Abstract
The mechanisms by which sphingosine kinase-1 (SK-1)/sphingosine 1-phosphate (S1P) activation contributes to imatinib resistance in chronic myeloid leukemia (CML) are unknown. We show herein that increased SK-1/S1P enhances Bcr-Abl1 protein stability, through inhibition of its proteasomal degradation in imatinib-resistant K562/IMA-3 and LAMA-4/IMA human CML cells. In fact, Bcr-Abl1 stability was enhanced by ectopic SK-1 expression. Conversely, siRNA-mediated SK-1 knockdown in K562/IMA-3 cells, or its genetic loss in SK-1−/− MEFs, significantly reduced Bcr-Abl1 stability. Regulation of Bcr-Abl1 by SK-1/S1P was dependent on S1P receptor 2 (S1P2) signaling, which prevented Bcr-Abl1 dephosphorylation, and degradation via inhibition of PP2A. Molecular or pharmacologic interference with SK-1/S1P2 restored PP2A-dependent Bcr-Abl1 dephosphorylation, and enhanced imatinib- or nilotinib-induced growth inhibition in primary CD34+ mononuclear cells obtained from chronic phase and blast crisis CML patients, K562/IMA-3 or LAMA4/IMA cells, and 32Dcl3 murine progenitor cells, expressing the wild-type or mutant (Y253H or T315I) Bcr-Abl1 in situ. Accordingly, impaired SK-1/S1P2 signaling enhanced the growth-inhibitory effects of nilotinib against 32D/T315I-Bcr-Abl1–derived mouse allografts. Since SK-1/S1P/S1P2 signaling regulates Bcr-Abl1 stability via modulation of PP2A, inhibition of SK-1/S1P2 axis represents a novel approach to target wild-type- or mutant-Bcr–Abl1 thereby overcoming drug resistance.
Introduction
CML is a clonal disorder of pluripotent hematopoietic stem cells characterized by the Philadelphia (Ph) chromosome, which results from the reciprocal translocation between the long arms of chromosomes 9 and 22.1-4 This hybrid B-cell receptor (BCR)–ABL1 gene encodes for a fusion protein Bcr-Abl1 with a constitutive tyrosine kinase activity.3,4
Despite high rates of clinical responses in early chronic phase CML (CML-CP) to the Bcr-Abl1 kinase inhibitor imatinib,5-8 development of resistance is a major problem in late CML-CP and in the treatment of blast crisis CML (CML-BC).9-12 Although Bcr-Abl1–independent mechanisms also exist,13-15 resistance in CML-CP is usually associated with the expression of mutant Bcr-Abl1 proteins, including T315I and Y253F/H mutations against which the second generation ABL tyrosine kinase inhibitors (TKI) such as nilotinib and/or dasatinib show limited effect.15-17 Nonetheless, BCR-ABL1 mutations may not account for all cases of drug resistance in CML (CP and BC); indeed, alternative Bcr-Abl1–dependent mechanisms including alterations of sphingolipid metabolism and signaling,18 might account for TKI resistance.
Sphingolipids, ceramide and sphingosine 1-phosphate (S1P) included, are a family of membrane lipids with important roles in the regulation of the fluidity and subdomain structure of membranes.19-21 Ceramide can be hydrolyzed by ceramidases to release sphingosine, which is phosphorylated by sphingosine kinases-1 or -2 (SK-1 or SK-2) to generate S1P.20 Ceramide plays proapoptotic roles21 whereas S1P mediates proliferation and/or resistance to apoptosis22,23 generally via G-protein–coupled S1P1-5 receptor signaling.24 However, receptor-independent intracellular functions of S1P were also reported.25 Recently, alteration of the balance between the proapoptotic ceramide and antiapoptotic S1P via up-regulation of SK-1 was shown to mediate imatinib resistance in K562 CML-BC patient-derived cells by an unknown mechanism.18
Here, we report the identification of a novel mechanism by which SK-1/S1P mediates imatinib resistance by regulation of the PP2A-dependent and SHP-1–mediated Bcr-Abl1 dephosphorylation and stability selectively via receptor 2 (S1P2) signaling in CML (CP and BC). In addition, our data suggest that targeting the SK-1/S1P2 signaling axis provides a novel strategy to modulate wild-type (wt) or mutant (T315I or Y253H) Bcr-Abl1 stability by restoring PP2A function, and attenuate drug resistance both in cell culture and in mice bearing 32D/T315I-Bcr-Abl1 allografts.
Methods
Cell lines and growth conditions
Human CML cell lines K562, LAMA4, and their imatinib-resistant derivatives K562/IMA-0.1, -1, -3, or LAMA4/IMA, were maintained as described.18 The Bcr-Abl–expressing 32Dcl3 cells, 32D-p210Bcr-Abl (wt), 32D-p210Bcr-Abl (Y253H) and (T315I) were maintained in RPMI containing 15% FBS, 2mM l-glutamine, and penicillin and streptomycin (P/S; 100 ng/mL each). MEFs (wt and SK-1−/−) were maintained in DMEM with 10% FBS and P/S. Human CD34+ primary cells from CML patients and normal donor were obtained under a protocol approved by the institutional review board and IACUC at the Medical University of South Carolina or The Ohio State University and were grown as described.26
Quantitative real-time PCR and Western blotting
Quantitative real-time PCR (Q-RT-PCR) was performed using TaqMan gene expression kit (Applied Biosystems) with ABI 7300 Q-PCR system. All primers and probes were obtained from ABI. The mRNA of GAPDH and rRNA were used as internal controls.18
Bcr-Abl1, P-Bcr–Abl1-Y245 or -Y177, SK1, PP2A, PP2A-pY307, SHP1, SHP1-pS591, V5, HA, GAPDH and β-actin protein was measured by Western blotting using mouse monoclonal anti–c-Abl antibody-3 (Calbiochem), rabbit polyclonal anti–Bcr-Abl1-pY245 or -pY177 (Cell Signaling), rabbit polyclonal anti-SK1,27 mouse monoclonal anti-PP2A (Millipore), rabbit polyclonal anti–P-PP2A-Y307 (Epitomics), mouse monoclonal anti-SHP1 (Santa Cruz), rabbit polyclonal anti–P-SHP1-S591 (ECM Biosciences), mouse monoclonal V5 (Invitrogen), mouse monoclonal anti-HA (Cell Signaling), mouse monoclonal anti-GAPDH (Millipore), rabbit polyclonal anti–β-actin (Sigma-Aldrich) and peroxidase-conjugated secondary anti–rabbit or anti–mouse antibodies (Jackson ImmunoResearch Laboratories).18 The quantification of blots were performed using ImageJ Version 1.44.
Measurement of SK activity, and S1P generation using LC/MS
The endogenous activity of SK for S1P generation was measured using C17-Sph (5μM) labeling in K562 and K562/IMA-3 cells followed by the detection of C17-S1P in cell extracts at various time points using LC/MS/MS as described.28
Plasmids and siRNAs
Overexpression of the V5-tagged SK-1, and HA-tagged PP2Ac was obtained by Amaxa nucleofection. I2PP2A (SET)-GFP was expressed in resistant K562 cells as described.29 The siRNAs were obtained from Dharmacon, and their specificity for target genes was confirmed by Q-PCR and/or Western blotting.29 Cells were transfected by siRNA (100nM for 48 hours) using Amaxa nucleofection. The siRNAs for SK-1, SK-2 and S1P1-4 receptors were obtained from Dharmacon (smart-pool siRNAs).
Determining the half-life of Bcr-Abl1 protein
The half-life of Bcr-Abl1 was determined using cycloheximide (CHX, 80 μg/mL) as described.30 Bcr-Abl1 stability was further examined by pulse-chase experiments, in which sensitive and resistant cells were pulsed with [35S]-methionine/cysteine, 150 μCi/mL for 18 hours, or [32P]-orthophosphate, 0.5 mCi/mL for 4 hours, (American Radiolabeled Chemicals) in amino acid– or phosphate-depleted media, respectively. Labeled proteins were then chased in intact media for various time points. Total and P-Bcr-Abl1 proteins were immunoprecipitated, resolved on SDS-PAGE, and visualized by autoradiography as described.30
Detection of cell growth and apoptosis
The growth of cells was measured using trypan blue exclusion and 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assays as described.18 Activation of caspases 3 and 9 in cell extracts was determined using their fluorescently labeled specific target peptides, as described by the manufacturer (Fluorometric Caspase Assay Kit; R&D Systems).29
Detection of the ubiquitination of Bcr-Abl1
The ubiquitination of Bcr-Abl1 was detected in the presence/absence of a proteasome inhibitor (lactacystin; AG Scientific), or expression of the exogenous ubiquitin, using an HA-ubiquitin vector, by immunoprecipitation followed by Western blotting using anti–Bcr-Abl1 or anti-HA antibodies, as described.31 Protein-antibody complexes were pulled down using protein A/G agarose beads for 60 minutes at 4°C, which were then washed and resuspended in 10 μL of 1× PBS and 10 μL of 2× loading buffer. After incubation at 95°C for 5 minutes, proteins were separated by SDS-PAGE and examined by Western blot analysis.
Measurement of PP2A activity
Phosphatase assays in cell extracts were carried out using the PP2A phosphatase assay kit (Upstate). Briefly, protein lysates (50 μg) in 100 μL of 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.0)/100mM NaCl, 5μg of PP2Ac antibody (Upstate), and 25 μL of Protein A-agarose were added to 400 μL of 50mM tris(hydroxymethyl)aminomethane (pH 7.0)/100mM CaCl2, and immunoprecipitations were carried out at 4°C for 2 hours. Immunoprecipitates were washed, and phosphatase activity was measured by malachite green reaction measuring free phosphate released from phosphothreonine peptide (K-R-pT-I-R-R), as described by the manufacturer.
