Abstract
Deregulation of the myeloid key transcription factor CEBPA is a common event in acute myeloid leukemia (AML). We previously reported that the chaperone calreticulin is activated in subgroups of AML patients and that calreticulin binds to the stem loop region of the CEBPA mRNA, thereby blocking CEBPA translation. In this study, we screened for additional CEBPA mRNA binding proteins and we identified protein disulfide isomerase (PDI), an endoplasmic reticulum (ER) resident protein, to bind to the CEBPA mRNA stem loop region. We found that forced PDI expression in myeloid leukemic cells in fact blocked CEBPA translation, but not transcription, whereas abolishing PDI function restored CEBPA protein. In addition, PDI protein displayed direct physical interaction with calreticulin. Induction of ER stress in leukemic HL60 and U937 cells activated PDI expression, thereby decreasing CEBPA protein levels. Finally, leukemic cells from 25.4% of all AML patients displayed activation of the unfolded protein response as a marker for ER stress, and these patients also expressed significantly higher PDI levels. Our results indicate a novel role of PDI as a member of the ER stress–associated complex mediating blocked CEBPA translation and thereby suppressing myeloid differentiation in AML patients with activated unfolded protein response (UPR).
Introduction
C/EBP-α (CEBPA) is a basic leucine zipper transcription factor crucial for normal neutrophil differentiation.1-3 Targeted disruption of CEBPA leads to a selective block in early granulocyte maturation resulting in the accumulation of myeloid blasts in the BM.4 Deregulation of CEBPA function by genomic mutations, transcriptional and posttranscriptional suppression, or phosphorylation-dependent inactivation is a common event in subgroups of acute myeloid leukemia (AML) patients, suggesting an important role of CEBPA both for normal myeloid differentiation and leukemogenesis.5-11
The endoplasmic reticulum (ER) is (among others) essential for protein synthesis and maturation, and for calcium storage. Impairment of these functions, such as disruption of Ca2+ homeostasis, inhibition of disulfide bond formation, or hypoxia, leads to ER stress which is characterized by the accumulation of unfolded or misfolded proteins that trigger the unfolded protein response (UPR).12 The UPR initiates transient attenuation of protein translation, degradation of misfolded proteins, and induction of molecular chaperones like calreticulin, GRP78, and CHOP.13
The protein disulfide isomerase (PDI) is a thiol-disulfide oxidoreductase resident in the ER lumen. The C-terminal peptide sequence -KDEL is crucial for ER retention of the protein. In addition, PDI contains a CXXC motif, with the 2 cysteine residues being responsible for the exchange of disulfide bonds with targeted peptides. Thereby, the enzyme catalyzes formation of disulfide bonds and isomerization of newly synthesized proteins.14-16 In addition, PDI acts as a chaperone and is a part of the quality-control system ensuring correct folding of proteins.17,18 Some reports also indicated a role of PDI in processes involving ER-associated degradation, trafficking, and calcium homeostasis.19,20
In this study, we identified PDI as a protein that binds to the stem loop region of the CEBPA mRNA, thereby blocking CEBPA expression at the translational level. We observed that induction of ER stress in leukemic cell lines resulted in activation of PDI thereby mediating CEBPA protein suppression. In addition, AML patient samples displaying activation of the UPR expressed significantly increased PDI levels. PDI also directly binds to the calreticulin protein, suggesting the formation of a protein complex ultimately regulating CEBPA translation. In conclusion, our results indicate a novel role of PDI during ER stress involving suppression of CEBPA protein and thereby blocking myeloid differentiation in AML patients with activation of the UPR.
Methods
Patient samples
Malignant cells were collected at diagnosis before initiation of treatment by BM aspiration from consecutive AML patients diagnosed between 2002 and 2009 using standard morphology and immunophenotype markers at the Department of Oncology, University Hospital (Berne, Switzerland). Informed consent was obtained according to the Declaration of Helsinki, approved by decisions of the local ethics committee of Berne, Switzerland. Mononucleated cells were isolated by Ficoll separation. Isolation of granulocytes and monocytes was performed on RoboSep (StemCell Technologies) with CD66b and CD14 selection kits, respectively. Detection of the XBP1 spliced variant was performed as described previously.21
Cell culture, transient transfection, and ER stress induction
The human leukemic cell lines HL60, U937, and K562, and the lung cancer cell line H1299 were grown at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% FBS (PAA Laboratories). For forced PDI expression, H1299 cells were transfected with pcDNA 3.1/V5-His-TOPO plasmid harboring a 1.5-kb KpnI/XbaI PDI or PDI/ΔKDEL fragment and pcDNA3 containing a 1.4-kb BamHI/EcoRI CEBPA or CEBPB cDNA using Lipofectamine 2000 (Invitrogen). HL60 and U937 cells were transfected with 2 μg of pcDNA 3.1/V5-His-TOPO containing the PDI cDNA by electroporation using nucleofection kit V (Lonza). For induction of ER stress, cells were incubated with 3 μg/mL tunicamycin or 3.7 μg/mL calcimycin (both from Sigma-Aldrich).
RNA probes and labeling
The CEBPA dsRNA oligonucleotides encoding the stem loop region had the sequence: A: 5′-ccgugggccccacgggcggcggcggcggcggcgacuu-3′, and B: 5′-ccgccgccgccgccgccgcccucgcacccgcacccgcacccg-3′, with the stem area shown in bold. Annealing was performed by heating equal amounts of oligonucleotides A and B at 95°C. The double-stranded CEBPA stem RNA was separated from single-stranded oligomers by gel electrophoresis and extraction. The CEBPA dsRNA oligo was labeled using T4 kinase and [γ-32P] ATP.
Protein fractionation by DEAE columns, ion exchange HPLC, and protein sequencing
To separate RNA and proteins, whole-cell extracts from K562 and HL60 cells were loaded on diethylaminoethyl (DEAE) sepharose in DEAE binding buffer (25mM Tris-HCl at pH 7.5 and 10mM β-mercapto-ethanol). After extensive washing, proteins were eluted using 0.1, 0.2, 0.3, 0.4, and 0.5M NaCl fractions. Elution fractions were desalted and cleared through centrifugal filter units (Millipore). Whole-cell extracts from mouse liver tissue were fractionated by HPLC-based ion exchange chromatography using UnoQ columns (Bio-Rad). The concentration of the proteins was performed using NaCl gradients (first step at 0.05-0.35M, second step at 0.1-0.25M, and third step at 0.12-0.16M). Proteins were then separated using SDS-PAGE, and the gel was stained by Coomassie blue. After tryptic digestion, mass spectrometry was performed on an Applied Biosystems 4700 Proteomics Analyzer.
