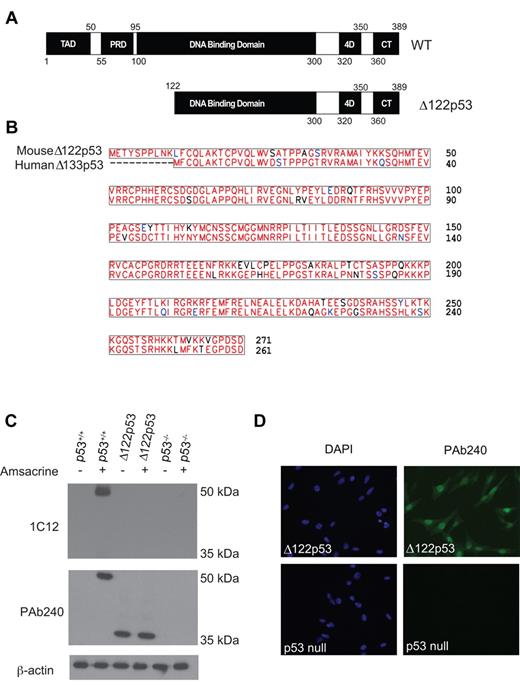
Mice deleted for the first 122 amino acids of p53 were constructed as outlined in the legend to supplemental Figure 1A. Sequencing showed correct splicing between exons 2 and 5 (supplemental Figure 1B), which with the use of an alternative translation start site in exon 2 generates a protein lacking the first 122 amino acid residues of mouse p53 (Figure 1A-B). Comparison of the predicted amino acid sequence shows it to be very similar to Δ133p53 (Figure 1B). Genotyping by PCR showed non-Mendelian inheritance of the Δ122p53 allele similar to the p53 null (p53−/−) allele (supplemental Figure 1C) in part because of exencephaly in female embryos as reported previously22 and confirmed in our cohort. Other developmental abnormalities were also noted, particularly enlarged brains in developing embryos, delayed fur development, and the absence of an optic nerve (data not shown); however, detailed analyses of these phenotypes have yet to be done.
Characterization of Δ122p53 mice. (A) Schematic diagram comparing full-length p53 and Δ122p53. Domains are identified as follows: transactivation (TAD), proline-rich (PRD), DNA binding, tetramerization (4D), and C-terminal regulatory (CT). (B) Protein alignment between human Δ133p53 and mouse Δ122p53 illustrating the high homology between the 2 protein sequences. (C) To detect Δ122p53 protein, adipocytes from p53+/+, p53−/−, and Δ122p53 mice were left untreated (−) or treated (+) with 1 μg/mL amsacrine for 5 hours, and expression was determined by Western blotting using antibodies PAb240 and 1C12. β-Actin (antibody AC-15) was used as a loading control. Experiments were repeated to investigate at least 3 mice per genotype. (D) Δ122p53 shows nuclear and cytoplasmic locations. Δ122p53-expressing fibroblasts or p53 null fibroblasts were fixed and then stained with DAPI and p53 antibody PAb240 and with an AlexaFluor-488 conjugated second antibody. Images were taken with an Olympus BX51-TRF fluorescent microscope with 20×/0.5 objective lens. They were captured using a SPOT Camera (SP 6.0) and Software Version 4.0.1. Images were cropped using Photoshop Version CS2 (Adobe).
Characterization of Δ122p53 mice. (A) Schematic diagram comparing full-length p53 and Δ122p53. Domains are identified as follows: transactivation (TAD), proline-rich (PRD), DNA binding, tetramerization (4D), and C-terminal regulatory (CT). (B) Protein alignment between human Δ133p53 and mouse Δ122p53 illustrating the high homology between the 2 protein sequences. (C) To detect Δ122p53 protein, adipocytes from p53+/+, p53−/−, and Δ122p53 mice were left untreated (−) or treated (+) with 1 μg/mL amsacrine for 5 hours, and expression was determined by Western blotting using antibodies PAb240 and 1C12. β-Actin (antibody AC-15) was used as a loading control. Experiments were repeated to investigate at least 3 mice per genotype. (D) Δ122p53 shows nuclear and cytoplasmic locations. Δ122p53-expressing fibroblasts or p53 null fibroblasts were fixed and then stained with DAPI and p53 antibody PAb240 and with an AlexaFluor-488 conjugated second antibody. Images were taken with an Olympus BX51-TRF fluorescent microscope with 20×/0.5 objective lens. They were captured using a SPOT Camera (SP 6.0) and Software Version 4.0.1. Images were cropped using Photoshop Version CS2 (Adobe).
To show that Δ122p53 is expressed at the protein level, adipocyte cells from Δ122p53 mice were treated with the topoisomerase II inhibitor amsacrine. Five hours later, cells were harvested, lysates prepared, and Western blotting carried out with an antibody to the N-terminus of p53 (1C12) and an antibody to the core domain beyond the deletion in Δ122p53 (PAb240). Results (Figure 1C) show that Δ122p53 is translated as a single protein species of approximately 37 kDa, detectable with antibody PAb240, but not with antibody 1C12. This result is consistent with the predicted size and absence of the N-terminus. Δ122p53 was readily detected in both untreated and amsacrine-treated cells, unlike wild-type p53 (p53+/+), which was only detectable after amsacrine treatment. This suggests that Δ122p53 is more stable than p53 in the absence of stress, presumably because of a lack of Mdm2-dependent degradation, which requires the N-terminus of p53.23 In addition to adipocytes (Figure 1C), Δ122p53 was also found to be expressed in several other tissues, including thymus, spleen, liver (supplemental Figure 2), lung, colon, and kidney (data not shown), but not in brain (supplemental Figure 2), indicating some tissue specific expression. Finally, immunofluorescence was carried out on Δ122p53-expressing fibroblasts using antibody PAb240, which shows Δ122p53 to be located in both the nucleus and the cytoplasm (Figure 1D) as has been shown for Δ133p53.5