Cell staining and sorting
Flow cytometry staining for analysis of hematologic cell types were performed using antibodies and lineage markers using flow cytometry as described.32
Analysis of 32D/wt- and 32D/T315I-Bcr-Abl1 allograft growth and TKI sensitivity in response to SK-1/S1P2 interference in mice
Severe combined immunodeficiency mice were (n = 4/group) injected subcutaneously with either 32D/wt Bcr-Abl cells, or 32D/T315I-Bcr-Abl cells (4 × 106 cells in each flank), and tumor growth was monitored weekly by calipers.29 Mice were treated with nilotinib at 10 mg/kg, in the absence/presence of SKI-II or JTE-013 (Cayman Chemicals; 15 or 0.5 mg/kg, respectively) at days 1, 8, 15, and tumor growth was measured at days 1, 6, 11, 14, and 18 after treatments. Independent experiments were performed at least twice in duplicates or triplicates. Statistical significance was examined using Student t test analysis, and P < .05 was considered significant.29
Results
SK-1/S1P mediates increased protein stability of Bcr-Abl1
Our previous data indicated that increased generation of S1P by SK-1 mediates drug resistance via up-regulation of Bcr-Abl1 protein but not mRNA levels in K562/IMA-1 cells.18 To define the role of SK-1/S1P signaling in the regulation of Bcr-Abl1, we compared total (Bcr-Abl1) and Y245 or Y17733 phosphorylated (P-Bcr-Abl1) proteins with regard to SK-1 expression and activity in sensitive K562 versus resistant K562/IMA-3 cells, which were derived by chronic exposure to 3μM imatinib. Bcr-Abl1 and P-Bcr-Abl1 levels were up-regulated approximately 2.0- and 2.5-fold, respectively, in K562/IMA-3 compared with K562 cells (Figure 1A). The mRNA, protein and the activity of SK-1 was also increased approximately 2.8-, 3.0-, and 3.5-fold, respectively, in K562/IMA-3 cells (supplemental Figure 1A-C, available on Blood Web site; see the Supplemental Materials link at the top of the online article). Because B-cell receptor-ABL1 mRNA did not significantly change in K562/IMA cells,18 we examined the protein stability of Bcr-Abl1 using cycloheximide (CHX) at 80 μg/mL. The half-life of Bcr-Abl1 was around 24 hours in K562 cells, and it was > 36 hours in K562/IMA-3 cells (Figure 1B). These data were also consistent when pulse-chase labeling with [35S]-methionine/cysteine was performed to examine the half-life of Bcr-Abl1 in these cells without CHX (data not shown). Moreover, the stability of P-Bcr-Abl1 was also measured using [32P]-orthophosphate labeling and immunoprecipitation, which revealed that the half-life of P-Bcr-Abl1 (at Y117) was around 16 hours in K562 versus > 24 hours in K562/IMA-3 cells (Figure 1C). Thus, stability of Bcr-Abl1 and activated P-Bcr-Abl1 is higher in imatinib-resistant than -sensitive K562 cells.
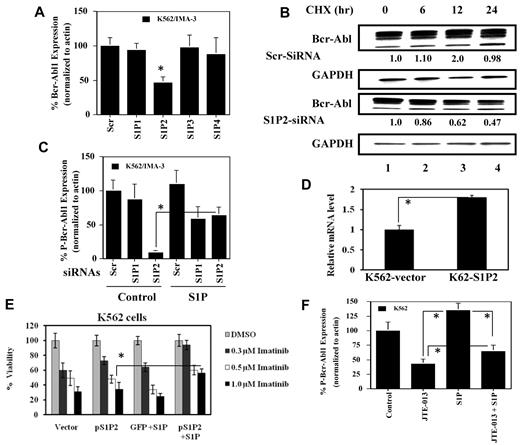
Regulation of the stability and expression of Bcr-Abl1 by SK-1/S1P signaling. (A) Total and P-Bcr-Abl1 (Y245) in K562/IMA-3 compared with K562 cells (lanes 2 and 1, respectively) were measured using Western blotting. β-actin was used as a loading control. (B) The protein stability of Bcr-Abl1 in K562 and K562/IMA-3 cells was measured by Western blotting at various time points (0-36 hours) using CHX. Long-lived protein GAPDH was used as a control. Relative expression levels of Bcr-Abl1 (normalized to GAPDH levels) at 0-36 hours after CHX treatment in K562 versus K562/IMA-3 cells are shown. (C) The stability of P-Bcr-Abl1 in K562 and K562/IMA-3 cells were also examined by pulse-chase experiments using [32P]-orthophosphate labeling without CHX using immunoprecipitation by anti–P-Bcr-Abl1 (Y177) antibody. (D) The protein stability of Bcr-Abl1 in response to overexpression of SK-1 with the V5 tag in K562 parental cells was determined at various time points (0-36 hours) compared with vector-transfected controls using Western blotting. GAPDH was used as a loading control (left panel). (E) The expression of SK-1 was down-regulated using siRNA, and its effect on the stability of Bcr-Abl1 was determined using CHX treatment at 12 hours compared with Scr-siRNA treated controls. (F) Effects of genetic loss of SK-1 on Bcr-Abl1 stability were examined in wt and SK-1−/− MEFs stably transfected with the human wt-Bcr-Abl1-GFP (p210) in the presence of CHX at 0, 18, 30 hours. The stability of P-Bcr-Abl1-GFP in wt versus SK-1−/− MEFs was measured using [32P]-orthophosphate labeling and immunoprecipitation (using the antibody that recognizes P-Bcr-Abl1 at Y177) as described in “Determining the half-life of Bcr-Abl1 protein.” These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
Regulation of the stability and expression of Bcr-Abl1 by SK-1/S1P signaling. (A) Total and P-Bcr-Abl1 (Y245) in K562/IMA-3 compared with K562 cells (lanes 2 and 1, respectively) were measured using Western blotting. β-actin was used as a loading control. (B) The protein stability of Bcr-Abl1 in K562 and K562/IMA-3 cells was measured by Western blotting at various time points (0-36 hours) using CHX. Long-lived protein GAPDH was used as a control. Relative expression levels of Bcr-Abl1 (normalized to GAPDH levels) at 0-36 hours after CHX treatment in K562 versus K562/IMA-3 cells are shown. (C) The stability of P-Bcr-Abl1 in K562 and K562/IMA-3 cells were also examined by pulse-chase experiments using [32P]-orthophosphate labeling without CHX using immunoprecipitation by anti–P-Bcr-Abl1 (Y177) antibody. (D) The protein stability of Bcr-Abl1 in response to overexpression of SK-1 with the V5 tag in K562 parental cells was determined at various time points (0-36 hours) compared with vector-transfected controls using Western blotting. GAPDH was used as a loading control (left panel). (E) The expression of SK-1 was down-regulated using siRNA, and its effect on the stability of Bcr-Abl1 was determined using CHX treatment at 12 hours compared with Scr-siRNA treated controls. (F) Effects of genetic loss of SK-1 on Bcr-Abl1 stability were examined in wt and SK-1−/− MEFs stably transfected with the human wt-Bcr-Abl1-GFP (p210) in the presence of CHX at 0, 18, 30 hours. The stability of P-Bcr-Abl1-GFP in wt versus SK-1−/− MEFs was measured using [32P]-orthophosphate labeling and immunoprecipitation (using the antibody that recognizes P-Bcr-Abl1 at Y177) as described in “Determining the half-life of Bcr-Abl1 protein.” These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
To assess whether SK-1/S1P plays a role in the stabilization of Bcr-Abl1, we investigated the effects of SK-1 overexpression on the half-life of Bcr-Abl1 in K562 cells. Ectopic SK-1-V5 expression (supplemental Figure 2A), strongly increased Bcr-Abl1 half-life from around 22 to 32 hours (Figure 1D). Accordingly, siRNA-mediated SK-1 knockdown (approximately 60% reduction by Q-PCR, supplemental Figure 2B), impaired SK activity of 40% (supplemental Figure 2C), thereby reducing Bcr-Abl1 levels of around 58% in K562/IMA-3 cells compared with same cells exposed to scrambled (Scr) siRNAs (Figure 1E). Moreover, the role of SK-1/S1P in the regulation of Bcr-Abl1 stability was also examined in MEFs isolated from wt or SK-1−/− knockout mice, transduced with a B-cell receptor-ABL1-GFP vector, in the absence/presence of CHX for 18 and 30 hours. As expected Bcr-Abl1-GFP was stable in wt-MEFs, whereas its expression in SK-1−/− MEFs was 40% and 65% decreased at 18 and 30 hours, respectively (Figure 1F), further strengthening the notion that SK-1 positively modulates Bcr-Abl1 stability. Furthermore, these data suggest that SK-1 decreases TKI sensitivity by increasing Bcr-Abl1 stability/expression, consistent with the reported ability of SK-1 to regulate imatinib-induced apoptosis in TKI-sensitive and -resistant K562 cells.18
SK-1/S1P-dependent Bcr-Abl1 stability is regulated selectively via S1P2 signaling
Levels of S1P2 and S1P4 mRNAs were found increased in K562/IMA-1 compared with K562 cells previously,18 however their specific involvement in drug resistance was not assessed. Note that the expression of S1P5 was undetectable in these cells.18 Therefore, roles of S1P1-4 signaling in the regulation of Bcr-Abl1 expression by SK-1/S1P were determined using siRNAs in K562/IMA-3 cells. The specificity of S1P1-4 siRNAs against their target genes (around 50%-80% knockdown) was confirmed using Q-PCR (supplemental Figure 3). Interestingly, knockdown of only S1P2, but not S1P1, 3 or 4, decreased Bcr-Abl1 approximately 50% compared with controls in K562/IMA-3 cells (Figure 2A). Down-regulation of S1P2 did not affect BCR-ABL1 mRNA levels (data not shown). Moreover, S1P2 knockdown (around 60% as shown by Q-PCR, supplemental Figure 3) in TKI resistant cells reduced Bcr-Abl1 half-life (around 24 hours; Figure 2B third panel compared with first panel lanes 1-4, respectively), making it similar to that of TKI sensitive cells.