UV cross-link and RNA gel-shift
Equal amounts of labeled RNA CEBPA stem probe were incubated with proteins for 50 minutes at room temperature in binding buffer (20mM Tris HCL pH 7.5, 150mM NaCl, 5mM DTT). Samples were exposed to UV treatment (3 times at 1500 μJ). The mixture was run on SDS-PAGE gels and transferred to nitrocellulose membrane with subsequent autoradiography. For RNA gel shift, the samples were incubated for 50 minutes in the presence of Abs and run on 6% native PAGE gels with subsequent autoradiography.
Subcellular fractionation and immunoblotting
One hundred micrograms of cell lysates in RIPA buffer were resolved by SDS-PAGE and transferred to nitrocellulose membrane. Cell fractionation (cytoplasm, membranous organelles, and nuclei) was performed as described.22 Membranes were incubated with rabbit anti-CEBPA (EP708Y; 1:10 000; Abcam), rabbit anti-CEBPB (C-19), rabbit anti-PDI (H-160), goat anti-calreticulin (C-17; sc-6467), mouse anti-V5 (sc-81775), mouse anti–lamin B (sc-6216; all 1:1000; Santa Cruz Biotechnology) or mouse anti–β-actin (1:10 000; Sigma-Aldrich) Abs. Anti–goat, anti–mouse, and anti–rabbit HRP-conjugated secondary Abs (all 1:10 000; GE Healthcare) were applied.
siRNA knockdown
For siRNA knockdown experiments, HL60 cells and U937 cells were transiently transfected with siRNA against PDI (P4HB Silencer Select ID:s439; Ambion) and negative control siRNA (Microsynth) by electroporation using nucleofection kit V for HL60 cells or nucleofection kit C for U937 cells (Lonza). Cells were harvested 48 hours after transfection.
Coimmunoprecipitation
Three hundred micrograms of U937 whole-cell lysates were incubated with 1 μg of rabbit anti-PDI (H-160; sc-20132) or mouse anti-calreticulin (1G6A7; sc-101436; Santa Cruz Biotechnology) at 4°C overnight. The mixture was bound to protein A sepharose beads (GE Healthcare) for 3 hours at 4°C. Washing was performed 3 times using RIPA buffer, and immunoprecipitates were eluted by adding 200mM glycine (pH 2.8) at room temperature. After elution, 80 μg of proteins were run on SDS-PAGE gels and detected by rabbit anti-PDI (1:1000; Abcam) or goat anti-calreticulin (C-17; sc-6467; 1:1000; Santa Cruz Biotechnology).
Quantitative RT-PCR
RNA was extracted using the Qiagen RNAeasy kit and transcribed with MMLV-reverse transcriptase (Promega) and random primers (Roche). RT-PCR was performed on the Applied Biosystems 7500 Ft Real-Time PCR System. Cycle threshold (Ct) values were normalized against the 3 housekeeping genes ABL, PBGD, and GAPDH and analyzed according to the 2-ΔΔCt method by Livak.23 Ct values derived from cell lines were normalized against PBGD. The mean ΔCt value of all AML-M0 patients served as reference (1-fold).
Statistical analysis
Mean values and SDs were calculated. Statistical analysis of RT-PCR results was performed by using the unpaired t test and Pearson correlation. Calculation was performed using Graphpad Prism 5 (2007) software.
Results
Protein disulfide isomerase binds to the stem loop region of CEBPA mRNA
Previously, we identified the chaperone calreticulin as a CEBPA mRNA binding protein in myeloid cells. The calreticulin-CEBPA mRNA interaction is mediated by 2 GC-rich repetitive sequences (nt 295-310 and nt 547-565) within the CEBPA coding sequence, and these 2 repetitive sequences form a stem loop secondary mRNA structure.24 We also showed that calreticulin is induced in certain types of AML—such as in the presence of the leukemogenic fusion protein CBFB-MYH11—and that calreticulin thereby blocks the translation of CEBPA mRNA in these leukemias. However, the analysis of complexes binding to the CEBPA dsRNA oligonucleotide consistently indicated a slowly migrating additional activity, besides the faster migrating complex which has been identified to be calreticulin.
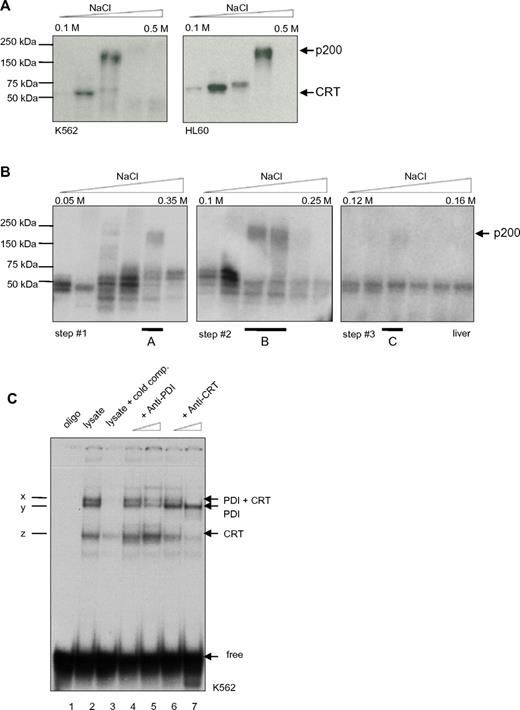
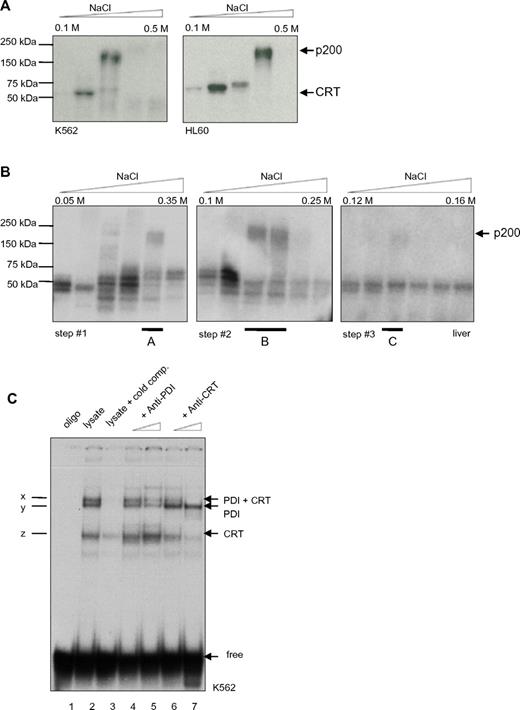
To further characterize this second binding complex, double-stranded radiolabeled CEBPA RNA oligonucleotides were designed encoding the repetitive stem loop sequences. Protein lysates from the leukemic cell lines K562 and HL60 were prepared for analysis of RNA-protein binding complexes by diethylaminoethyl (DEAE) fractionation. Then, we cross-linked the RNA-protein interaction using UV treatment and separated the RNA-protein complexes by SDS-PAGE. Consistently, we detected 2 binding activities, at 55 kDa and at 200 kDa (Figure 1A). In accordance with previous reports, we expected the faster migrating complex to be calreticulin.