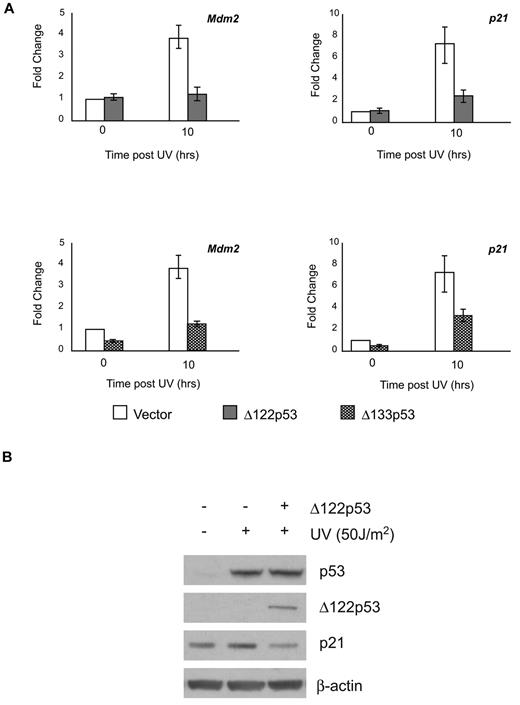
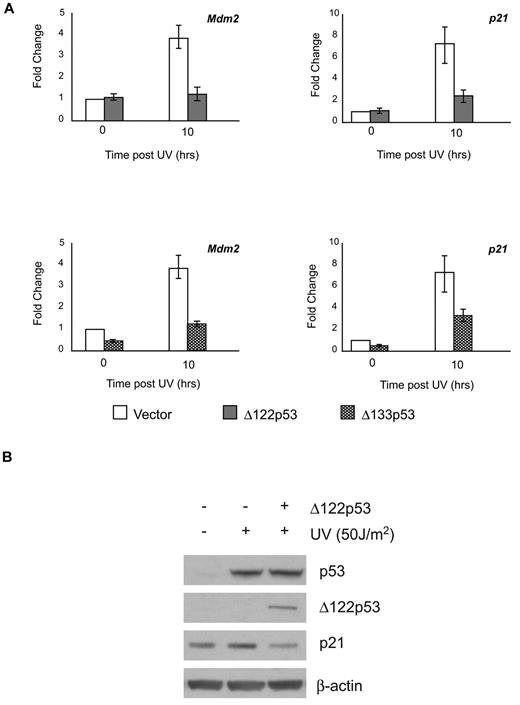
Several reports have shown that Δ133p53 can modulate full-length p53 functions.5,13,24 To determine whether this is also the case for Δ122p53, the ability of Δ122p53 to affect wild-type p53-dependent transactivation ability was determined. NIH3T3 cells were transduced with retroviruses containing Δ122p53 or Δ133p53. The resulting cell lines were exposed to ultraviolet irradiation and the ability of p53 to up-regulate cyclin dependent kinase inhibitor p21 and Mdm2 genes in the presence or absence of the isoforms was measured. This was done using quantitative RT-PCR for both genes and by Western blotting for p21. Results (Figure 2A upper panels) show that Δ122p53 attenuated wild-type p53 transactivation of both Mdm2 and p21 genes, which in the case of p21, also resulted in a lower level of p21 protein (Figure 2B). In addition, coexpression of Δ133p53 in these cells also attenuated the transactivation of Mdm2 and p21, indicating the functional parallel of the isoforms (Figure 2A lower panels).
Δ122p53 modulates p53 transactivation similar to Δ133p53. To demonstrate inhibition of p53-induced transcription by the Δ122p53 isoform, NIH3T3 cells were transduced with retrovirus vectors expressing Δ122p53 and Δ133p53. Vector-only control cells and Δ122p53- and Δ133p53-expressing cells were exposed to 50 J/m2 ultraviolet. (A) RNA was extracted after 10 hours, and quantitative RT-PCR was carried out for the p21 and Mdm2 genes. (B) Proteins were extracted after 6 hours, and p21 expression was determined by Western blotting. Experiments were repeated 3 times.
Δ122p53 modulates p53 transactivation similar to Δ133p53. To demonstrate inhibition of p53-induced transcription by the Δ122p53 isoform, NIH3T3 cells were transduced with retrovirus vectors expressing Δ122p53 and Δ133p53. Vector-only control cells and Δ122p53- and Δ133p53-expressing cells were exposed to 50 J/m2 ultraviolet. (A) RNA was extracted after 10 hours, and quantitative RT-PCR was carried out for the p21 and Mdm2 genes. (B) Proteins were extracted after 6 hours, and p21 expression was determined by Western blotting. Experiments were repeated 3 times.
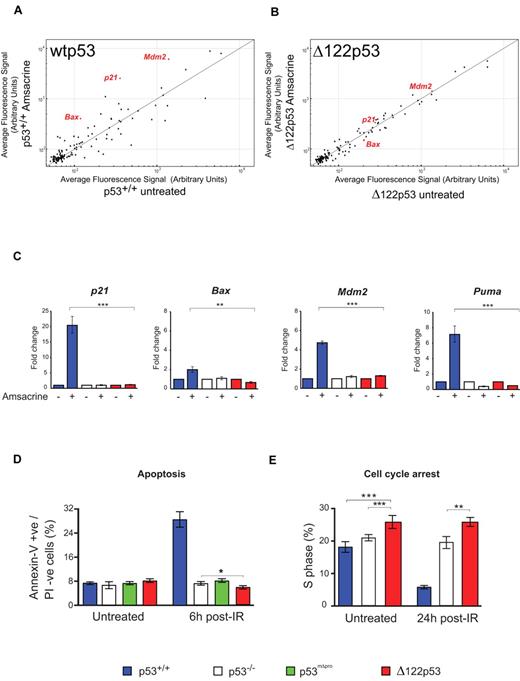
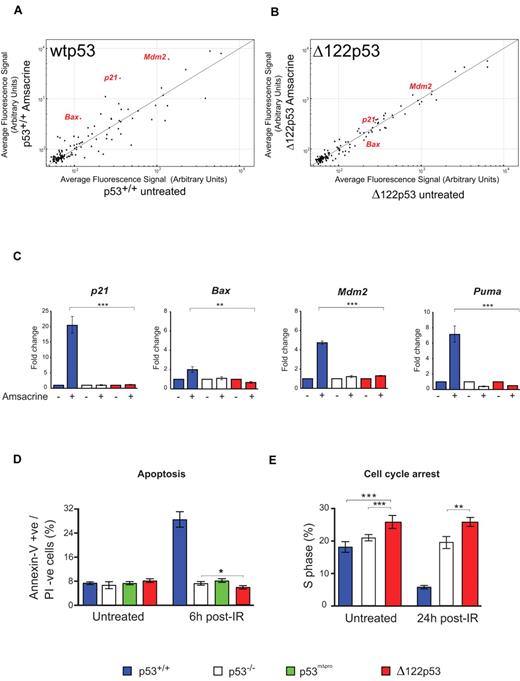
To test whether Δ122p53 can carry out the well-established functions of p53-dependent transactivation, cell cycle arrest, and apoptosis, splenocytes from mice of different genotypes were treated with amsacrine and the levels of p53 target transcripts determined. Microarray analysis of 125 putative p53 target genes25 showed Δ122p53 to be defective in regulation of many genes (Figure 3A-B), as expected for a p53 protein without the transactivation domain. This defect was confirmed using quantitative RT-PCR. Δ122p53 did not transactivate p21, Mdm2, or the proapoptotic Bax and Puma genes, after amsacrine treatment (Figure 3C). Next, apoptosis was measured after DNA damage. Thymocytes were treated with γ-irradiation, and apoptosis was measured by the annexin-V inclusion/propidium iodide exclusion method. Results (Figure 3D) showed a 4-fold increase in apoptosis in cells expressing wild-type p53 (p53+/+) after irradiation, but no increase in apoptosis was observed in p53−/− cells, cells expressing a p53 proline domain deletion (mΔpro) as previously reported,29,31 or in cells expressing Δ122p53. Indeed, the level of apoptosis in Δ122p53 cells was consistently lower than in p53−/− cells (P = .044). Reduced apoptosis compared with p53−/− cells was also found for Δ122p53 thymocytes treated with amsacrine (P = .009, data not shown). The absence of p53-mediated apoptosis and cell cycle arrest explains why Δ122p53 mice are not embryonic lethal, as would be expected in the absence of Mdm2 binding.32,33 Finally, the ability of Δ122p53 to arrest cell cycle progression was investigated. Mice were pulsed-labeled with BrdU and after γ-irradiation, bone marrow was isolated, and BrdU incorporation was measured by flow cytometry. Cells from wild-type (p53+/+) mice showed clear evidence of cell cycle arrest 24 hours after irradiation (Figure 3E), whereas cells from Δ122p53 mice did not. Instead, Δ122p53-expressing cells showed a reproducibly higher proportion of S phase than either p53+/+ or p53−/− cells irrespective of treatment (Figure 3E; P = .011 untreated Δ122p53 vs untreated p53−/−, and P = .005 γ-irradiated Δ122p53 vs treated p53−/−). Thus, in general, the data show that Δ122p53 has not retained the normal tumor suppressor functions of wild-type p53, similar to the human Δ133p53 isoform.5,13,16
Δ122p53 is defective for p53 functions. Splenocytes were treated with 1 μg/mL amsacrine, and RNA was isolated after 5 hours; p53 transcriptional profiling was carried out by microarray. (A) Comparison of the average fluorescence of 125 p53 target gene transcripts25 from p53+/+ mice with and without amsacrine was performed by plotting transcript levels from amsacrine-treated p53+/+ cells (y-axis) against untreated p53+/+ cells (x-axis). Transcripts above the line indicate up-regulation by wild-type p53; transcripts below the line, down-regulation; and transcripts on the line, the same or not regulated by this stress. (B) Comparison of the average fluorescence of the 125 p53 target gene transcripts from Δ122p53 mice; amsacrine-treated Δ122p53 cells (y-axis) are plotted against untreated Δ122p53 cells (x-axis). (C) Selected p53 target genes were validated by quantitative RT-PCR. Treated and untreated splenocytes from each of the genotypes were analyzed. Untreated samples were assigned a value of 1.0. **P < .001. ***P < .0001. (D) Thymocytes from p53+/+, p53−/−, p53mΔpro (mice homozygous for a deletion in the p53 proline domain), and Δ122p53 mice were irradiated with 4 Gy γ-radiation and harvested after 6 hours; apoptosis was assayed by staining for annexin V and propidium iodide. *P < .05. (E) Mice were treated with 2 Gy γ-radiation and 24 hours later pulsed with BrdU to label cells in S phase. Bone marrow was harvested, and the proportion of BrdU positive cells was determined by flow cytometry. **P < .01. ***P < .001. All experiments were performed using cells from 3 mice per genotype.
Δ122p53 is defective for p53 functions. Splenocytes were treated with 1 μg/mL amsacrine, and RNA was isolated after 5 hours; p53 transcriptional profiling was carried out by microarray. (A) Comparison of the average fluorescence of 125 p53 target gene transcripts25 from p53+/+ mice with and without amsacrine was performed by plotting transcript levels from amsacrine-treated p53+/+ cells (y-axis) against untreated p53+/+ cells (x-axis). Transcripts above the line indicate up-regulation by wild-type p53; transcripts below the line, down-regulation; and transcripts on the line, the same or not regulated by this stress. (B) Comparison of the average fluorescence of the 125 p53 target gene transcripts from Δ122p53 mice; amsacrine-treated Δ122p53 cells (y-axis) are plotted against untreated Δ122p53 cells (x-axis). (C) Selected p53 target genes were validated by quantitative RT-PCR. Treated and untreated splenocytes from each of the genotypes were analyzed. Untreated samples were assigned a value of 1.0. **P < .001. ***P < .0001. (D) Thymocytes from p53+/+, p53−/−, p53mΔpro (mice homozygous for a deletion in the p53 proline domain), and Δ122p53 mice were irradiated with 4 Gy γ-radiation and harvested after 6 hours; apoptosis was assayed by staining for annexin V and propidium iodide. *P < .05. (E) Mice were treated with 2 Gy γ-radiation and 24 hours later pulsed with BrdU to label cells in S phase. Bone marrow was harvested, and the proportion of BrdU positive cells was determined by flow cytometry. **P < .01. ***P < .001. All experiments were performed using cells from 3 mice per genotype.