Involvement of S1P2 signaling in SK-1/S1P-induced stability of Bcr-Abl1 in drug resistant compared with sensitive K562 cells. (A) Effects of the knockdown of S1P1, S1P2, S1P3 and S1P4 using siRNAs on Bcr-Abl1 protein in K562/IMA-3 cells were examined using Western blotting compared with nontargeting scrambled (Scr) controls. Actin was used as a loading control. (B) Effects of knockdown of S1P2 using siRNA on the stability of Bcr-Abl1 protein compared with controls (Scr siRNA transfected cells) in the K562/IMA-3 drug resistant cells was measured at 0, 6, 12, and 24 hours after CHX treatments using Western blotting (first and third panels lanes 1-4, respectively). GAPDH was used as a loading control (second and fourth panels). Relative expression levels of Bcr-Abl1 (normalized to GAPDH levels) at 0-24 hours post-CHX treatment in cells transfected with Scr or S1P2 siRNAs are shown below each lane. (C) Effects of knockdown of S1P1, or S1P2 using siRNAs in the absence/presence of exogenous BSA-conjugated S1P (1.0μM, added every 3 hours for 24 hours) on P-Bcr-Abl1 were assessed in K562/IMA-3 cells using Western blotting. Actin was used as a loading control. (D-E) Effects of overexpression of human S1P2-GFP using a lentiviral expression vector, as confirmed by Q-PCR (D) on imatinib-induced growth inhibition in sensitive K562 cells at 0.3, 0.5, and 1.0μM for 48 hours in the absence/presence of exogenous S1P (1μM) were examined using trypan blue exclusion (E). Vector-transduced cells were used as controls. (F) Regulation of P-Bcr-Abl1 (Y245) by S1P/S1P2 signaling was determined after treatment of K562 cells with 0.5μM BSA-conjugated S1P for 24 hours in the absence/presence of JTE-013 compared with untreated controls or JTE-013–treated cells using Western blotting. Actin was used as a loading control. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
Involvement of S1P2 signaling in SK-1/S1P-induced stability of Bcr-Abl1 in drug resistant compared with sensitive K562 cells. (A) Effects of the knockdown of S1P1, S1P2, S1P3 and S1P4 using siRNAs on Bcr-Abl1 protein in K562/IMA-3 cells were examined using Western blotting compared with nontargeting scrambled (Scr) controls. Actin was used as a loading control. (B) Effects of knockdown of S1P2 using siRNA on the stability of Bcr-Abl1 protein compared with controls (Scr siRNA transfected cells) in the K562/IMA-3 drug resistant cells was measured at 0, 6, 12, and 24 hours after CHX treatments using Western blotting (first and third panels lanes 1-4, respectively). GAPDH was used as a loading control (second and fourth panels). Relative expression levels of Bcr-Abl1 (normalized to GAPDH levels) at 0-24 hours post-CHX treatment in cells transfected with Scr or S1P2 siRNAs are shown below each lane. (C) Effects of knockdown of S1P1, or S1P2 using siRNAs in the absence/presence of exogenous BSA-conjugated S1P (1.0μM, added every 3 hours for 24 hours) on P-Bcr-Abl1 were assessed in K562/IMA-3 cells using Western blotting. Actin was used as a loading control. (D-E) Effects of overexpression of human S1P2-GFP using a lentiviral expression vector, as confirmed by Q-PCR (D) on imatinib-induced growth inhibition in sensitive K562 cells at 0.3, 0.5, and 1.0μM for 48 hours in the absence/presence of exogenous S1P (1μM) were examined using trypan blue exclusion (E). Vector-transduced cells were used as controls. (F) Regulation of P-Bcr-Abl1 (Y245) by S1P/S1P2 signaling was determined after treatment of K562 cells with 0.5μM BSA-conjugated S1P for 24 hours in the absence/presence of JTE-013 compared with untreated controls or JTE-013–treated cells using Western blotting. Actin was used as a loading control. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
Then, we examined the effects of S1P1, or S1P2 siRNAs in the absence/presence of exogenous S1P (1μM) on Bcr-Abl1 expression in K562/IMA-3 cells. Down-regulation of S1P2, but not S1P1, reduced P-Bcr-Abl1 (approximately 50%) and total Bcr-Abl1 (data not shown) compared with scrambled siRNA-treated controls (Figure 2C). Addition of S1P almost completely prevented the down-regulation of P-Bcr-Abl1 in response to S1P2 siRNA compared with controls (Figure 2C). Thus, these data suggest that knockdown of S1P2, but not S1P1, mediates the down-regulation of Bcr-Abl1, which is prevented by exogenous S1P.
In reciprocal experiments, lentiviral-mediated stable S1P2-GFP expression, approximately 2-fold (measured by Q-PCR, Figure 2D), in sensitive K562 cells using a lenti-viral transduction, decreased imatinib-induced (0.3, 0.5, and 1.0μM) growth suppression of 40% only in the presence of exogenous S1P (0.5μM) compared with vector-transfected control cells (Figure 2E). Over-expression of S1P2 alone (without S1P) did not have any significant effect on imatinib-induced cell death in K562 cells (Figures 2E). These data suggest that over-expression of S1P2 mediates imatinib resistance only in the presence of an additional S1P in sensitive K562 cells, indicating the involvement of increased S1P (exogenously supplied, or endogenously generated by SK-1) upstream of S1P2 signaling in Bcr-Abl1 expression and imatinib resistance.
Accordingly, inhibition of S1P2 by JTE-01334 at 10 μM (24 hours) decreased P-Bcr-Abl1 (approximately 50%) in sensitive K562 cells, and addition of S1P (0.5μM) increased P-Bcr-Abl1 approximately 30% compared with controls (Figure 2F). Importantly, JTE-013 treatment prevented up-regulation of P-Bcr-Abl1 in response to S1P (Figure 2F). Note that inhibition of S1P1 or S1P1/3 by VPC23019, or W146, respectively, did not significantly change P-Bcr-Abl1 in the presence of S1P (data not shown). Thus, these data support that inhibition of S1P2, but not S1P1 or S1P3, prevents S1P-mediated P-Bcr-Abl1 stability.
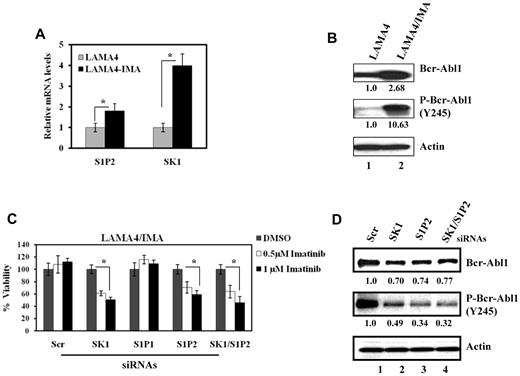
Then, the expression levels of SK-1 and S1P2 were examined in LAMA-4 and their independently developed drug-resistant variant LAMA-4/IMA cells. SK-1 and S1P2 mRNAs were up-regulated approximately 4- and 2-fold, respectively, in LAMA-4/IMA compared with parental cells (Figure 3A). LAMA-4/IMA cells over-expressed both Bcr-Abl1 and P-Bcr-Abl1 approximately 2.5 and 10-fold, respectively, compared with controls (Figure 3B lanes 2 and 1, respectively). Importantly, siRNA-mediated down-regulation of SK-1 and/or S1P2, but not S1P1, which decreased expression of their mRNAs of 60%-70% compared with scrambled siRNA-treated controls (data not shown), significantly enhanced (40%-50% increase) imatinib-induced growth inhibition at 0.5 and 1.0 μM (Figure 3C). Knockdown of SK-1 and/or S1P2 also decreased Bcr-Abl1 and P-Bcr-Abl1 expression by approximately 23%-30% or 50%-68%, respectively, compared with controls (Figures 3D). Thus, these data indicate that SK-1/S1P/S1P2-mediated regulation of Bcr-Abl1 stability, phosphorylation/activation, and drug resistance is also conserved in human LAMA-4/IMA cells.
Roles of SK-1/S1P2 signaling in the regulation of Bcr-Abl1 expression and drug resistance in human LAMA-4 CML cells. (A) The mRNA levels of S1P2 and SK-1 were measured (and normalized to the levels of rRNA) using Q-PCR in parental LAMA-4 and drug-resistant LAMA-4/IMA cells. (B) Bcr-Abl1 (total) and P-Bcr-Abl1 (Y245) were examined in LAMA-4 and LAMA-4/IMA (first and second panels lanes 1 and 2, respectively) cells using Western blotting. Actin was used as a loading control (third panel). (C-D). Effects of knockdown of SK-1 and/or S1P2 using siRNAs on imatinib-induced growth inhibition (C) or Bcr-Abl1 (total) and P-Bcr-Abl1 expression (D first and second panels lanes 2-4) compared with controls (lane 1) in LAMA-4/IMA cells were determined using trypan blue exclusion and Western blotting, respectively. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels B and D, relative expression levels of Bcr-Abl1 or P-Bcr-Abl1 (normalized to actin levels) are shown below each lane.