PDI binds to the CEBPA mRNA. (A) UV cross-link analysis. Radiolabeled CEBPA dsRNA oligo (encoding the RNA stem motif) was incubated with DEAE elution fractions (from 0.1M to 0.5M NaCl) of K562 or HL60 whole-cell lysates, linked by UV treatment, and separated by SDS-PAGE electrophoresis. Position of calreticulin (CRT) and the 200-kDa complex are shown on the right. (B) Whole-cell lysates from mouse liver were analyzed by UV cross-link to the CEBPA dsRNA oligonucleotide and subsequently separated by ion exchange chromatography using UnoQ columns. Proteins were eluted along a linear gradient from 0.12M to 0.16M NaCl. Fraction A of step 1, fraction B of step 2, and fraction C of step 3 were each loaded on UnoQ column, with adjustment of the linear gradient, and again analyzed by UV cross-link. The 200-kDa complex in fraction C was excised and analyzed by mass spectrometry. (C) RNA gel-shift assay. The radiolabeled CEBPA dsRNA oligonucleotide was incubated with the cytoplasmic fraction of K562 cell lysates. Oligo: no lysates (lane 1); cold competition: unlabeled oligonucleotides were added in excess (lane 3); increasing amounts of PDI (lanes 4 and 5) or calreticulin (lanes 6 and 7) Ab were added; complex x indicates presence of PDI and calreticulin (CRT) protein, y: only PDI; and z: only CRT; free: position of the unbound RNA-stem probe added in excess.
PDI binds to the CEBPA mRNA. (A) UV cross-link analysis. Radiolabeled CEBPA dsRNA oligo (encoding the RNA stem motif) was incubated with DEAE elution fractions (from 0.1M to 0.5M NaCl) of K562 or HL60 whole-cell lysates, linked by UV treatment, and separated by SDS-PAGE electrophoresis. Position of calreticulin (CRT) and the 200-kDa complex are shown on the right. (B) Whole-cell lysates from mouse liver were analyzed by UV cross-link to the CEBPA dsRNA oligonucleotide and subsequently separated by ion exchange chromatography using UnoQ columns. Proteins were eluted along a linear gradient from 0.12M to 0.16M NaCl. Fraction A of step 1, fraction B of step 2, and fraction C of step 3 were each loaded on UnoQ column, with adjustment of the linear gradient, and again analyzed by UV cross-link. The 200-kDa complex in fraction C was excised and analyzed by mass spectrometry. (C) RNA gel-shift assay. The radiolabeled CEBPA dsRNA oligonucleotide was incubated with the cytoplasmic fraction of K562 cell lysates. Oligo: no lysates (lane 1); cold competition: unlabeled oligonucleotides were added in excess (lane 3); increasing amounts of PDI (lanes 4 and 5) or calreticulin (lanes 6 and 7) Ab were added; complex x indicates presence of PDI and calreticulin (CRT) protein, y: only PDI; and z: only CRT; free: position of the unbound RNA-stem probe added in excess.
To analyze the high molecular complex, we performed ion exchange HPLC protein separation. Protein lysates derived from mouse liver tissue were used because large amounts of material were needed for the various steps of separation and also because the calreticulin-CEBPA RNA interaction was first described in liver tissue.24
Whole-cell liver lysate fractions were UV cross-linked with the radiolabeled CEBPA dsRNA oligonucleotide and separated by SDS-PAGE (Figure 1B). The fraction containing the 200-kDa complex was isolated and separated by several steps with adjusted salt gradients. Finally, the isolated 200-kDa complex was analyzed using mass spectrometry. We obtained the amino acid sequence VHSFPTLKFFPASADRT, which corresponded to PDI.
To verify the mass spectrometry result, we performed RNA gel-shift assays. The CEBPA dsRNA oligonucleotide was incubated with whole-cell lysates from the leukemic cell line K562 and native PAGE was performed. Three major binding activities (arbitrarily denoted x, y, and z) were observed (Figure 1C). We applied increasing amounts of PDI or calreticulin Abs to the reaction mix for competition. We found that the PDI Ab abolished binding activities x and y, whereas the calreticulin Abs decreased bands x and z, respectively. This confirms that both PDI and calreticulin are in fact binding to the CEBPA dsRNA oligo.