Increased proliferation in Δ122p53-expressing cells
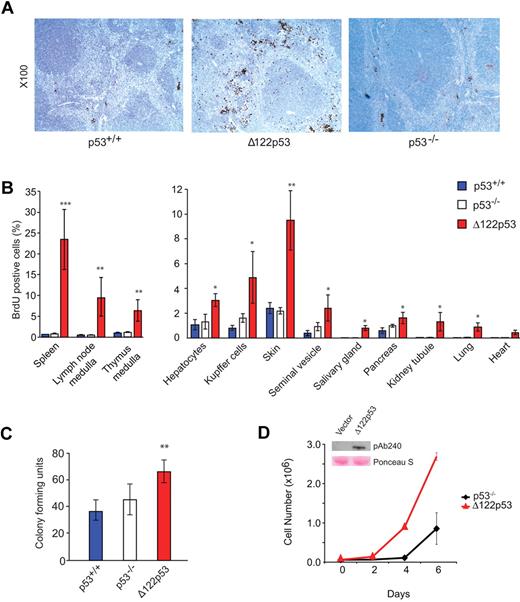
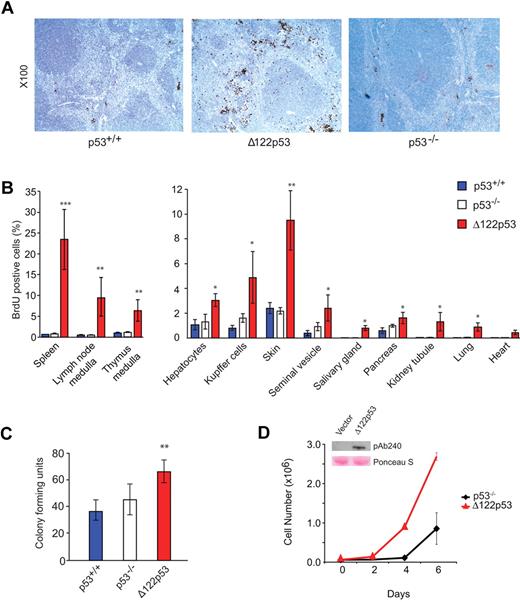
To explore further the elevated S phase observed in Δ122p53 bone marrow, the analysis was extended to other tissues. Wild-type p53, Δ122p53, and p53 null mice were pulse-labeled with BrdU and different tissues harvested, fixed, sectioned, and stained with an antibody to BrdU. The proportion of cells that stained positive for BrdU was then determined by direct counting. An example for spleen is shown in Figure 4A, and quantitation of this and for other tissues is shown in Figure 4B. In all cases, a higher proportion of BrdU+ cells was found in tissues from Δ122p53 mice compared with other genotypes but was particularly noticeable in the spleen in the red pulp region, and in the medulla region of the thymus, and to a lesser extent in the skin. In the case of the spleen, there was also notable organ enlargement. The details of this have yet to be investigated. Next, colony-forming assays were done using granulocyte and macrophage cells from the bone marrow of wild-type p53, p53 null, and Δ122p53 mice. Results (Figure 4C) show more colonies in cells from Δ122p53 mice than cells from mice of other genotypes. To exclude this phenotype being the result of other factors in the mice and not a direct effect of Δ122p53, the p53 null mouse fibroblast line 10–1 was transduced with a retrovirus expressing Δ122p53, and the growth rate of these cells was compared with the vector control. Results (Figure 4D) showed that Δ122p53-expressing cells proliferate faster than the control. Similar results were obtained with 2 additional transduced cell lines (data not shown). Thus, the increased proliferation observed here and in the tissues is a direct effect of Δ122p53.
Increased proliferative capacity of Δ122p53. (A) Photomicrographs illustrate more cells in S phase of spleens from Δ122p53 mice compared with p53+/+ and p53−/− mice. Mice were pulse-labeled with BrdU for 100 minutes to label proliferating cells. Organs were harvested, and BrdU+ cells were detected with an horseradish peroxidase-labeled antibody and light microscopy. Images were taken with a Leica DM 2000 microscope, Leica DFC 295 camera, and Leica Version 3.5.0 Application Suite (Software). Lens aperture was 0.3 with a 10× objective. Images were cropped using Photoshop Version CS2 (Adobe). (B) Quantitation of BrdU+ cells in different tissues. BrdU was administered for 100 minutes (thymus) or 24 hours (other organs). The proportion of BrdU+ cells per 2000 to 5000 total cells was determined. (C) Colony-forming assays show that Δ122p53 mice have more granulocyte and macrophage colonies compared with p53+/+ and p53−/− mice. Bone marrow was cultured in methylcellulose-based media, and colonies were counted at day 10. *P < .05. **P < .01. ***P < .001. All experiments were performed using at least 3 mice per genotype. Results are mean plus or minus SD. (D) The p53 null 10–1 fibroblasts transduced with the Δ122p53 retrovirus (red triangles) or vector control (black diamonds) were seeded at a density of 0.6 × 105 cells/35-mm dish, and viable cell numbers were determined over time by trypan blue counting. (Inset) Expression of Δ122p53 protein by Western blotting (PAb 240 antibody) in Δ122p53-transduced cells but not in the vector control. Lower (pink) bands illustrate equal loading by Ponceau S staining.
Increased proliferative capacity of Δ122p53. (A) Photomicrographs illustrate more cells in S phase of spleens from Δ122p53 mice compared with p53+/+ and p53−/− mice. Mice were pulse-labeled with BrdU for 100 minutes to label proliferating cells. Organs were harvested, and BrdU+ cells were detected with an horseradish peroxidase-labeled antibody and light microscopy. Images were taken with a Leica DM 2000 microscope, Leica DFC 295 camera, and Leica Version 3.5.0 Application Suite (Software). Lens aperture was 0.3 with a 10× objective. Images were cropped using Photoshop Version CS2 (Adobe). (B) Quantitation of BrdU+ cells in different tissues. BrdU was administered for 100 minutes (thymus) or 24 hours (other organs). The proportion of BrdU+ cells per 2000 to 5000 total cells was determined. (C) Colony-forming assays show that Δ122p53 mice have more granulocyte and macrophage colonies compared with p53+/+ and p53−/− mice. Bone marrow was cultured in methylcellulose-based media, and colonies were counted at day 10. *P < .05. **P < .01. ***P < .001. All experiments were performed using at least 3 mice per genotype. Results are mean plus or minus SD. (D) The p53 null 10–1 fibroblasts transduced with the Δ122p53 retrovirus (red triangles) or vector control (black diamonds) were seeded at a density of 0.6 × 105 cells/35-mm dish, and viable cell numbers were determined over time by trypan blue counting. (Inset) Expression of Δ122p53 protein by Western blotting (PAb 240 antibody) in Δ122p53-transduced cells but not in the vector control. Lower (pink) bands illustrate equal loading by Ponceau S staining.
In confirmation of increased proliferation, unsupervised analyses of microarray data (Figure 3) showed that Δ122p53-expressing spleen cells had a distinct gene expression profile compared with p53+/+ and p53−/− cells, irrespective of treatment (supplemental Figure 3). Functional classification showed that differentially expressed transcripts between the Δ122p53 and other genotypes were enriched for biologic themes representing antigen processing and presentation (eg, Fcgrt, H2-Oa, H2-DMa); regulation of the immune system (eg, Cd37, Cd79b); leukocyte activation (eg, Coro1a, Lst1, Thy1); cytoskeleton organization (eg, Flna, Was); cell adhesion (eg, Itgb7, Vcam1); inflammation mediated by chemokine and cytokine signaling (eg, Stat1, Junb); cell cycle (Cdk1, Cdkn1a, Mki67); and regulation of apoptosis (Birc5, Traf1) (supplemental Table 1). Of importance, the 5 top-scoring networks from biologic network analysis identified were enriched with ontologies for positive regulation of cell cycle and cell proliferation and negative regulation of apoptosis (supplemental Table 1).