Roles of SK-1/S1P2 signaling in the regulation of Bcr-Abl1 expression and drug resistance in human LAMA-4 CML cells. (A) The mRNA levels of S1P2 and SK-1 were measured (and normalized to the levels of rRNA) using Q-PCR in parental LAMA-4 and drug-resistant LAMA-4/IMA cells. (B) Bcr-Abl1 (total) and P-Bcr-Abl1 (Y245) were examined in LAMA-4 and LAMA-4/IMA (first and second panels lanes 1 and 2, respectively) cells using Western blotting. Actin was used as a loading control (third panel). (C-D). Effects of knockdown of SK-1 and/or S1P2 using siRNAs on imatinib-induced growth inhibition (C) or Bcr-Abl1 (total) and P-Bcr-Abl1 expression (D first and second panels lanes 2-4) compared with controls (lane 1) in LAMA-4/IMA cells were determined using trypan blue exclusion and Western blotting, respectively. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels B and D, relative expression levels of Bcr-Abl1 or P-Bcr-Abl1 (normalized to actin levels) are shown below each lane.
SK-1/S1P signaling regulates Bcr-Abl1 stability via inhibition of PP2A
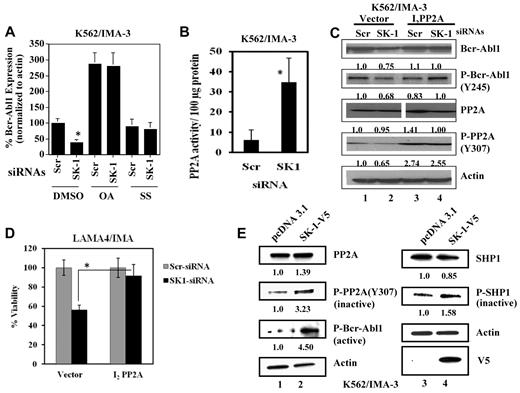
The ubiquitination/degradation of Bcr-Abl1 protein is controlled via its dephosphorylation by SHP1, a tyrosine protein phosphatase, whose activation is regulated by PP2A.35 Therefore, we examined the involvement of PP2A and SHP1 in the regulation of Bcr-Abl1 stability using 10nM okadaic acid (OA) or 10μM sodium stibogluconate (SS), inhibitors of PP2A or SHP1, respectively. In K562/IMA-3 cells, inhibition of PP2A by OA increased Bcr-Abl1 protein approximately 3-fold compared with untreated controls, however, SS alone did not increase basal levels of Bcr-Abl1 (Figure 4A). As expected, siRNA-mediated SK-1 knockdown decreased Bcr-Abl1 expression of 40% (Figure 4A), and suppression of PP2A or SHP-1 activity by OA or SS, respectively, rescued Bcr-Abl1 expression in K562-IMA3 cells in response to SK-1 siRNAs (Figure 4A). These results highlight the importance of the PP2A/SHP1/SK-1 interplay in the regulation of Bcr-Abl1 expression/stability. Importantly, siRNA-mediated 80% down-regulation of SK-2 did not alter Bcr-Abl1 expression (supplemental Figure 4 left and right panels, respectively). Thus, SK-1–, but not SK-2–generated S1P controls Bcr-Abl1 protein stability through modulation of PP2A activity. Indeed, SK-1 down-regulation by siRNAs rescued PP2A activity in K562/IMA-3 cells, and decreased Bcr-Abl1 and P-Bcr-Abl1 levels (Figure 4B and C, respectively). It was reported previously that I2PP2A (SET) suppressed PP2A (approximately 50% compared with controls) activity in vitro.29 Consistent with these data, ectopic I2PP2A (SET) expression resulted in accumulation of the tyrosine 307 phosphorylated form of PP2Ac (Figure 4C fourth panel lanes 3-4 and 1-2), which, as reported,36 is indicative of PP2A inactivation. Importantly, while SK-1 siRNA mediated Bcr-Abl1 down-regulation (both total and phosphorylated forms of 25%-32%, respectively), I2PP2A (SET)-mediated inhibition of PP2A almost completely prevented P-Bcr-Abl1 down-regulation in response to SK-1 siRNA (Figure 4C first and second panels lanes 4-3 and 2-1, respectively). Moreover, while knockdown of SK-1 reduced the viability of LAMA-4/IMA resistant cells (approximately 50%), I2PP2A blunted the growth inhibitory effects of SK-1 siRNA (Figure 4D).
Regulation of Bcr-Abl1 expression levels by SK-1/S1P signaling via the modulation of PP2A. (A) Effects of the down-regulation of SK-1 using siRNA on Bcr-Abl1 protein in the absence/presence of okadaic acid (OA; 10nM) or SS (10μM), inhibitors of PP2A or SHP1, respectively, for 24 hours were determined by Western blotting, and the Scr-siRNA transfected samples were used as controls. Actin was used as a loading control. (B) Activation of PP2A in response to knockdown of SK-1 using siRNA in K562/IMA-3 cell extracts was determined ex vivo using the PP2A activity assay kit. (C) Roles of PP2A activation in the regulation of Bcr-Abl1 and P-Bcr-Abl1 in response to Scr or SK-1 siRNAs were examined in vector controls vs I2PP2A-GFP over-expressing K562/IMA-3 cells using Western blotting (first and second panels lanes 1-2 and 3-4, respectively). Effects of vector-only and I2PP2A overexpression on total and P-PP2A at Y307 (inactive form) in Scr or SK-1 siRNA treated K562/IMA-3 cells were also determined using Western blotting (third and fourth panels lanes 1-2 and 3-4, respectively). Actin was used as a loading control (fifth panel). (D) Role of I2PP2A overexpression in the regulation of cell viability in response to SK-1 siRNA was examined using trypan blue exclusion in drug resistant LAMA-4/IMA cells. (E) Effects of SK-1-V5 overexpression compared with vector-transfections (lanes 2 and 1, respectively) on the total PP2A or SHP1 vs inactive P-PP2A at Y307, or P-SHP1 at S591 (top left and right first and second panels, respectively) were measured by Western blotting. Effects of overexpression of SK-1 compared with vector transfections on the levels of P-Bcr-Abl1 at Y245 were determined by Western blotting (bottom left third panel lanes 2 and 1, respectively). Expression of SK-1-V5 was confirmed using the anti-V5 antibody in Western blotting (bottom right panel), and β-actin was used as a loading control (bottom left third panel). These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels C and E, relative expression levels of proteins (normalized to actin levels) are shown below each lane.
Regulation of Bcr-Abl1 expression levels by SK-1/S1P signaling via the modulation of PP2A. (A) Effects of the down-regulation of SK-1 using siRNA on Bcr-Abl1 protein in the absence/presence of okadaic acid (OA; 10nM) or SS (10μM), inhibitors of PP2A or SHP1, respectively, for 24 hours were determined by Western blotting, and the Scr-siRNA transfected samples were used as controls. Actin was used as a loading control. (B) Activation of PP2A in response to knockdown of SK-1 using siRNA in K562/IMA-3 cell extracts was determined ex vivo using the PP2A activity assay kit. (C) Roles of PP2A activation in the regulation of Bcr-Abl1 and P-Bcr-Abl1 in response to Scr or SK-1 siRNAs were examined in vector controls vs I2PP2A-GFP over-expressing K562/IMA-3 cells using Western blotting (first and second panels lanes 1-2 and 3-4, respectively). Effects of vector-only and I2PP2A overexpression on total and P-PP2A at Y307 (inactive form) in Scr or SK-1 siRNA treated K562/IMA-3 cells were also determined using Western blotting (third and fourth panels lanes 1-2 and 3-4, respectively). Actin was used as a loading control (fifth panel). (D) Role of I2PP2A overexpression in the regulation of cell viability in response to SK-1 siRNA was examined using trypan blue exclusion in drug resistant LAMA-4/IMA cells. (E) Effects of SK-1-V5 overexpression compared with vector-transfections (lanes 2 and 1, respectively) on the total PP2A or SHP1 vs inactive P-PP2A at Y307, or P-SHP1 at S591 (top left and right first and second panels, respectively) were measured by Western blotting. Effects of overexpression of SK-1 compared with vector transfections on the levels of P-Bcr-Abl1 at Y245 were determined by Western blotting (bottom left third panel lanes 2 and 1, respectively). Expression of SK-1-V5 was confirmed using the anti-V5 antibody in Western blotting (bottom right panel), and β-actin was used as a loading control (bottom left third panel). These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels C and E, relative expression levels of proteins (normalized to actin levels) are shown below each lane.
In contrast, overexpression of SK-1 in sensitive K562 cells increased P-Bcr-Abl1 (Y245) of 4-fold compared with controls (Figure 4E). This effect was concomitant with increased levels of inactive forms of P-PP2A (Y307) and P-SHP1 (S591) around 3- and 1.5-fold, respectively, without increasing total PP2A or SHP1 protein expression (Figure 4E left and right panels lanes 2-1, and 4-3, respectively). Overall, these data indicate that activation of SK-1/S1P signaling regulates Bcr-Abl1 stability and drug resistance via inhibition of PP2A/SHP1 axis, and knockdown of SK-1 mediates down-regulation of Bcr-Abl1 by activation of PP2A.
Role of PP2A in the regulation of Bcr-Abl1 in response to targeting SK-1/S1P/S1P2 signaling
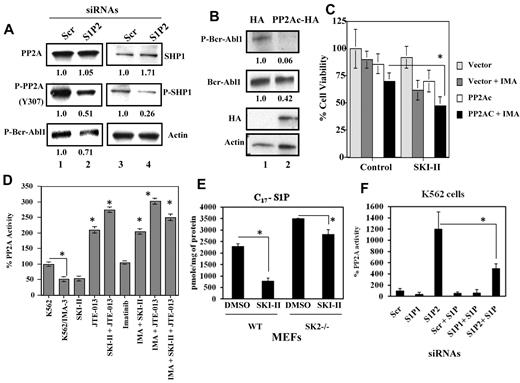
The involvement of PP2A and SHP1 in the regulation of Bcr-Abl1 by S1P2 signaling was assessed. S1P2 knockdown in K562/IMA-3 cells led to a 60%-70% decrease in inactive forms of PP2A (P-PP2A) and SHP1 (P-SHP1), respectively (Figure 5A second left and right panels lanes 2-1, respectively). Accordingly, around 30% decrease in P-Bcr-Abl1 was observed on S1P2 knockdown (Figure 5A bottom left panel lanes 2-1, respectively). Thus, these data suggest that knockdown of S1P2 activates PP2A/SHP1 axis, concomitant with down-regulation of Bcr-Abl1.