PDI blocks CEBPA translation
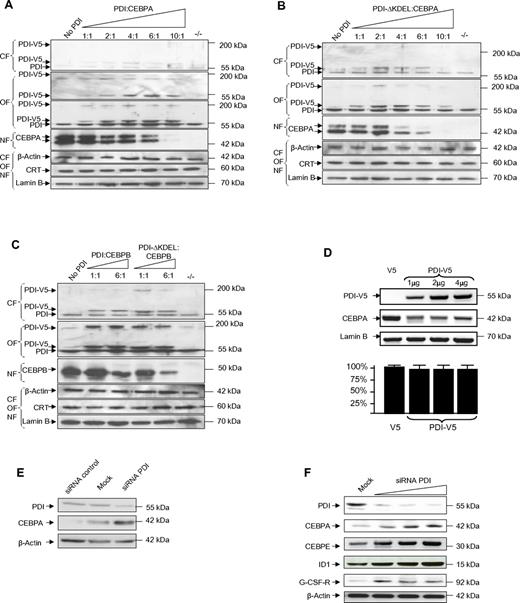
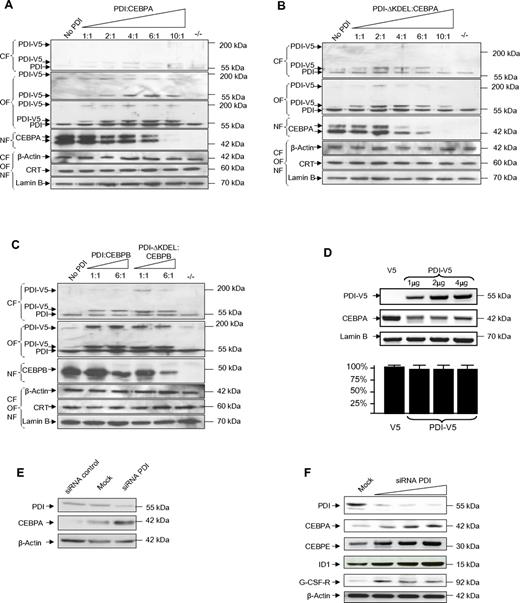
Because binding of calreticulin to the CEBPA mRNA blocked CEBPA translation, we analyzed whether PDI binding similarly affected CEBPA expression. We transfected CEBPA-deficient H1299 cells with increasing amounts of the expression vectors pcDNA3/V5-His encoding for PDI, and with constant amounts of pcDNA3 encoding for CEBPA. Subcellular fractions from cytoplasm, organelles, or nuclei were analyzed by Western blot. Besides its size of 55 kDa, PDI protein is also detectable in Western blot analysis as a tetrameric complex of roughly 200 kDa. We observed that the fusion protein PDI-V5, detectable with both V5- and PDI-specific Abs, was predominantly localized in the organelle fraction and only to a minor part in the cytoplasm. Transfection with increasing amounts of PDI-V5 and stable amounts of CEBPA-expressing vectors indicated significant suppression of CEBPA protein (Figure 2A).
Forced expression of PDI suppresses CEBP proteins. (A-C) Western blot analyses of subcellular fractions from lung cancer cells H1299 cells transiently transfected with pcDNA3/PDI-V5-His and pcDNA3/CEBPA (A), pcDNA3/PDI-ΔKDEL-V5-His and pcDNA3/CEBPA (B), and pcDNA3/PDI-V5-His or pcDNA3/PDI-ΔKDEL-V5-His and pcDNA3/CEBPB (C) as indicated. The membranes were stained with V5 (top panel of the organelle fraction in panel A only), with PDI, CEBPA, and CEBPB Abs. The loading controls were β-actin for the cytoplasmic fraction (CF), calreticulin for the organelle fraction (OF), and lamin B for the nuclear fraction (NF). (D top panel) Western blot of whole-cell lysates from leukemic HL60 cells transfected with pcDNA3/V5-His (lane 1) or increasing amounts of pcDNA3/PDI-V5-His (lanes 2-4) and stained with PDI and CEBPA Abs. Relative densitometric quantitation of CEBPA bands indicated: 100%, 26%, 24%, and 28% (lanes 1-4). Lamin B was used as loading control. (Bottom panel) Results from 3 independent experiments of quantitative RT-PCR for CEBPA mRNA, normalized against ABL expression. (E) Western blot of whole-cell extracts from HL60 cells after transfection of unspecific siRNA as a control, of siRNA against PDI (each 900nM), and of untransfected (mock) cells, stained with PDI and CEBPA Abs. β-actin was used as loading control. (F) Western blot of whole-cell lysates from U937 cells after transfection with increasing amounts of siRNA against PDI at concentrations of 300nM, 600nM, 900nM, and of untransfected (mock) cells. Relative densitometric quantitation of CEBPA bands indicated: 18%, 46%, 92%, and 100% (lanes 1-4).
Forced expression of PDI suppresses CEBP proteins. (A-C) Western blot analyses of subcellular fractions from lung cancer cells H1299 cells transiently transfected with pcDNA3/PDI-V5-His and pcDNA3/CEBPA (A), pcDNA3/PDI-ΔKDEL-V5-His and pcDNA3/CEBPA (B), and pcDNA3/PDI-V5-His or pcDNA3/PDI-ΔKDEL-V5-His and pcDNA3/CEBPB (C) as indicated. The membranes were stained with V5 (top panel of the organelle fraction in panel A only), with PDI, CEBPA, and CEBPB Abs. The loading controls were β-actin for the cytoplasmic fraction (CF), calreticulin for the organelle fraction (OF), and lamin B for the nuclear fraction (NF). (D top panel) Western blot of whole-cell lysates from leukemic HL60 cells transfected with pcDNA3/V5-His (lane 1) or increasing amounts of pcDNA3/PDI-V5-His (lanes 2-4) and stained with PDI and CEBPA Abs. Relative densitometric quantitation of CEBPA bands indicated: 100%, 26%, 24%, and 28% (lanes 1-4). Lamin B was used as loading control. (Bottom panel) Results from 3 independent experiments of quantitative RT-PCR for CEBPA mRNA, normalized against ABL expression. (E) Western blot of whole-cell extracts from HL60 cells after transfection of unspecific siRNA as a control, of siRNA against PDI (each 900nM), and of untransfected (mock) cells, stained with PDI and CEBPA Abs. β-actin was used as loading control. (F) Western blot of whole-cell lysates from U937 cells after transfection with increasing amounts of siRNA against PDI at concentrations of 300nM, 600nM, 900nM, and of untransfected (mock) cells. Relative densitometric quantitation of CEBPA bands indicated: 18%, 46%, 92%, and 100% (lanes 1-4).
Remarkably, larger amounts of a mutant PDI protein (PDI-ΔKDEL) lacking the C-terminal KDEL sequence (lysine-aspartate-glutamate-leucin) were detectable in the cytoplasm compared with the experiments with the wild-type PDI protein. This observation underlines the previously described function of the KDEL motif as an ER retention signal. In addition, the PDI-ΔKDEL protein was more efficient in blocking CEBPA translation (Figure 2B). These data suggest that PDI binding to the CEBPA mRNA in fact blocks CEBPA translation.