Δ122p53 is an oncogene
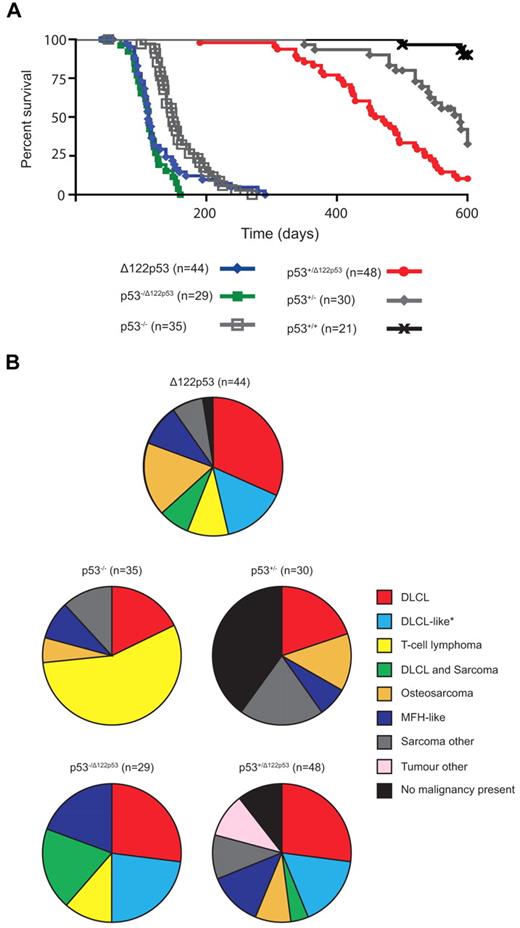
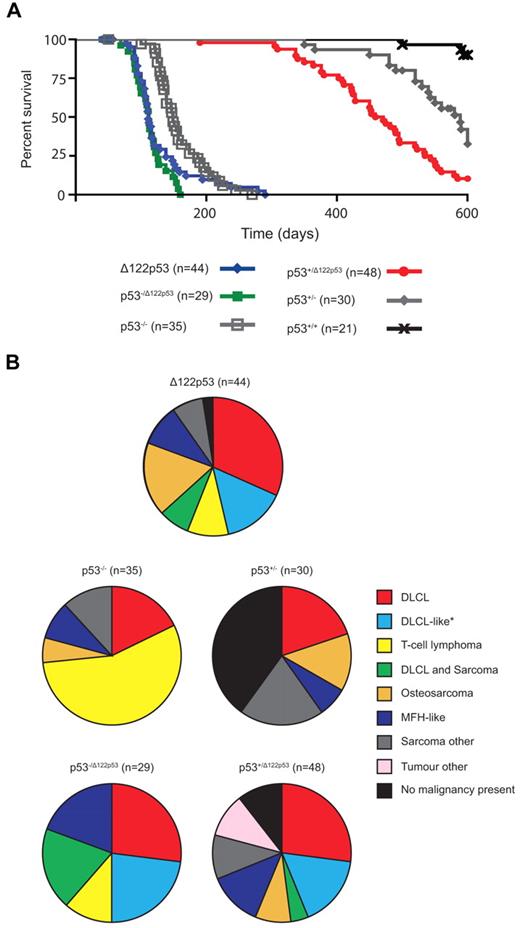
To address whether Δ122p53 mice are tumor prone, cohorts were followed over time and compared with p53+/+ and p53−/− mice. Δ122p53 mice had reduced survival compared with p53−/− mice with a median survival of 111 days compared with 149 days, respectively (P = .008; Figure 5A). The Δ122p53 allele was also associated with poorer survival in heterozygotes p53−/Δ122p53 (Figure 5A) and p53+/Δ122p53 (Figure 5A). The p53−/Δ122p53 mice had a significantly reduced survival to p53−/− mice (P < .0001), but similar survival to Δ122p53 homozygotes (median survival, 112 days; P = .3 comparison with Δ122p53). The p53+/Δ122p53 mice had reduced survival compared with p53+/− mice (median survival, 465 vs 589 days; P = .0001), with 90% of p53+/Δ122p53 mice developing a malignancy compared with only 60% of p53+/− mice. The median survival times for our p53−/− and p53+/− cohorts were similar to previous reports for such mice on a predominantly C57/BL6 background (p53−/−, ∼ 135-160 days).1,26,30
The Δ122p53 allele is associated with a reduced life span and altered tumor spectrum compared with the p53 null allele. (A) Mice of various genotypes were observed for 600 days and Kaplan-Meier curves plotted. (B) Relative frequencies of tumors identified by histopathologic examination are displayed as pie charts. n = cohort size − mice censored because of postnatal exencephaly.
The Δ122p53 allele is associated with a reduced life span and altered tumor spectrum compared with the p53 null allele. (A) Mice of various genotypes were observed for 600 days and Kaplan-Meier curves plotted. (B) Relative frequencies of tumors identified by histopathologic examination are displayed as pie charts. n = cohort size − mice censored because of postnatal exencephaly.
In addition to a higher tumor incidence, the Δ122p53 mice also had a different tumor spectrum compared with p53−/− mice (Figure 5B). Diffuse large B-cell lymphoma (DLBCL), positive for the B-cell marker B220, was the most prevalent tumor type (32%), followed by osteosarcoma (17%), and a DLBCL-like tumor (classified as DLBCL by morphology but negative for the B220 marker, 15%), and T-cell lymphoma (10%), common in p53−/− mice.26 Most of our p53−/− mice (56%) developed a T-cell lymphoma positive for the T-cell markers CD4 and CD8, with fewer developing DLBCL (18%, all B220+) and osteosarcoma (6%). Like Δ122p53 mice, p53−/Δ122p53 mice predominantly developed DLBCL: 24% were B220+ and 21% were the B220− DLBCL-like tumor, with fewer T-cell lymphomas (12%) (Figure 5B). The cell type responsible for the DLBCL-like tumor classification is currently under further investigation but it is negative for the T-cell markers CD4 and CD8. Three other differences specific to mice carrying the Δ122p53 allele, characteristic of a more aggressive tumor phenotype, were noted: multiple malignancies in the same animal, for example, DLBCL and sarcoma (19% of p53−/Δ122p53, 7% of Δ122p53, and 4% of p53+/Δ122p53); metastasized osteosarcomas (7 of 9 Δ122p53 and 4 of 4 p53+/Δ122p53); and malignant fibrous histiocytomas with angiogenesis (4 of 4 Δ122p53, 3 of 4 p53−/Δ122p53, and 5 of 7 p53+/Δ122p53 tumors). A range of other rare tumors was also observed in Δ122p53 mice, but not in mice of other genotypes (data not shown). Thus, these and the data in Figure 4 demonstrate that Δ122p53 has “gained” function and is indeed a dominant oncogene. Further to this point, no additional mutations were identified from sequencing the Trp53 gene from 10 tumors from Δ122p53 mice, and only 2 of 15 p53+/Δ122p53 tumors had lost the wild-type allele compared with 7 of 10 p53+/− tumors. This lack of loss of heterozygosity in p53+/Δ122p53 tumors suggests that loss of the wild-type p53 allele is not essential for tumor development.