Role of PP2A activation in the regulation of Bcr-Abl1 in response to SK-1/S1P2 targeting. (A) Effects of knockdown of S1P2 in K562/IMA-3 cells using siRNA compared with Scr-transfected controls (lanes 2 and 1, respectively) on total and P-PP2A and P-SHP1 (at Y307 and S591, respectively), were assessed by Western blotting (left and right top and middle panels, respectively). Effects of knockdown of S1P2 on the expression of P-Bcr-Abl1 at Y245 were also determined by Western blotting compared with Scr controls (bottom left panel lanes 2 and 1, respectively). β-actin was used as a loading control (bottom right panel). (B) Effects of overexpression of PP2Ac-HA on the levels of P-Bcr-Abl1 (Y245), or total Bcr-Abl1 (first and second panels) were determined by Western blotting, compared with controls (lanes 2-1, respectively). Expression of PP2Ac-HA was also confirmed in these experiments (third panel). (C) Inhibition of growth in response to imatinib (0.5μM for 48 hours) in the absence/presence of PP2Ac-HA overexpression and/or SKI-II in K562/IMA-3 cells was examined using trypan blue exclusion. (D) Effects of targeting SK and S1P2 using small molecule inhibitors SKI-II and JTE-013, respectively, on PP2A activity levels in extracts isolated from K562 and K562/IMA-3 cells in the absence or presence of imatinib compared with untreated controls were measured as described in “Measurement of PP2A activity.” (E) Inhibitory effects of SKI-II on SK-1 and SK-2 enzyme activities in wt MEFs, versus SK-1 activity on SK-2−/− MEFs were measured using C17-Sph labeling followed by measurement of C17-S1P generation in cells by LC/MS/MS. (F) Effects of knockdown of S1P1 or S1P2 using siRNAs on PP2A activity in the absence/presence of exogenous S1P (1μM) in sensitive K562 cell extracts were measured using the PP2A activity assay kit. Scr siRNA was used as a control. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels A and B, relative expression levels of proteins (normalized to actin levels) are shown below each lane.
Role of PP2A activation in the regulation of Bcr-Abl1 in response to SK-1/S1P2 targeting. (A) Effects of knockdown of S1P2 in K562/IMA-3 cells using siRNA compared with Scr-transfected controls (lanes 2 and 1, respectively) on total and P-PP2A and P-SHP1 (at Y307 and S591, respectively), were assessed by Western blotting (left and right top and middle panels, respectively). Effects of knockdown of S1P2 on the expression of P-Bcr-Abl1 at Y245 were also determined by Western blotting compared with Scr controls (bottom left panel lanes 2 and 1, respectively). β-actin was used as a loading control (bottom right panel). (B) Effects of overexpression of PP2Ac-HA on the levels of P-Bcr-Abl1 (Y245), or total Bcr-Abl1 (first and second panels) were determined by Western blotting, compared with controls (lanes 2-1, respectively). Expression of PP2Ac-HA was also confirmed in these experiments (third panel). (C) Inhibition of growth in response to imatinib (0.5μM for 48 hours) in the absence/presence of PP2Ac-HA overexpression and/or SKI-II in K562/IMA-3 cells was examined using trypan blue exclusion. (D) Effects of targeting SK and S1P2 using small molecule inhibitors SKI-II and JTE-013, respectively, on PP2A activity levels in extracts isolated from K562 and K562/IMA-3 cells in the absence or presence of imatinib compared with untreated controls were measured as described in “Measurement of PP2A activity.” (E) Inhibitory effects of SKI-II on SK-1 and SK-2 enzyme activities in wt MEFs, versus SK-1 activity on SK-2−/− MEFs were measured using C17-Sph labeling followed by measurement of C17-S1P generation in cells by LC/MS/MS. (F) Effects of knockdown of S1P1 or S1P2 using siRNAs on PP2A activity in the absence/presence of exogenous S1P (1μM) in sensitive K562 cell extracts were measured using the PP2A activity assay kit. Scr siRNA was used as a control. These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant. In panels A and B, relative expression levels of proteins (normalized to actin levels) are shown below each lane.
To define the role of PP2A in the regulation of Bcr-Abl1, we over-expressed PP2Ac in K562/IMA-3 cells, and determined its effects on Bcr-Abl1 expression. Overexpression of the catalytic domain of PP2A (PP2Ac)-HA, confirmed by Western blotting (Figure 5B third panel lanes 2-1, respectively), reduced the levels of P-Bcr–Abl1 and total Bcr-Abl1 compared with vector-transfected controls (Figure 5B first and second panels lanes 2-1, respectively). In addition, ectopic PP2Ac-HA expression decreased the viability of K562/IMA-3 cells in response to imatinib (0.5μM; around 40%; Figure 5C). Importantly, when cells that over-express PP2Ac were treated with an inhibitor of SK, SKI-II,37 growth inhibitory effect of imatinib was further enhanced (around 65%), compared with controls (Figure 5C). Thus, these data indicate that activation of PP2A signaling reduces Bcr-Abl1 and induces imatinib-mediated growth inhibition in the absence/presence of SKI-II in K562/IMA-3 cells.
Then we examined the effect of pharmacologic inhibition of SK-1 and S1P2 on PP2A activity. As expected, PP2A activity was lower (around 50%) in resistant compared with parental K562 cells (Figure 5D). Inhibition of S1P2 using JTE-013,34 alone or in combination with an SK inhibitor, SKI-II37 increased PP2A activity approximately 2.5- or 3-fold, respectively in K562/IMA-3 cells compared with controls in the absence/presence of imatinib (Figure 5D). Interestingly, when SKI-II or JTE/013 were combined with imatinib, PP2A activity was increased of approximately 2- or 3-fold, respectively (Figure 5D). Moreover, since SKI-II can inhibit both SK-1 and SK-2, we measured its inhibitory activity against these enzymes selectively using SK-1+/+ (wt) and SK-1−/− MEFs and C17-sphingosine labeling by LC/MS/MS. Treatment with SKI-II resulted in the inhibition of C17-S1P generation by SK-1 and/or SK-2 (around 70%) in wt MEFs, and inhibited SK-1–generated C17-S1P approximately 30% in SK-2−/− MEFs (Figure 5E), confirming the inhibitory effects of SKI-II on SK-1 and SK-2. However, since knockdown of SK-2 did not alter Bcr-Abl1 expression in K562/IMA-3 cells (see supplemental Figure 4), effects of SKI-II on drug-induced cell death, and PP2A activation are most likely because of inhibition of SK-1 in these cells. Thus, results shown in Figure 5D suggest that inhibition of SK-1 and/or S1P2 activate PP2A in the absence/presence of imatinib.
In addition, effects of S1P1 or S1P2 knockdown using siRNAs on PP2A activity in the absence/presence of S1P were examined in parental K562 cells (Figure 5F). Down-regulation of S1P2, but not S1P1, increased PP2A activity approximately 12-fold. Moreover, exogenous S1P inhibited PP2A activity by approximately 50%, and it partially prevented increased PP2A activity in response to S1P2 knockdown (Figure 5F), consistent with the protective effects of S1P on the Bcr-Abl1 down-regulation induced by S1P2 siRNA (see Figure 2C). Thus, these data suggest that SK-1/S1P/S1P2 signaling regulates Bcr-Abl1 stability via modulating PP2A activity in K562 cells. These data also suggest that targeting/inhibition of SK-1/S1P2 decreases the stability of P-Bcr-Abl1 via induction of PP2A activity, which is prevented by exogenous S1P.
Inhibition of SK-1/S1P2 signaling increases imatinib-induced cell death in drug resistant CML cells
Because targeting/inhibition of SK-1/S1P2 axis increased PP2A activity, thereby decreasing Bcr-Abl1 and P-Bcr-Abl1 stability, this should also enhance imatinib-induced apoptosis and attenuate drug resistance. Thus, K562, K562/IMA-1, or K562/IMA-3 cells were treated with increasing concentrations of imatinib in the absence/presence of SKI-II (20-30μM) and/or JTE-013 (10μM), and their effect on PP2A activity, Bcr–Abl1 expression and growth inhibition, and/or apoptosis was assessed. Cotreatment with SKI-II and JTE-013 (48 hours) significantly (≥ 80% induction) increased imatinib-induced apoptosis in K562 (0.1μM), K562/IMA-1 (1μM) and/or K562/IMA-3 cells (0.5μM; supplemental Figures 5A-B, and Figure 6A, respectively). Similar effect was observed on SK-1 knockdown, which enhanced imatinib-induced cell death in resistant K562/IMA-3 cells (Figure 6B).