Calreticulin mRNA binding and its translation block of target mRNAs are not exclusive for CEBPA because we previously reported that calreticulin similarly affected CEBPB translation. Consequently, we also observed that PDI and PDI-ΔKDEL proteins decreased CEBPB protein expression, with the mutant PDI protein again being more potent in blocking CEBP translation than the wild-type protein (Figure 2C).
In accordance to these observations in the lung cancer cell line H1299, we also found in leukemic HL60 cells that expression of exogenous PDI inhibited CEBPA protein whereas CEBPA mRNA remained unchanged (Figure 2D). Finally, we depleted PDI protein levels by specific siRNA in leukemic HL60 (Figure 2E) and U937 cells (Figure 2F). In a dose-dependent manner, we observed enhancement of CEBPA protein after knockdown of PDI expression. Increasing CEBPA levels were associated with higher levels of the CEBPA target proteins CEBPE, ID1, and G-CSF–receptor. These data suggest that PDI can efficiently inhibit CEBPA protein expression in human myeloid leukemic cells.
PDI directly interacts with calreticulin
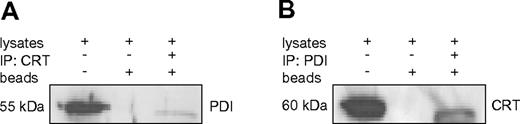
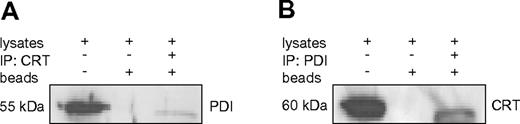
Our experiments above indicated that the chaperone PDI binds to the stem loop region of the CEBPA mRNA. Together with our previous report that calreticulin targets this same mRNA region, we evaluated a direct interaction between these 2 RNA binding proteins. We performed coimmunoprecipitation with specific Abs against PDI and calreticulin. Staining of the immunoprecipitates with PDI and then calreticulin Abs (and vice versa), in fact, indicated a direct interaction between these 2 proteins (Figures 3).
PDI interacts with calreticulin in U937 cells. (A-B) Western blot of coimmunoprecipitation samples from U937 cell lysates. Whole-cell lysates, sepharose beads without Ab incubation, and immunoprecipitates were loaded and probed with PDI and calreticulin (CRT) Abs as indicated.
PDI interacts with calreticulin in U937 cells. (A-B) Western blot of coimmunoprecipitation samples from U937 cell lysates. Whole-cell lysates, sepharose beads without Ab incubation, and immunoprecipitates were loaded and probed with PDI and calreticulin (CRT) Abs as indicated.
Induction of ER stress is associated with activation of PDI
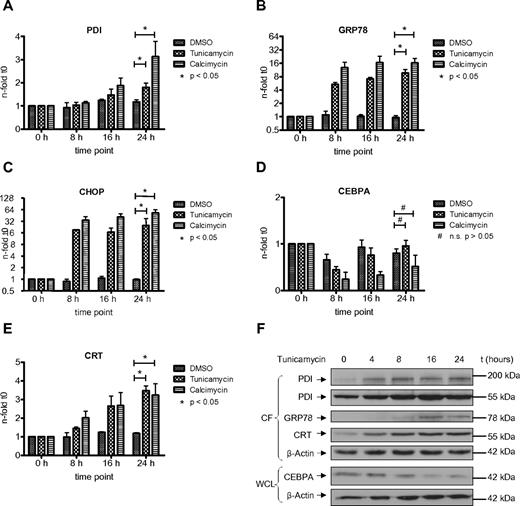
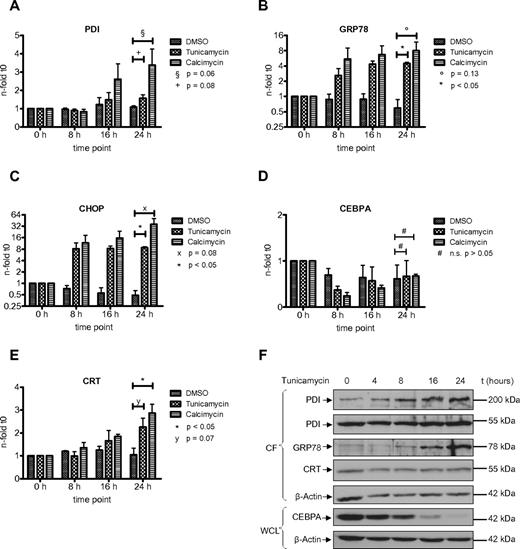
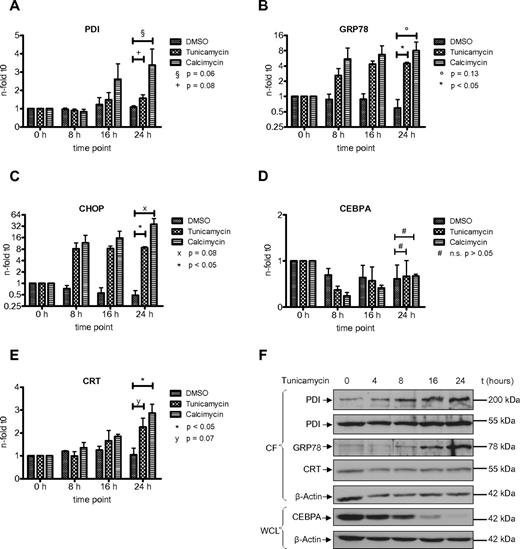
The UPR is an adaptive response of the cell toward different kind of stress conditions accompanied by the accumulation of unfolded or misfolded proteins within the endoplasmic reticulum.25 Previous reports have indicated the involvement of PDI in the unfolded protein response in various benign and malignant solid tumor cells.26,27 In AML, we have shown that the UPR is activated in a subgroup of patients.21 However, the role of PDI during the UPR has not been investigated so far in AML cells. We therefore studied whether PDI expression is activated during ER stress and UPR activation. We exposed HL60 and U937 cells to tunicamycin, an inhibitor of N-acetylglucosamine transferases, or the ionophore calcimycin. Both compounds are efficiently inducing UPR. After 24 hours of treatment, we detected a significant increase in PDI mRNA and protein expression in HL60 and U937 cells (Figures 4,5). As positive control, we analyzed the GRP78 and CHOP genes, both important effectors of the ER stress complex.