Δ122p53 is associated with a proinflammatory phenotype
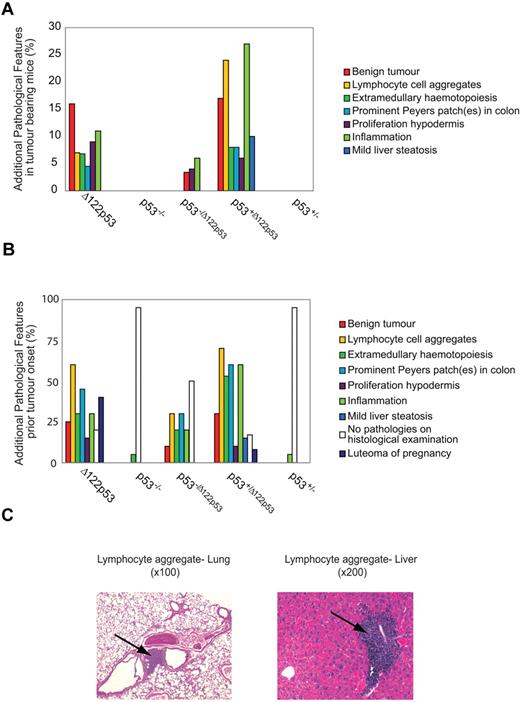
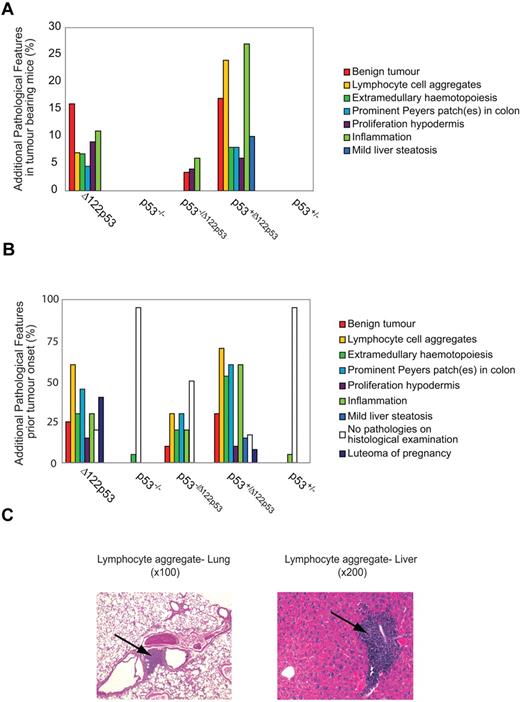
In addition to tumors, Δ122p53 mice displayed a wide range of inflammatory conditions, including lymphocyte aggregation in different organs (predominantly liver, lung, and adipose tissue), extramedullary hematopoiesis, prominent Peyer patches in the colon, and generalized inflammation. The frequencies of these additional pathologies are shown in Figure 6A as well as 2 examples of lymphocyte aggregation (Figure 6C). In addition, other examples of inflammatory pathologies are shown in supplemental Figure 4, including hamartomas and luteoma of pregnancy. These conditions are present only in mice with at least one Δ122p53 allele; 6% of Δ122p53 and 24% of p53+/Δ122p53 mice exhibited lymphocyte aggregations in different organs, extramedullary hematopoiesis, prominent Peyer patches in the colon, or generalized inflammation. Five Δ122p53 mice (11%) and 13 p53+/Δ122p53 mice (27%) showed inflammation: vasculitis affecting the skin, heart, aorta, kidney, or seminal vesicles vessels and/or lobulitis, hepatitis, or cirrhosis of the liver. At necropsy, 7 Δ122p53 mice (16%) had a benign tumor: 6 mice had benign and malignant tumors and 1 mouse had a large mass on the skin that met the death criteria for tumor burden. In p53+/Δ122p53 mice, 8 (17%) animals had benign masses. The most prominent benign mass for Δ122p53 and p53+/Δ122p53 mice was skin hamartoma (7), followed by colon hamartoma (3), lung adenoma (2), colon adenoma (2), and hibernoma (1).
The Δ122p53 allele is associated with nonmalignant proliferative pathologies and inflammation. The 8 most common additional pathologies found with the Δ122p53 allele are listed with their corresponding frequencies compared with other genotypes. Pathologies were identified from hematoxylin and eosin-stained tissue sections. (A) Additional pathologies in the tumor cohorts from Figure 5. Mice that developed DLBCLs and DLBCL-like lymphomas are not included. (B) Additional pathologies in mice killed before tumor development (5-7 weeks old: Δ122p53, p53−/Δ122p53, and p53−/−; 7-10 months old: p53+/Δ122p53 and p53+/−); female mice in the breeding cohort that developed luteoma were killed. Results were from 20 Δ122p53, 10 p53−/Δ122p53, 20 p53−/−, 30 p53+/Δ122p53, and 20 p53+/− mice. (C) Photomicrographs of hematoxylin and eosin-stained sections show lymphocyte aggregations in the lung and liver present in Δ122p53 mice. Magnifications are indicated. Photomicrographs were taken with a Zeiss Axioplan Universal Microscope with 10×/0.3 and 20×/0.8 objective lenses. Images were captured using a SPOT Camera (SP 6.0) and Software Version 4.6.1. Images were cropped using Photoshop Version CS2 (Adobe).
The Δ122p53 allele is associated with nonmalignant proliferative pathologies and inflammation. The 8 most common additional pathologies found with the Δ122p53 allele are listed with their corresponding frequencies compared with other genotypes. Pathologies were identified from hematoxylin and eosin-stained tissue sections. (A) Additional pathologies in the tumor cohorts from Figure 5. Mice that developed DLBCLs and DLBCL-like lymphomas are not included. (B) Additional pathologies in mice killed before tumor development (5-7 weeks old: Δ122p53, p53−/Δ122p53, and p53−/−; 7-10 months old: p53+/Δ122p53 and p53+/−); female mice in the breeding cohort that developed luteoma were killed. Results were from 20 Δ122p53, 10 p53−/Δ122p53, 20 p53−/−, 30 p53+/Δ122p53, and 20 p53+/− mice. (C) Photomicrographs of hematoxylin and eosin-stained sections show lymphocyte aggregations in the lung and liver present in Δ122p53 mice. Magnifications are indicated. Photomicrographs were taken with a Zeiss Axioplan Universal Microscope with 10×/0.3 and 20×/0.8 objective lenses. Images were captured using a SPOT Camera (SP 6.0) and Software Version 4.6.1. Images were cropped using Photoshop Version CS2 (Adobe).