Effects of the inhibition of SK-1 and S1P2 on imatinib- or nilotinib-induced apoptosis in K562/IMA-3 and MNCs obtained from a CML patient at BC with the T315I-Bcr-Abl1 expression. (A) Imatinib-induced apoptosis (at 0.5μM for 24 hours) in the absence/presence of SKI-II, JTE-013 or SKI-II/JTE-013 in combination was measured using the caspase 3 activity assay in K562/IMA-3 cells. (B) Effects of knockdown of SK-1 using siRNA on the inhibition of survival in response to imatinib treatment (at 0.3, 0.5, and 1.0μM at 48 hours) in K562/IMA-3 cells were determined using trypan blue exclusion. (C) Effects of knockdown of SK-1 using siRNA on total and P-Bcr-Abl1 expression in the absence/presence of LACT (5 μM) in K562/IMA-3 cells were assessed by Western blotting compared with Scr siRNA treated controls (top and middle panels lanes 2-4 and 1-3, respectively). Actin levels were used as loading controls (bottom panel). Relative expression levels of Bcr-Abl1 and P-Bcr-Abl1 (normalized to actin levels) are shown below each lane. (D) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013, respectively, alone or in combination, on the survival of MNCs isolated from a CML patient at BC (patient 4), who expressed the T315I-Bcr-Abl1 who did not respond to any treatment in the clinic, were determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay in the absence/presence of nilotinib (0.5, 1.0, or 3.0μM for 48 hours). The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
Effects of the inhibition of SK-1 and S1P2 on imatinib- or nilotinib-induced apoptosis in K562/IMA-3 and MNCs obtained from a CML patient at BC with the T315I-Bcr-Abl1 expression. (A) Imatinib-induced apoptosis (at 0.5μM for 24 hours) in the absence/presence of SKI-II, JTE-013 or SKI-II/JTE-013 in combination was measured using the caspase 3 activity assay in K562/IMA-3 cells. (B) Effects of knockdown of SK-1 using siRNA on the inhibition of survival in response to imatinib treatment (at 0.3, 0.5, and 1.0μM at 48 hours) in K562/IMA-3 cells were determined using trypan blue exclusion. (C) Effects of knockdown of SK-1 using siRNA on total and P-Bcr-Abl1 expression in the absence/presence of LACT (5 μM) in K562/IMA-3 cells were assessed by Western blotting compared with Scr siRNA treated controls (top and middle panels lanes 2-4 and 1-3, respectively). Actin levels were used as loading controls (bottom panel). Relative expression levels of Bcr-Abl1 and P-Bcr-Abl1 (normalized to actin levels) are shown below each lane. (D) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013, respectively, alone or in combination, on the survival of MNCs isolated from a CML patient at BC (patient 4), who expressed the T315I-Bcr-Abl1 who did not respond to any treatment in the clinic, were determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay in the absence/presence of nilotinib (0.5, 1.0, or 3.0μM for 48 hours). The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. *P < .05 was considered significant.
Mechanistically, the ubiquitination of Bcr-Abl1 was reduced in K562/IMA-3 compared with K562 cells, and treatment of these cells with a proteasome inhibitor lactacystin (LACT) increased the levels of ubiquitinated Bcr-Abl1 (supplemental Figure 6A lanes 1-2 and 3-4, respectively). In fact, ubiquitination of Bcr-Abl1 was reduced in resistant compared with sensitive K562 cells, transduced with an HA-ubiquitin construct, and LACT treatment restored the levels of HA-ubiquitinated Bcr-Abl1 in these cells (supplemental Figure 6B lanes 1-2 and 3-4, respectively). Treatment of drug resistant K562/IMA-1 cells with SKI-II and/or JTE-013 (alone or in combination) also increased the ubiquitination of Bcr-Abl1 compared with untreated controls (supplemental Figure 6C lanes 4-1). Importantly, knockdown of SK-1 decreased Bcr-Abl1 and P-Bcr-Abl1 around 40% and 65%, respectively, and proteasome inhibition by LACT almost completely prevented down regulation of Bcr-Abl1 in response to SK-1 siRNA in K562/IMA-3 cells (Figure 6C lanes 2 and 4 top and middle panels). Interestingly, LACT did not have any detectable effect on down-regulation of P-Bcr-Abl1 (Y245) in response to SK-1 knockdown (Figure 6C middle panel lanes 2 and 4, respectively), indicating that proteasomal degradation is regulated down-stream of dephosphorylation of Bcr-Abl1. Taken together, these data suggest that pharmacologic and molecular targeting of SK-1/S1P2 enhance imatinib-induced apoptosis, and attenuate drug resistance by induction of PP2A-dependent Bcr-Abl1 dephosphorylation, and subsequent ubiquitination and proteasomal degradation in drug-resistant cells.
Targeting of SK-1 and S1P2 signaling in MNCs isolated from bone marrow of TKI-sensitive and -resistant CML patients
To validate the data obtained in K562 and LAMA4 derivative cell lines, the proapoptotic effects of targeting SK1/S1P2 signaling were evaluated in MNCs, isolated from bone marrow of CML-CP (patient 1) and CML-BC (patients 2 and 3) patients. CML cells were exposed to increasing concentrations (0.5-1μM) of imatinib in the presence/absence of SKI-II and/or JTE-013. As shown in supplemental Figure 7A through C, SKI-II and JTE-013, alone or in combination, significantly enhanced the growth inhibitory effects of imatinib. The ability of SKI-II and JTE-013 to increase sensitivity to TKIs was also evaluated in CD34+ progenitors isolated from a CML-BC patient 4, bearing the imatinib-, nilotinib-, and dasatinib-resistant T315I-BCR-ABL1 mutant (Figure 6D). Note that SK-1 and S1P2 mRNA levels were 2.5- and 10-fold higher, respectively, in MNCs from CML-BC (patient 4) than healthy individuals (supplemental Figure 7D). Treatment with SKI-II or JTE-013 to inhibit SK-1/S1P2 signaling in the absence/presence of nilotinib (1 and 3 μM)38 enhanced nilotinib-induced growth inhibition of CD34+ T315I+ CML-BC progenitors compared with controls (Figure 6D). Thus, these data suggest that targeting SK-1/S1P2 signaling allow, at least in part, nilotinib-induced apoptosis in CML-BC progenitors expressing the wt- and/or T315I mutant of Bcr-Abl1.
Inhibition of SK1 and S1PR2 overcomes resistance to imatinib or nilotinib in 32Dcl3 cells that express Y253H or T315I mutants of Bcr-Abl1
To examine whether inhibition of SK1/S1PR2 signaling overcomes imatinib resistance in cells that express mutant forms of Bcr-Abl1, murine myeloid 32Dcl3 (32D) cells,39 which stably express wt-, Y253H- or T315I-Bcr-Abl1 were treated with nilotinib (0.05-0.5μM) in the absence/presence of SKI-II and JTE-013 in combination, and cell survival was examined using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assays. Vector-transfected cells were used as controls. The 32D/Y253H-, or 32D/T315I-Bcr-Abl1 (supplemental Figures 8B, or Figure 7A, respectively) cells exhibited resistance to nilotinib when compared with 32D/wt-Bcr-Abl1 cells (supplemental Figure 8A), with IC50 values around 0.5 and > 0.5 compared with around 0.03μM, respectively. The 32D/wt- 32D/Y253H-, or 32D/T315I-Bcr-Abl1 cells overexpressed SK-1 and S1P2 around 20, 45, 50, or 25, 73, 220%, respectively, compared with 32D/vector cells (supplemental Figure 8C). Remarkably, treatment with SKI-II and/or JTE-013 enhanced TKI-induced growth inhibition against 32D/wt-, 32D/Y253H-Bcr-Abl1, and 32D/T315I-Bcr-Abl1 cells (supplemental Figures 8A-B, and Figure 7A, respectively). These data were also consistent with decreased P-T315I-Bcr-Abl1 (at Y177) in response to SKI-II and/or JTE-013 treatment (Figure 7A right panel). Moreover, SK-1 down-regulation using siRNA also enhanced nilotinib-induced growth inhibition in 32D/T315I-Bcr-Abl1 cells at 0.05, 0.1, and 0.5μM by approximately 30%, 45% and 60%, respectively (Figure 7B).
Effects of the inhibition of SK-1 and S1P2 on nilotinib-induced apoptosis in drug resistant 32D cells, expressing the T315I-Bcr-Abl1 in situ and in severe combined immunodeficiency mice in vivo. (A) 32D/T315I-Bcr-Abl1 cells were treated with increasing concentrations of nilotinib (0.05, 0.1, and 0.5μM for 48 hours) in the absence/presence of SKI-II or JTE-031 (alone or in combination), and then their effects on cell survival were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (left panel). Effects of nilotinib, SKI-II, JTE-013, or SKI-II/JTE-013 treatments on the levels of P-T315I-Bcr-Abl1, normalized to actin levels, were determined by Western blotting using the antibody that recognizes P-Bcr-Abl1 at Y177 compared with untreated cells. Relative expression levels of P-Bcr-Abl1 (normalized to actin levels) detected in Western blots are shown (right panel). (B) Role of SK-1 in the regulation of nilotinib-mediated growth inhibition (at 0.05, 0.1, and 0.5μM for 48 hours) was examined using trypan blue exclusion in the absence/presence of Scr or SK-1 siRNAs in 32D/T315I-Bcr-Abl1 cells. The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. (C) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013 in the absence/presence of nilotinib on the growth of 32D/T315I-Bcr-Abl1–derived tumors (n = 8) were determined in severe combined immunodeficiency/Balb/c mice. Mice containing 32D/wt-Bcr-Abl1– or 32D/T315I-Bcr-Abl1–derived allografts were treated with nilotinib at 10 mg/kg (every 3 days for 18 days), and untreated mice were used as a control (left panel). Mice containing the 32D/T315I-Bcr-Abl1–derived allografts were treated with 10 mg/kg nilotinib in the absence/presence of SKI-II and/or JTE-013 at 15, or 0.5 mg/kg,34,37 respectively (right panel). Treatments were performed at days 1, 8, 15, and tumor growth was measured at days 1, 6, 11, 14, and 18 after treatments. Error bars represent SD, and *P < .05 was considered significant. (D) The proposed mechanism by which SK-1/S1P/S1P2 signaling regulates drug resistance. Regulation of drug resistance by SK-1/S1P involves the modulation of PP2A-dependent and SHP1-mediated degradation of Bcr-Abl1 (wt and mutants) selectively via S1P2 signaling. In addition, targeting SK-1/S1P2 signaling using pharmacologic (SKI-II and/or JTE-013) or molecular (siRNAs against SK-1 or S1P2) tools induces PP2A activity, leading to degradation of Bcr-Abl1, and attenuate resistance to TKIs (imatinib or nilotinib).