Induction of ER stress in HL60 cells activates PDI and thereby suppresses CEBPA protein. (A-E) Real-time PCR assessing mRNAs for PDI (A), GRP78 (B), CHOP (C), CEBPA (D), and calreticulin (CRT; E) from HL60 cells treated for 24 hours with 3 μg/mL tunicamycin, 3.7 μg/mL calcimycin, or DMSO as control. Error bars indicate SD based on 3 independent experiments. (F) Western blot of the cytoplasmic fraction (CF) and whole-cell lysates (WCL) from HL60 cells treated with 3 μg/mL tunicamycin for 24 hours. Membranes were stained with Abs against PDI, GRP78, calreticulin (CRT), and CEBPA. β-actin was used as loading control.
Induction of ER stress in HL60 cells activates PDI and thereby suppresses CEBPA protein. (A-E) Real-time PCR assessing mRNAs for PDI (A), GRP78 (B), CHOP (C), CEBPA (D), and calreticulin (CRT; E) from HL60 cells treated for 24 hours with 3 μg/mL tunicamycin, 3.7 μg/mL calcimycin, or DMSO as control. Error bars indicate SD based on 3 independent experiments. (F) Western blot of the cytoplasmic fraction (CF) and whole-cell lysates (WCL) from HL60 cells treated with 3 μg/mL tunicamycin for 24 hours. Membranes were stained with Abs against PDI, GRP78, calreticulin (CRT), and CEBPA. β-actin was used as loading control.
PDI is up-regulated in U937 cells after induction of ER stress and suppresses CEBPA protein. (A-E) Real-time PCR assessing mRNAs for PDI (A), GRP78 (B), CHOP (C), CEBPA (D), and calreticulin (CRT; E) from U937 cells treated for 24 hours with 3 μg/mL tunicamycin, 3.7μg/mL calcimycin, or DMSO as control. Error bars indicate SD based on 3 independent experiments. (F) Western blot of the cytoplasmic fraction (CF) and whole-cell lysates (WCL) from U937 cells treated with 3 μg/mL tunicamycin for 24 hours. Membranes were stained with Abs against PDI, GRP78, calreticulin (CRT), and CEBPA. β-actin was used as loading control.
PDI is up-regulated in U937 cells after induction of ER stress and suppresses CEBPA protein. (A-E) Real-time PCR assessing mRNAs for PDI (A), GRP78 (B), CHOP (C), CEBPA (D), and calreticulin (CRT; E) from U937 cells treated for 24 hours with 3 μg/mL tunicamycin, 3.7μg/mL calcimycin, or DMSO as control. Error bars indicate SD based on 3 independent experiments. (F) Western blot of the cytoplasmic fraction (CF) and whole-cell lysates (WCL) from U937 cells treated with 3 μg/mL tunicamycin for 24 hours. Membranes were stained with Abs against PDI, GRP78, calreticulin (CRT), and CEBPA. β-actin was used as loading control.
Consistently, we observed a transient and early decrease in CEBPA mRNA expression after induction of ER stress, particularly after exposure to calcimycin (Figures 4D,5D). This decrease in CEBPA mRNA was not observed after isolated forced expression of exogenous PDI (or calreticulin), and it is related to initial unspecific effects after induction of ER stress involving a transient transcriptional suppression of transcription factors driving differentiation or cell-cycle progression. After 24 hours, CEBPA mRNA levels reached again starting levels. However, Figures 4F and 5F indicate that CEBPA protein levels remain suppressed after ER stress induction. This is paralleled by continuous up-regulation of the 55-kDa and the 200-kDa PDI complex as well as continued calreticulin induction. We therefore conclude that ER stress first mediates a transient decrease in CEBPA mRNA, whereas the up-regulation of the CEBPA mRNA binding proteins PDI and calreticulin sustains a prolonged block of CEBPA translation.
PDI expression correlates with induction of the UPR in AML patients
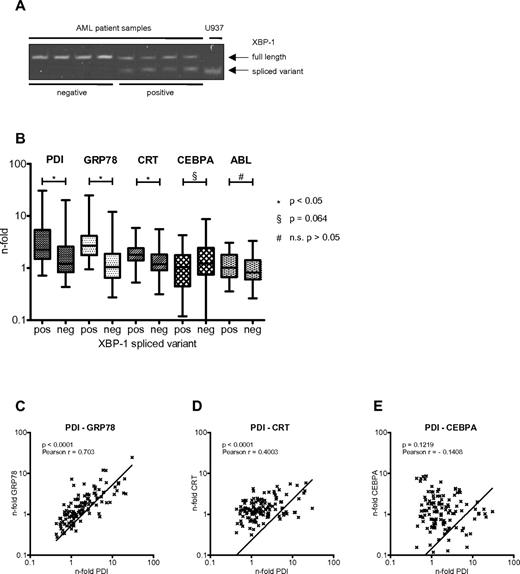
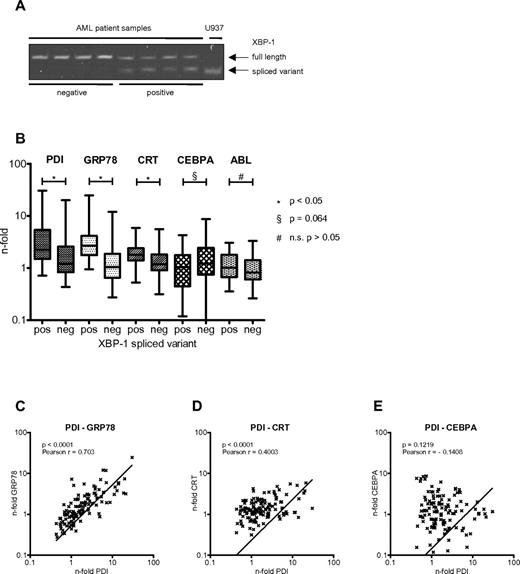
The various pathways mediating the UPR involve the activation of the ER membrane-bound kinases PERK, ATF6, and IRE1. Activation of IRE1 leads—through its RNase domain—to splicing of the transcription factor XBP1.28 Detection of the spliced variant of XBP1 has therefore become a useful marker to demonstrate UPR activation.29 We screened leukemic cells from 122 AML patients for the presence of the XBP1 spliced variant (Figure 6A). We found 31 of 122 (25.4%) AML patient samples (Table 1) to express detectable amounts of the XBP1 spliced variant, with the clinical characteristics summarized in Table 2. This suggests that the UPR is activated in roughly one-quarter of AML patients at diagnosis. We also assessed the expression of PDI mRNA and other UPR-associated genes such as GRP78 and calreticulin in this cohort of AML patients (Figure 6B). We found that PDI, GRP78, and calreticulin mRNAs were significantly increased in the XBP1 spliced variant group (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In particular, PDI expression strongly correlated with expression of GRP78 and calreticulin (Figure 6C-D), but not CEBPA (Figure 6E). These results indicate that PDI is up-regulated in AML samples with activation of the UPR.