To determine whether the inflammatory conditions occurred only in tumor-bearing animals (Figure 6A) or also in animals without tumors, young mice (5-7 weeks of age) of the Δ122p53, p53−/Δ122p53, and p53−/− genotypes and older mice (7-10 months of age) of the p53+/Δ122p53 and p53+/− genotypes were killed and examined histologically for inflammatory phenotypes. Results (Figure 6B) again show evidence of widespread inflammation associated with the presence of the Δ122p53 allele. Thirty percent of Δ122p53, 60% of p53+/Δ122p53, and 20% of p53−/Δ122p53 developed inflammation, most frequently vasculitis in at least one organ and/or liver lobulitis. The most frequent pathology before the onset of malignancy was aggregates of nonmalignant lymphocytes in at least one organ. This presented in 60% of Δ122p53, 70% of p53+/Δ122p53, and 30% of p53−/Δ122p53. In most cases, lymphocytes aggregated in the lung and liver (Figure 6C), followed by adipose tissue, kidney (25% of Δ122p53, and 30% of p53+/Δ122p53, and 15% of p53−/Δ122p53), and in p53+/Δ122p53 salivary glands (10%). Increased lymphocytes in Peyer patches presented in 45% of Δ122p53, 60% of p53+/Δ122p53, and 30% of p53−/Δ122p53. The “prominent” Peyer patch phenomenon was documented separately as it also met the criterion for hamartoma classification. Before malignancy, benign tumors presented in 15 (25%) of mice with a Δ122p53 allele but were absent in p53−/− and p53+/− mice. The most frequent masses were skin and colon hamartomas (8), followed by lung and colon adenomas (6). Also documented in Figure 6B is the development of luteomas: 40% of Δ122p53 and 8% of p53+/Δ122p53 female mice used in the breeding cohort.
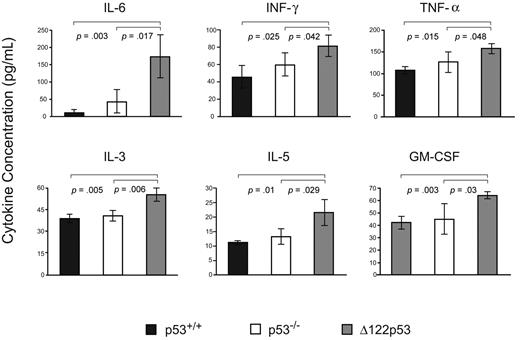
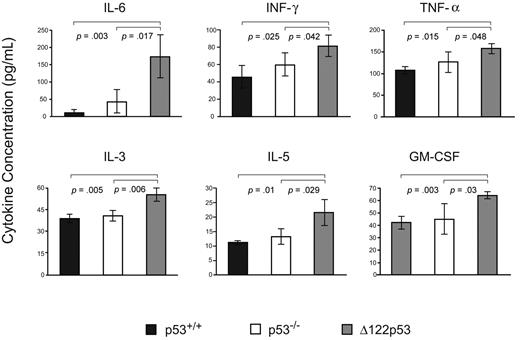
To explore the inflammatory phenotype further, sera from Δ122p53 mice were assayed for cytokines indicative of an inflammatory immune response and compared with sera from p53+/+ and p53−/− mice. Results (Figure 7) show elevated levels of interleukin-6 (IL-6), interferon-γ, tumor necrosis factor-α, IL-3, IL-5, and granulocyte-macrophage colony-stimulating factor compared with wild-type p53 mice and p53 null mice. Other cytokines measured in this assay that showed no significant changes between wild-type p53 or p53 null mice compared with Δ122p53 mice included IL-2, IL-13, IL-12p70, IL-12p40, IL-9, IL-10, IL-17, granulocyte colony-stimulating factor, keratinocyte derived chemokine (KC), and macrophage inflammatory protein-1α.
Inflammatory cytokine/chemokine profile associated with Δ122p53. Sera from 5- to 6-week-old male Δ122p53 and p53+/+ mice were assayed for 16 cytokines/chemokines using the Bio-Plex Pro Mouse Cytokine 23-Plex Panel plate and the Bio-Plex suspension array system (Bio-Rad). Cytokines/chemokines shown are those with significant differences between p53+/+ and p53−/− compared with Δ122p53 mice. Measurements were made from 3 serum samples per genotype. Cytokines levels are shown in picograms per milliliter.
Inflammatory cytokine/chemokine profile associated with Δ122p53. Sera from 5- to 6-week-old male Δ122p53 and p53+/+ mice were assayed for 16 cytokines/chemokines using the Bio-Plex Pro Mouse Cytokine 23-Plex Panel plate and the Bio-Plex suspension array system (Bio-Rad). Cytokines/chemokines shown are those with significant differences between p53+/+ and p53−/− compared with Δ122p53 mice. Measurements were made from 3 serum samples per genotype. Cytokines levels are shown in picograms per milliliter.
Discussion
The discovery that several human p53 isoforms are frequently present at elevated levels in several types of human tumor, combined with the ability of some isoforms to moderate full-length p53 functions, raises the possibility that one or more might be oncogenic. This was shown not to be the case for the Δ40p53 isoform9 using transgenic mice, but it has not been tested for other isoforms. We constructed a mutant p53 mouse that expresses a protein deleted for the first 122 amino acids, which is very similar to the human Δ133p53 isoform (Figure 1B). A series of experiments showed that Δ122p53 is unable to carry out any of the well-known functions associated with full-length p53 of transactivation, cell cycle arrest, and apoptosis but instead showed that Δ122p53 is able to confer a marked growth advantage on cells in vivo and in cell culture (Figure 4) and also showed a modest but consistent reduction in basal apoptosis (Figure 3).
Mice expressing Δ122p53 are highly tumor prone. The median survival of Δ122p53 mice is approximately 40 days shorter than p53 null mice, with all Δ122p53 mice displaying one or more tumors. Most tumors were of B-cell origin (∼ 50%) but approximately 20% were bone cancers and the remainder were a mixture of other tumor types. There was also a significant proportion of benign growths or hamartomas. These data, in conjunction with the lack of need for loss of heterozygosity in heterozygous p53+/Δ122p53 mice, imply that Δ122p53 is a dominantly acting oncogene.