Effects of the inhibition of SK-1 and S1P2 on nilotinib-induced apoptosis in drug resistant 32D cells, expressing the T315I-Bcr-Abl1 in situ and in severe combined immunodeficiency mice in vivo. (A) 32D/T315I-Bcr-Abl1 cells were treated with increasing concentrations of nilotinib (0.05, 0.1, and 0.5μM for 48 hours) in the absence/presence of SKI-II or JTE-031 (alone or in combination), and then their effects on cell survival were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (left panel). Effects of nilotinib, SKI-II, JTE-013, or SKI-II/JTE-013 treatments on the levels of P-T315I-Bcr-Abl1, normalized to actin levels, were determined by Western blotting using the antibody that recognizes P-Bcr-Abl1 at Y177 compared with untreated cells. Relative expression levels of P-Bcr-Abl1 (normalized to actin levels) detected in Western blots are shown (right panel). (B) Role of SK-1 in the regulation of nilotinib-mediated growth inhibition (at 0.05, 0.1, and 0.5μM for 48 hours) was examined using trypan blue exclusion in the absence/presence of Scr or SK-1 siRNAs in 32D/T315I-Bcr-Abl1 cells. The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. (C) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013 in the absence/presence of nilotinib on the growth of 32D/T315I-Bcr-Abl1–derived tumors (n = 8) were determined in severe combined immunodeficiency/Balb/c mice. Mice containing 32D/wt-Bcr-Abl1– or 32D/T315I-Bcr-Abl1–derived allografts were treated with nilotinib at 10 mg/kg (every 3 days for 18 days), and untreated mice were used as a control (left panel). Mice containing the 32D/T315I-Bcr-Abl1–derived allografts were treated with 10 mg/kg nilotinib in the absence/presence of SKI-II and/or JTE-013 at 15, or 0.5 mg/kg,34,37 respectively (right panel). Treatments were performed at days 1, 8, 15, and tumor growth was measured at days 1, 6, 11, 14, and 18 after treatments. Error bars represent SD, and *P < .05 was considered significant. (D) The proposed mechanism by which SK-1/S1P/S1P2 signaling regulates drug resistance. Regulation of drug resistance by SK-1/S1P involves the modulation of PP2A-dependent and SHP1-mediated degradation of Bcr-Abl1 (wt and mutants) selectively via S1P2 signaling. In addition, targeting SK-1/S1P2 signaling using pharmacologic (SKI-II and/or JTE-013) or molecular (siRNAs against SK-1 or S1P2) tools induces PP2A activity, leading to degradation of Bcr-Abl1, and attenuate resistance to TKIs (imatinib or nilotinib).
These results were also confirmed in animal studies, in which treatment of severe combined immunodeficiency mice bearing 32D/T315I-Bcr-Abl1–derived allografts with nilotinib in combination with JTE-013 and/or SKI-II significantly (around 50%-70%, P < .05, n = 8) reduced tumor growth compared with untreated or SKI-II/JTE-013-only–treated controls (Figure 7C right panel). Moreover, expression of the T315I-Bcr-Abl1 resulted in resistance to nilotinib-induced growth suppression (approximately 50%) when compared with wt-Bcr-Abl1 expressing 32D-derived allografts in these mice (Figure 7C left panel). Thus, these data show that targeting SK1 and S1P2 signaling using pharmacologic or molecular tools mediate T315I-Bcr-Abl1 dephosphorylation and instability, and enhance nilotinib-induced growth inhibition in situ and in vivo.
Discussion
The data presented here revealed that SK-1/S1P-mediated drug resistance is transduced selectively via S1P2-dependent modulation of PP2A, which attenuates ubiquitination and proteasomal degradation of Bcr-Abl1, enhancing its stability, and resulting in subsequent drug resistance (Figure 7D). Moreover, these results also demonstrate that targeting the SK-1/S1P/S1P2 axis using SKI-II/JTE/013 or siRNAs presents a novel strategy for overcoming drug resistance (Figure 7D), associated with decreased stability of Bcr-Abl1 (wt or Y253H and T315I mutants) in CML cells or progenitors both in situ and in vivo.
Recently, roles of SK-1/S1P signaling in imatinib resistance were shown in various CML cell lines and MNCs obtained from CML patients.18,40,41 However, mechanisms of this process, especially the involvement of S1P2 signaling via activation of PP2A in the regulation of Bcr-Abl1 (wt and mutants) and drug resistance were unknown, which is revealed for the first time in this study. Previously, increased ceramide generation was implicated in imatinib-induced apoptosis in K562 cells, and alterations of ceramide/S1P rheostat was reported to mediate TKI resistance. However, the role of ceramide in the modulation of Bcr-Abl1 stability, and/or PP2A activation in response to TKI treatment remains unknown. Since ceramide activates PP2A partially by prevention of I2PP2A/SET,29 whose overexpression causes drug resistance in CML,35 it is likely that imatinib-induced ceramide might mediate PP2A-dependent dephosphorylation and inactivation of Bcr-Abl1, leading to its degradation and increased cell death in CML cells. This view, however, needs further evaluation.
Inhibition of SK-1 by small molecule inhibitors, such as SKI-II,37 or neutralizing S1P using a monoclonal antibody42 effectively kill cancer cells, and inhibit tumor growth. Indeed, SKI-II mediates the proteasome-dependent degradation of SK-1, leading to the inhibition of S1P generation.43 Consistent with these data, our results indicate that pharmacologic and/or molecular inhibition of SK-1 help overcome drug resistance in CML cells via inducing PP2A-dependent dephosphorylation/inactivation and degradation of Bcr-Abl1.
In this study, we also showed that S1P2 signaling contributes to drug resistance in CML via attenuation of Bcr-Abl1 dephosphorylation/degradation. Overexpression of S1P2 was not sufficient to induce drug resistance, and it required the addition of S1P in K562 cells, suggesting that SK-1–generated S1P is necessary for the inhibition of PP2A and regulation of imatinib resistance upstream of S1P2 signaling. Importantly, in our studies, targeting S1P2 using JTE-013 or siRNA resulted in the activation of PP2A and degradation of Bcr-Abl1, which were partially prevented by exogenous S1P, leading to increased imatinib-induced apoptosis in drug resistant CML cells. To this end, the role of S1P2 in CML drug resistance was supported recently by global siRNA screening studies, in which S1P2 siRNA was found as one of siRNAs to sensitize drug resistant CML cells.44 In addition to its involvement in drug resistance, S1P2 also plays an essential role in hypoxia-triggered pathologic angiogenesis of the mouse retina, which was suppressed in S1P2−/− knock-out mice.45 Moreover, transforming growth factor-β-induced migration and invasion of esophageal cancer cells was shown to involve SK-1-generated S1P via S1P2 signaling.46 It should be noted, however, that disruption of the S1P2 gene led to the development of diffuse large B-cell lymphoma (DLBCL) in approximately half of S1P2−/− KO mice within 1.5 to 2 years of age.47 Thus, long-term targeting of S1P2 might have some unwanted consequences.
The role of PP2A/SHP1-dependent degradation of Bcr-Abl1 has been well documented previously.35 Our data show that SK-1/S1P/S1P2 signaling inhibits PP2A activity, and thus prevents PP2A-dependent regulation of Bcr-Abl1 dephosphorylation and subsequent proteasomal degradation. Interestingly, PP2A was reported to inhibit SK-1 activity, suggesting an interesting negative regulatory loop between SK-1/S1P and PP2A activities.48 However, SK-1/S1P/S1P2–dependent inhibition of PP2A has not been described previously, and specific signaling cascade involved in this process remain unknown.
Importantly, targeting SK-1/S1P2 axis was efficacious to overcome imatinib or nilotinib resistance in 32D/Y253H-Bcr-Abl1 or 32D/T315I-Bcr-Abl1 cells. In addition, MNCs isolated from CML/BC patients expressing the 3T315I-Bcr-Abl1 were sensitized to nilotinib in response to SKI-II and/or JTE-013. However, nilotinib was found to have limited38 or no effect against the T315I-Bcr-Abl1 mutant previously.49,50 Therefore, there might be Bcr-Abl1–independent mechanisms of nilotinib-induced cell death in the absence/presence of SKI-II/JTE-013.
In summary, these data suggest that SK-1/S1P plays important roles in the regulation of drug resistance in CML selectively via S1P2-dependent modulation of PP2A, leading to the stability of wt- and mutant forms of Bcr-Abl1, and drug resistance. Moreover, we show here that targeting the SK-1/S1P/S1P2 axis presents a novel therapeutic approach to induce PP2A-dependent dephosphorylation/inactivation and subsequent degradation of Bcr-Abl1 (wt and the T315I or Y253H mutants), and overcome TKI resistance both in situ and in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr U. Gunduz (Middle East Technical University, Ankara, Turkey) for providing us with the K562/IMA-1 cells. We also thank Dr Jennifer Schnellmann (Medical University of South Carolina) for her editorial review of this manuscript.
This work was supported by research grants from the National Institutes of Health (NIH; CA088932, DE016572, and CA097132 to B.O., and CA095512 to D.P.) and the US Army (CML Research Program, W81XWH-07-1-0270 to D.P.). D.P. is a Scholar of The Leukemia & Lymphoma Society. A.S. was supported by the Minority Fellowship grant from the NIH. Lipid measurements and animal studies were conducted in facilities constructed with support from the NIH (C06 RR015455), at the Extramural Research Facilities Program of the National Center for Research Resources.