PDI expression is induced in AML patients with activated UPR. (A) XBP1 mRNA spliced variant-specific PCR for negative (lanes 1-4) and positive (lanes 5-8) AML patient samples. U937 cells harvested 24 hours after ER stress induction after treatment with 3 μg/mL tunicamycin served as positive control. (B) Real-time PCR results for mRNAs of PDI, GRP78, calreticulin (CRT), CEBPA, and ABL from AML patient samples stratified according to being positive (n = 31; 25%) or negative (n = 91; 75%) for the XBP1 spliced variant as a marker for activated UPR. (C-E) Correlation between differential gene expression of PDI and GRP78 (C), of PDI and calreticulin (CRT; D), and of PDI and CEBPA mRNAs (E) in 122 AML patients.
PDI expression is induced in AML patients with activated UPR. (A) XBP1 mRNA spliced variant-specific PCR for negative (lanes 1-4) and positive (lanes 5-8) AML patient samples. U937 cells harvested 24 hours after ER stress induction after treatment with 3 μg/mL tunicamycin served as positive control. (B) Real-time PCR results for mRNAs of PDI, GRP78, calreticulin (CRT), CEBPA, and ABL from AML patient samples stratified according to being positive (n = 31; 25%) or negative (n = 91; 75%) for the XBP1 spliced variant as a marker for activated UPR. (C-E) Correlation between differential gene expression of PDI and GRP78 (C), of PDI and calreticulin (CRT; D), and of PDI and CEBPA mRNAs (E) in 122 AML patients.
PDI protein expression in AML patients
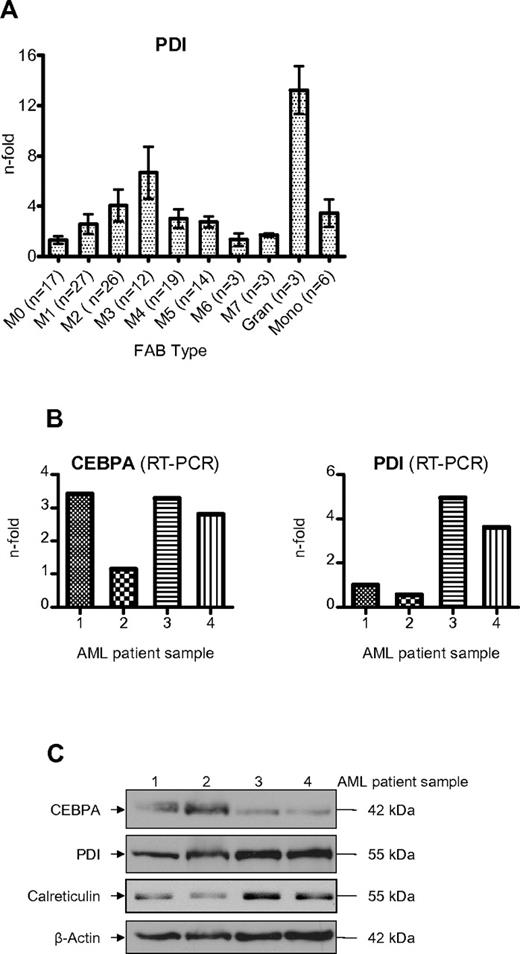
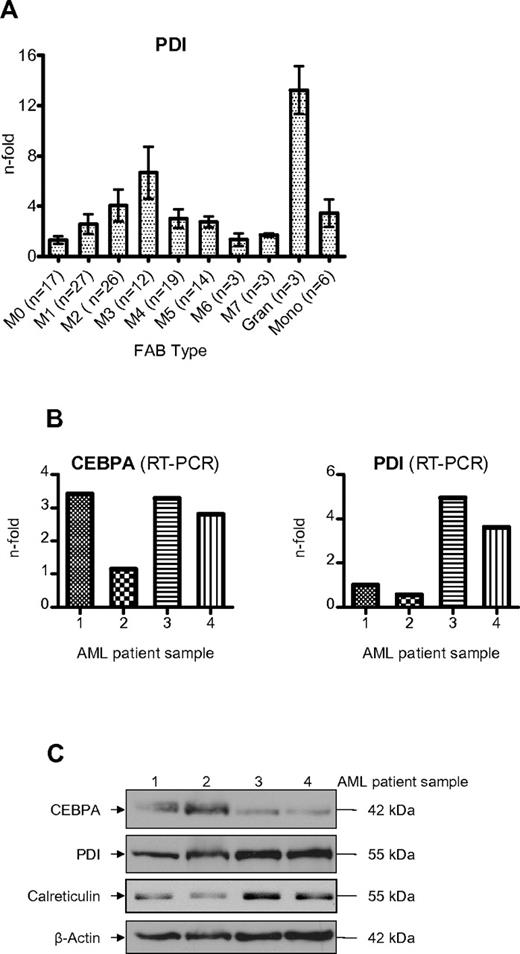
We observed that the UPR is activated more often in the myeloid FAB subtypes M2 and M3 compared with the monocytic subtypes M4 and M5 (Table 2). Consistently, PDI mRNA was also stronger expressed in FAB M2 and M3 patients (Figure 7A). Finally, we analyzed whether elevated PDI expression was associated with suppressed CEBPA protein in AML patients. As depicted in Figure 7B, we observed that AML patients with high levels of PDI expression in fact had suppressed CEBPA protein even if CEBPA mRNA was clearly present in these cells (Figure 7C). We therefore conclude that activation of PDI can block CEBPA translation in AML patients.
Induction of PDI in AML patients suppresses CEBPA protein. (A) Real-time PCR of PDI mRNA levels in AML patients according to the FAB classification. Mature granulocytes and monocytes from 6 healthy donors are indicated for comparison. Error bars indicate SD. (B) Real-time PCR results of leukemic cells from 4 AML patients for CEBPA and PDI mRNAs. The patients had AML-M2 (#1), -M0 (#2), -M5 (#3), and -M4 (#4). (C) Western blot analysis using whole-cell lysates from blasts of the same 4 AML patients. Membranes were stained with Abs against CEBPA, PDI, and calreticulin. β-actin was used as loading control.
Induction of PDI in AML patients suppresses CEBPA protein. (A) Real-time PCR of PDI mRNA levels in AML patients according to the FAB classification. Mature granulocytes and monocytes from 6 healthy donors are indicated for comparison. Error bars indicate SD. (B) Real-time PCR results of leukemic cells from 4 AML patients for CEBPA and PDI mRNAs. The patients had AML-M2 (#1), -M0 (#2), -M5 (#3), and -M4 (#4). (C) Western blot analysis using whole-cell lysates from blasts of the same 4 AML patients. Membranes were stained with Abs against CEBPA, PDI, and calreticulin. β-actin was used as loading control.
Discussion
We previously reported that calreticulin, a chaperone molecule also involved in calcium homeostasis, can bind to the stem loop secondary structure of the CEBPA mRNA and thereby blocks its translation in myeloid cells. In particular, AML patients with inv(16) consistently showed activation of calreticulin mediating CEBPA suppression. Studying protein lysates from leukemic cell lines, we consistently observed 2 complexes binding to the CEBPA dsRNA oligonucleotide in UV cross-link experiments. Whereas the faster migrating complex of 55 kDa was caused by calreticulin, the identity of the high molecular complex of around 200 kDa remained to be elucidated. We report in this study that this complex was found by mass spectrometry to be PDI. Further analysis in myeloid leukemic cells revealed that the ER-resident chaperone PDI blocks the translation of the myeloid key transcription factor CEBPA by binding to its mRNA.
The PDI protein is the founding member of the PDI family of proteins and it is mainly located in the ER.20 It establishes disulfide bonds and acts as a chaperone in nascent proteins. However, reports on non-ER localizations of PDI family members identified PDI also in the cytosol suggesting additional roles for PDI.30,31
To further characterize the interaction between mRNA binding proteins and the CEBPA mRNA, we performed RNA gel-shift experiments. Using specific Abs against PDI and calreticulin, the RNA-protein binding was abolished. However, no supershift activities were detectable. We assume this is either because of a conformational change of the PDI and calreticulin proteins after binding of the Ab, disabling the RNA-protein interaction, or because of masking of the RNA binding site by the Ab. Interestingly, addition of the PDI Ab slightly strengthened the binding activity of calreticulin, and vice versa, indicating that the 2 proteins are present at this binding site in a noncompetitive manner. In addition and consistent with previous reports in solid cancer cell types, we were able to demonstrate in leukemic cell lines the physical interaction between the 2 mRNA binding proteins calreticulin and PDI.32,33
We found that forced expression of PDI blocked CEBPA protein, but not transcription in leukemic myeloid cells. Fractionation of cellular lysates revealed that PDI was mainly localized in the organelle fraction and to a minor part also in the cytosol. Expressing a mutant PDI protein lacking the C-terminal KDEL motif—which acts as an ER retention signal—enhances the amount of PDI protein in the cytosol. Interestingly, the KDEL-deficient PDI protein leads to a more prominent block of CEBPA protein expression. These results indicate that the cytosol PDI fraction is critical for the block of CEBPA translation. It remains to be investigated whether aberrant cellular localization of PDI proteins can be observed in AML cells.
Block of CEBPA translation through the upstream untranslated open reading frame of the CEBPA mRNA in myeloid leukemic cells has been first reported to be mediated by hnRNP E2.34 Calreticulin has been shown to block both CEBPA and CEBPB translation because both contain a GC-rich stem loop region within their mRNAs.24 Consequently, we also observed this target variety for PDI. Forced expression of PDI had the same dose-dependent effect on CEBPA and CEBPB expression. Other targets of PDI remain to be demonstrated. One study indicated that PDI blocked NF-kB–dependent gene expression in the mouse leukemic cell line RAW 264.7.35 Because CEBPA and NF-kB proteins can interact in target gene activation, suppressed levels of CEBPA protein—mediated by activation of PDI—might be involved in blocked NF-kB–dependent target gene expression.36,37
We previously showed that the UPR is activated in a considerable subset of AML patients indicating a role of the UPR in the pathogenesis of these leukemias.21,38 Here, we report that PDI is activated in AML patients with induced UPR. Apart from that, activation of PDI during ER stress was described so far in plants and in brain tissue of mice with Alzheimer disease.39,40 These reports have not addressed the question of whether the 55 kDa or the 200 kDa (or both) represent the active PDI form. Studying the leukemic cell lines HL60 and U937, we found that ER stress preferentially induced the oligomeric 200-kDa PDI complex paralleled by a decrease in CEBPA protein.
Consistent with our data in leukemic cell lines, we observed in a comprehensive cohort of AML patients that UPR can be activated in 25% of the patients, and that PDI expression is strongly correlated with activation of the UPR.21 However, it remains to be elucidated why the UPR is activated in this quarter of all AML patients. Studies investigating the leukemogenic fusion proteins BCR-ABL or PML-RARA suggest that expression and accumulation of specific fusion proteins can trigger the UPR.41,42 Consistently, we observed in our cohort that blasts from patients with AML-M3 showed a high degree of UPR activation and that they expressed the highest PDI mRNA levels.
Increased levels of effectors of the UPR—such as GRP78, CHOP, calreticulin and the spliced variant of XBP1—strongly correlated with increased levels of PDI. Activation of PDI in leukemic cells of AML patients did not correlate with CEBPA mRNA expression. However, AML patients with induced PDI protein expression showed suppressed CEBPA protein, confirming our in vitro data in cell lines. These data identify a novel role of PDI during the UPR in leukemic cells of AML patients by blocking the important myeloid differentiation factor CEBPA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation SF 310000-109388 and Oncosuisse OCS-01833-02-2006 (T.P.). The funding source had no role in publication of these data.
Authorship
Contribution: S.H. and C.K. performed research and wrote the paper; K.S. performed research; J.S. contributed vital material; N.T. contributed vital material; B.U.M. analyzed data; and T.P. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Pabst, MD, Associate Professor, Department of Medical Oncology, University Hospital, 3010 Berne, Switzerland; e-mail: thomas.pabst@insel.ch.
References
Author notes
S.H. and C.K. contributed equally to this work and appear in alphabetical order.