Tumors were not observed in the Δ40p53 mice9 and not reported for another N-terminally truncated p53 mouse,28 but mice containing small deletions or point mutations in the N-terminus of p53 are tumor prone,29-31 predominantly developing B-cell tumors. In these cases, the tumors appeared much later than in Δ122p53 mice and are the result of impaired p53 function. Another N-terminal p53 mutant was embryonic lethal34 because of a marked increase in apoptosis in utero and possibly because of hypoxia. However, gain-of-function effects have been observed for missense p53 mutations in the DNA binding domain,27 which are the equivalent to commonly found mutations in human cancers. Mice heterozygous for the p53R172H mutation and wild-type p53 were associated with an altered tumor spectrum and/or an increased number of metastases compared with p53+/− mice,27 but no significant differences in survival time were found. Similarly, in a separate study, another gain-of-function tumor mutant mouse was constructed, p53R270H and a separate construction of p53R172H.35 Again, altered tumor spectrum was reported and the survival curves for heterozygous mice (both p53m/− and p53m/+) were indistinguishable from p53+/− mice. In 2 of these mutant strains, the frequency of B-cell lymphomas was markedly elevated compared with p53+/−35 mice; and in all cases, there was a marked increase in lymphomas and hematologic tumors in general, and bone cancers in some heterozygous mice.27,35 Some of the differences between these studies can be accounted for by background strain. As well as an altered tumor spectrum, cells derived from these mice had increased proliferative capacity similar to the Δ122p53 mice.27,35 Thus, the effects of Δ122p53 and the missense mutants have some common features. The biggest difference is the survival kinetics where, as an oncogene, Δ122p53 is much more aggressive than the other mutants.
Although we do not yet know the molecular basis of the increased proliferative capacity caused by Δ122p53, it could be in part the result of Δ122p53 binding and inactivating the other p53 family members, p63 and p73.36 Such a mechanism seems to underscore the proliferative gain of function observed for one of the p53R172H mutants.27 Alternatively, Δ122p53 might directly regulate genes involved in cell proliferation as tissues expressing Δ122p53 showed enrichment for transcripts in proliferation pathways. However, these 2 mechanisms are not necessarily mutually exclusive. Whatever the mechanism, increased proliferation plus the reduced basal levels of apoptosis would be sufficient to satisfy the basic requirements for oncogenic signaling, by analogy with Myc and Bcl2. Increased proliferation, reduced apoptosis, and impaired DNA repair (no p53) implies increased replicative stress, allowing a higher frequency of mutations to accumulate followed by genetic instability.37 Thus, the increased proliferative capacity of Δ122p53-expressing cells, combined with the tendency of B cells to undergo hypermutation,38 would be the simplest explanation for the tumor susceptibility of the Δ122p53 mice. Interestingly, we observed elevation of Birc5 and Plk1 in Δ122p53 mice, which are associated with poor prognosis in human DLBCL,39,40 suggesting that a pattern of gene expression that shares features of the transcriptional profile of human DLBCL precedes the development of measurable tumors in relatively young Δ122p53 mice. Thus, Δ122p53 mice may prove useful for modeling the initiation and progression of human DLBCL.
As well as the tumor phenotype, Δ122p53 mice also exhibit widespread inflammation that precedes the onset of malignancy, and this is accompanied by increased levels of proinflammatory cytokines in the serum, notably IL-6, interferon-γ, and IL-1β. Inflammation is not a widely documented pathology for p53−/− mice, nor is it documented for other mutant Trp53 mice.3,9,30,35 However, small increases in the levels of proinflammatory cytokines, including IL-6 and IL-1β, have been reported in p53 null mice,41,42 although this was much more evident after mitogen stimulation of B cells in vitro.42 Thus, wild-type p53 may suppress proinflammatory cytokine production and Δ122p53 elevates their levels. Further, our expression profiling data showed significant enrichment for transcripts implicated in immune system function and autoimmune disorders, such as systemic lupus erythematosus in Δ122p53 splenocytes compared with p53−/− and wild-type mice, which suggests that Δ122p53-driven disruption of normal immune function may underpin the inflammatory phenotype observed in Δ122p53 mice.
Consistent with wild-type p53 suppressing inflammation, functionally inactive p53 mutants have been implicated in the pathogenesis of some autoimmune diseases, including systemic lupus erythematosus,43 diabetes,44 and rheumatoid arthritis45 and in the last case has been specifically associated with increased levels of IL-6.45 This is not surprising as p53 is known to repress the IL-6 promoter.46 Interestingly, IL-6 is also elevated in a range of human cancers47 and is known to play diverse roles in oncogenicity, including increasing tumor cell proliferation and promoting angiogenesis48 and metastases.49 Other inflammatory pathways have also been linked with oncogenesis.50 Thus, it is plausible that, in addition to the proliferative advantage of Δ122p53, the release of proinflammatory cytokines into the serum of Δ122p53 mice could also contribute to tumorigenesis. This possibility is currently being tested.
In conclusion, given the structural and functional similarities between Δ122p53 and Δ133p53, we propose that the Δ133p53 isoform is an oncogene that also plays an important role in inflammation, which if deregulated, might contribute to inflammatory diseases and cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Wiles, M. Fisher, B. Li, N. Bennett, J. North, and M. Schultz for technical assistance and advice; C. Fitzpatrick and S. Young for assistance with the Bio-Plex analysis; C. Stocking (Hamburg) for the retroviral vector; and I. Morison and S. Cohen for commentary on the study.
This work was supported by the Royal Society of New Zealand Marsden Fund, the Health Research Council of New Zealand, the Dunedin School of Medicine Dean's Bequest Fund, and the Cancer Institute NSW.
Authorship
Contribution: T.L.S. designed and conducted the experiments and cowrote the paper; N.H. and G.W. carried out the histopathologic analyses; H.C. designed and conducted experiments, analyzed the microarray data, and cowrote the manuscript; A.J. was involved in microarray analysis and editing of the manuscript; M.W. assisted with flow cytometric analyses; P.R. assisted with gene expression experiments; A.E. constructed the retroviruses and transduced cell lines; C.R. carried out experiments with retrovirus Δ122p53 and performed immunoblotting experiments; R.M. carried out most of the gene expression experiments; J.A.R. and M.A.B. designed experiments, commented on the manuscript, and are principal investigators; and A.W.B. designed the mΔpro mice, designed experiments, cowrote the paper, and is a principal investigator.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Antony W. Braithwaite, Children's Medical Research Institute, University of Sydney, Locked Bag 23, Wentworthville NSW 2145, Australia; e-mail: abraithwaite@cmri.usyd.edu.au.