National Institutes of Health
Authorship
Contribution: A.S., S.P., C.E.S., M.M.-N. S.P.D., S.A.S., E.A., R.D.S., Y.B., and H.M.E. performed key experiments and analyzed data; C.S. provided inhibitors of SK and helped draft the manuscript; C.G. and L.M.O. performed experiments for the development of SK-1−/− MEFs, and performed analysis of SK activity assays; J.O., R.S., G.M., and D.P. performed experiments regarding PP2A activity and cell growth assays using MNCs obtained from CML patients and helped draft and revise the manuscript, S.M., D.F., and R.S. helped isolate MNCs; and B.O. designed experiments, analyzed data, and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of E.A. is Inonu University, Faculty of Science, Department of Biology, Malatya, Turkey. The current affiliation of Y.B. is Izmir Institute of Technology, Department of Molecular Biology and Genetics, Izmir, Turkey.
Correspondence: Dr Besim Ogretmen, 86 Jonathan Lucas St, Rm HO512A-B, Charleston, SC 29425; e-mail: ogretmen@musc.edu.
References
Author notes
A.S. and S.P. contributed equally to this study.

![Figure 1. Regulation of the stability and expression of Bcr-Abl1 by SK-1/S1P signaling. (A) Total and P-Bcr-Abl1 (Y245) in K562/IMA-3 compared with K562 cells (lanes 2 and 1, respectively) were measured using Western blotting. β-actin was used as a loading control. (B) The protein stability of Bcr-Abl1 in K562 and K562/IMA-3 cells was measured by Western blotting at various time points (0-36 hours) using CHX. Long-lived protein GAPDH was used as a control. Relative expression levels of Bcr-Abl1 (normalized to GAPDH levels) at 0-36 hours after CHX treatment in K562 versus K562/IMA-3 cells are shown. (C) The stability of P-Bcr-Abl1 in K562 and K562/IMA-3 cells were also examined by pulse-chase experiments using [32P]-orthophosphate labeling without CHX using immunoprecipitation by anti–P-Bcr-Abl1 (Y177) antibody. (D) The protein stability of Bcr-Abl1 in response to overexpression of SK-1 with the V5 tag in K562 parental cells was determined at various time points (0-36 hours) compared with vector-transfected controls using Western blotting. GAPDH was used as a loading control (left panel). (E) The expression of SK-1 was down-regulated using siRNA, and its effect on the stability of Bcr-Abl1 was determined using CHX treatment at 12 hours compared with Scr-siRNA treated controls. (F) Effects of genetic loss of SK-1 on Bcr-Abl1 stability were examined in wt and SK-1−/− MEFs stably transfected with the human wt-Bcr-Abl1-GFP (p210) in the presence of CHX at 0, 18, 30 hours. The stability of P-Bcr-Abl1-GFP in wt versus SK-1−/− MEFs was measured using [32P]-orthophosphate labeling and immunoprecipitation (using the antibody that recognizes P-Bcr-Abl1 at Y177) as described in “Determining the half-life of Bcr-Abl1 protein.” These experiments were performed in duplicates, and error bars represent SD. *P < .05 was considered significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/22/10.1182_blood-2010-08-300772/4/m_zh89991173130001.jpeg?Expires=1767716075&Signature=vO-~MnDWPFby97pZq5fHrpWAMVLchc7wS7tMN5FkKLVDePUpMT0Yu83lTiBdWkLeVag7urDRmFEcYT8u9zdcLQ-~DBM5XGeqVcsmu9Wn8X0Bkxdw5T36jbUDuNmNqG3ig7B2cqTjAom95G0ZnxfNMmPYjXeRfLVvdy-~DpKQjTw5vkj4CG6~VT4qTccuHiShVq4WJ-kh3ub1oXfb0Ur6chv5FcuU08P-ZVLShk44ynfGaL0uw2mg5pqDqz1KHAOodPZf7cAnmPNoNGsEgN6c2cjLUkTfQl-tzfqXtnpZDGAg1ReynMj9MpbAclc0N0uxgn0CyOTB6GQSF03ohVAmog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Effects of the inhibition of SK-1 and S1P2 on imatinib- or nilotinib-induced apoptosis in K562/IMA-3 and MNCs obtained from a CML patient at BC with the T315I-Bcr-Abl1 expression. (A) Imatinib-induced apoptosis (at 0.5μM for 24 hours) in the absence/presence of SKI-II, JTE-013 or SKI-II/JTE-013 in combination was measured using the caspase 3 activity assay in K562/IMA-3 cells. (B) Effects of knockdown of SK-1 using siRNA on the inhibition of survival in response to imatinib treatment (at 0.3, 0.5, and 1.0μM at 48 hours) in K562/IMA-3 cells were determined using trypan blue exclusion. (C) Effects of knockdown of SK-1 using siRNA on total and P-Bcr-Abl1 expression in the absence/presence of LACT (5 μM) in K562/IMA-3 cells were assessed by Western blotting compared with Scr siRNA treated controls (top and middle panels lanes 2-4 and 1-3, respectively). Actin levels were used as loading controls (bottom panel). Relative expression levels of Bcr-Abl1 and P-Bcr-Abl1 (normalized to actin levels) are shown below each lane. (D) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013, respectively, alone or in combination, on the survival of MNCs isolated from a CML patient at BC (patient 4), who expressed the T315I-Bcr-Abl1 who did not respond to any treatment in the clinic, were determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay in the absence/presence of nilotinib (0.5, 1.0, or 3.0μM for 48 hours). The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. *P < .05 was considered significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/22/10.1182_blood-2010-08-300772/4/m_zh89991173130006.jpeg?Expires=1767716075&Signature=CffB6jMPk4O3ijN0gZ3OGlv3-NtJgKOdyM4eCTCDtsvcoLvB6AhvJ-xIuvWsWOqwj~EIDLRgplaIgprdCdyYdz-a-BCEx9ZxXzCLGwkRolJDpPRC86NfKX1RXgIX-7rl-fQZjkHpbH80UjejRcv-71geKm38QJbB2e0pyQFHSiCOQ6GJjrbax5tXIahkAQDS0ifbhQ84kqgVTq9U-kWs5L62GMSKP1FcEi1AotEQbi-p-nPMzRNSNvp2HEGOXcXOxpZEOsjnwpDRGt03voxp4UC2XkdhHtGkwiMrbXEVliAVFA6RUZkjxm10bmY6eJnojnXzgYiVhO2kVPwAofcwSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Effects of the inhibition of SK-1 and S1P2 on nilotinib-induced apoptosis in drug resistant 32D cells, expressing the T315I-Bcr-Abl1 in situ and in severe combined immunodeficiency mice in vivo. (A) 32D/T315I-Bcr-Abl1 cells were treated with increasing concentrations of nilotinib (0.05, 0.1, and 0.5μM for 48 hours) in the absence/presence of SKI-II or JTE-031 (alone or in combination), and then their effects on cell survival were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (left panel). Effects of nilotinib, SKI-II, JTE-013, or SKI-II/JTE-013 treatments on the levels of P-T315I-Bcr-Abl1, normalized to actin levels, were determined by Western blotting using the antibody that recognizes P-Bcr-Abl1 at Y177 compared with untreated cells. Relative expression levels of P-Bcr-Abl1 (normalized to actin levels) detected in Western blots are shown (right panel). (B) Role of SK-1 in the regulation of nilotinib-mediated growth inhibition (at 0.05, 0.1, and 0.5μM for 48 hours) was examined using trypan blue exclusion in the absence/presence of Scr or SK-1 siRNAs in 32D/T315I-Bcr-Abl1 cells. The data represent at least 2 independent trials performed in duplicates, and error bars represent SD. (C) Effects of the inhibition of SK-1 and S1P2 using SKI-II and JTE-013 in the absence/presence of nilotinib on the growth of 32D/T315I-Bcr-Abl1–derived tumors (n = 8) were determined in severe combined immunodeficiency/Balb/c mice. Mice containing 32D/wt-Bcr-Abl1– or 32D/T315I-Bcr-Abl1–derived allografts were treated with nilotinib at 10 mg/kg (every 3 days for 18 days), and untreated mice were used as a control (left panel). Mice containing the 32D/T315I-Bcr-Abl1–derived allografts were treated with 10 mg/kg nilotinib in the absence/presence of SKI-II and/or JTE-013 at 15, or 0.5 mg/kg,37,34 respectively (right panel). Treatments were performed at days 1, 8, 15, and tumor growth was measured at days 1, 6, 11, 14, and 18 after treatments. Error bars represent SD, and *P < .05 was considered significant. (D) The proposed mechanism by which SK-1/S1P/S1P2 signaling regulates drug resistance. Regulation of drug resistance by SK-1/S1P involves the modulation of PP2A-dependent and SHP1-mediated degradation of Bcr-Abl1 (wt and mutants) selectively via S1P2 signaling. In addition, targeting SK-1/S1P2 signaling using pharmacologic (SKI-II and/or JTE-013) or molecular (siRNAs against SK-1 or S1P2) tools induces PP2A activity, leading to degradation of Bcr-Abl1, and attenuate resistance to TKIs (imatinib or nilotinib).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/22/10.1182_blood-2010-08-300772/4/m_zh89991173130007.jpeg?Expires=1767716075&Signature=Ebi0QbzGwLHZOd7sCqIG8x8wgf~giaxdzBdZ24IsAFPVWQo94kPhb~vEqh30ZzL1GSCMMYfyK93pg2y3vGVbALe7hcH42CdhAlIymBOH5QRq38bvQBHoxzQsnLXxl6GPMYFs4REIXaKkYPOU4gnbGFX0OHq2p8WH3MQ67C3~ZqO8cljCrRdCN1LceC7ZcZ070imLeltraXgJZZ2yrYYNoJBXwhkRJse2bv-VxS-MbQ8KqqcgxIgKl01KOdWHILepURtlLsrudtu1QLeVd422C92ET35OP27RQDP2UBxtVw8o8EKREMd24qyrIFshSV~tXAnCOGXZHG34jOWoLGJ27g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal