To the editor:
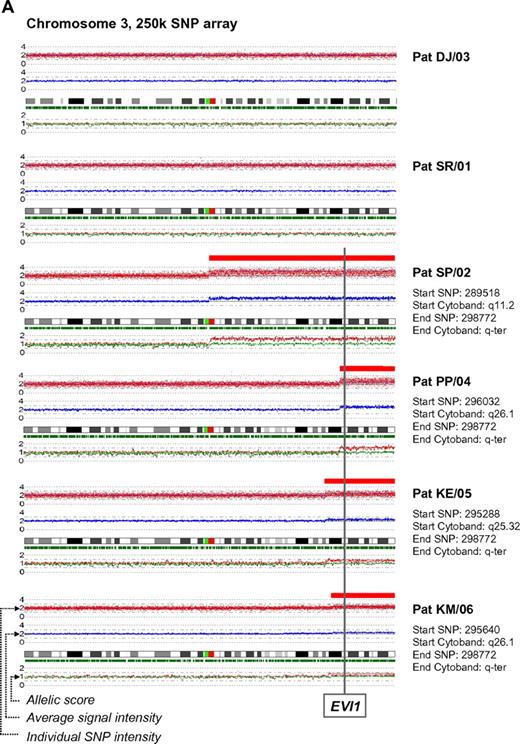
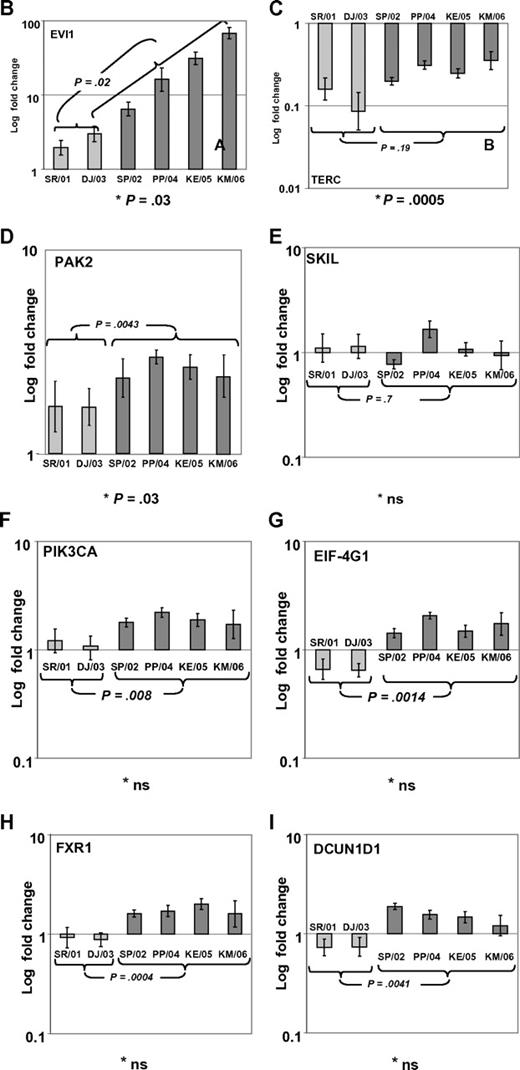
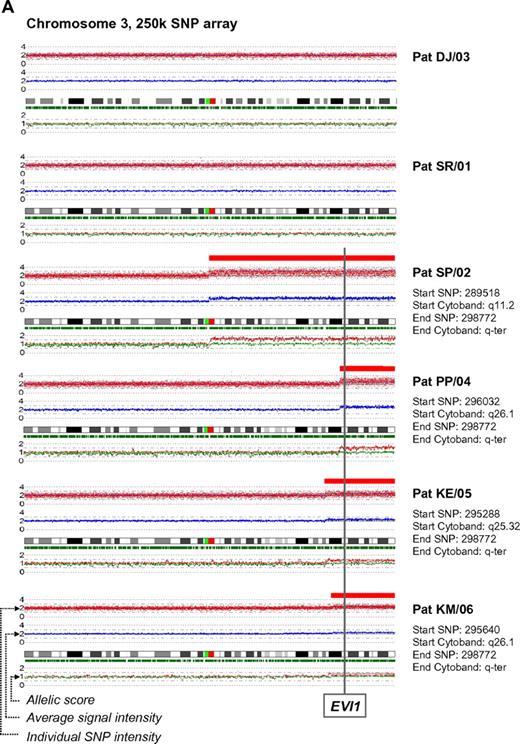
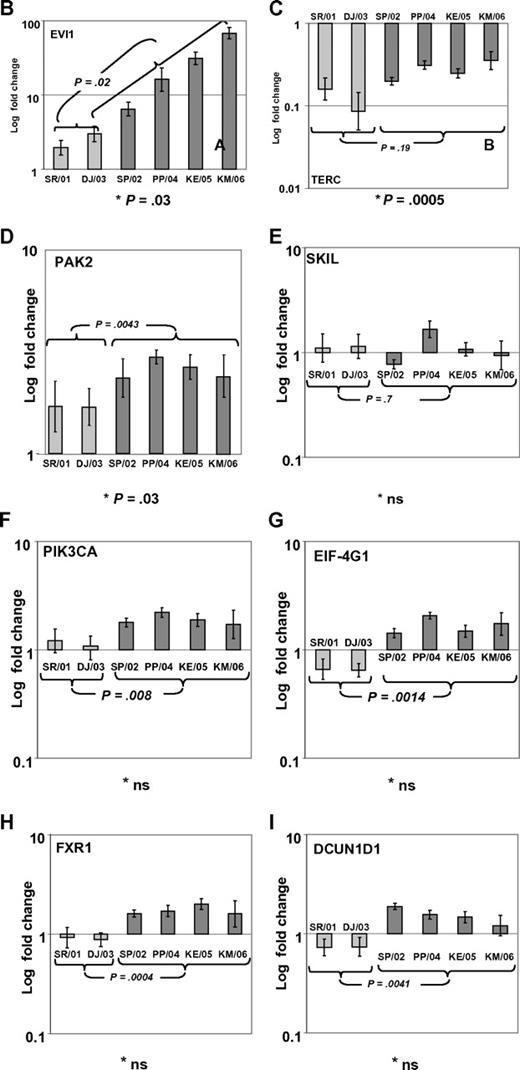
In a recent issue of Blood, Quentin et al1 reported on genomic abnormalities in Fanconi anemia (FA)–associated leukemic transformation. They confirmed 3q gains as an FA-characteristic chromosomal aberration and discussed the potential role of EVI1. Complementing this comprehensive study, we have analyzed gene expression patterns of FA-associated 3q26q29 gains using mononuclear bone marrow cells from backup harvests from 6 FA patients (FA-BM; clinical details in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online letter), and 2 healthy child-donor marrow harvests as controls. Approval was obtained from the Charité institutional review board for these studies. Informed consent was provided in accordance with the Declaration of Helsinki. We characterized 3q26q29 gains in 4 FA-BMs by high-resolution analysis with 250K SNP array (Affymetrix). Two FA-BM patients did not have 3q gains (Figure 1A). Expression patterns were determined with HG-U133plus2 arrays (Affymetrix) and quantitative real-time PCR (qRT-PCR; supplemental Methods and supplemental Tables 2-3). Transcripts encoded at 3q26q29 are targeted by 491 probesets on the U133plus2arrays, of which 134 were called “absent” in at least 7 of the 8 samples. Present in 2 or more samples, but mean expression levels not different comparing FA-BM with or without 3q gains with normal controls, were 231 probesets. Only 2 probesets detected significantly lower transcript levels in FA-BM. Twenty-five probe sets were more highly expressed in FA-BM, irrespective of 3q gains. Changing from absent to present, or significantly more highly expressed in samples with 3q gains, were 99 probesets, of which 32 were also significantly more highly expressed in all FA-BM. The highest fold change of these had probe set 226420_at, which targets EVI1. A greater than 10-fold increase in mean EVI1 transcripts in FA-BM samples was confirmed by qRT-PCR (Figure 1B). U133plus2 arrays do not detect telomerase complex RNA component (TERC), which is encoded in close proximity to EVI1. As TERC has an important role in hematopoiesis, and TERC overexpression is commonly associated with 3q gains,2-4 we analyzed TERC expression by qRT-PCR. We detected an up to 10-fold lower TERC level in all FA-BM (Figures 1C; P = .0005), irrespective of 3q gains or bone marrow morphology. Validation of expression analysis of other genes in the 3q26q29 region with a role in malignant transformation by qRT-PCR confirmed a small increase in transcript levels for PIK3CA, EIF-4G1, DCUN1D, and FXR1, while PAK2 was also more highly expressed comparing all FA-BM to normal BM. SKIL/SNON expression was not different in FA-BM or affected by 3q gains (Figure 1D-I).
SNP characterization and RT-PCR analysis of 3q gains in FA. (A) SNP array characterization of 3q gains in all FA bone marrow harvests. SNP array data are visualized using CNAG 3.0 software. Analysis of call intensity was conducted using standard Affymetrix analysis software algorithms provided in GCOS and GTYPE 4.0. Chromosomal aberrations were analyzed using Copy Number Analyzer for Affymetrix GeneChip Mapping arrays (CNAG) 3.0 software employing the AsCNAR algorithm. In 4 patients with 3q gain, the extension of aberration is depicted using red bars and the position coordinates are shown on the right side. Position of EVI1 is indicated. (B-I) qRT-PCR analysis of selected genes as indicated (B: EVI1; C: TERC; D: PAK2; E: SKIL; F: PIK3CA; G: EIF-4G1; H: FXR1; I: DCUN1D1) in FA bone marrow samples with 3q-gains (darker-shaded right 4 columns) and without (brighter-shaded left 2 columns). Transcripts levels are shown as 2−ΔΔCt normalized to normal BM (*P below graph) using β-actin and SDHA as housekeeper genes. Comparison also between mean transcript levels of FA marrows with and without 3q gains (t test).
SNP characterization and RT-PCR analysis of 3q gains in FA. (A) SNP array characterization of 3q gains in all FA bone marrow harvests. SNP array data are visualized using CNAG 3.0 software. Analysis of call intensity was conducted using standard Affymetrix analysis software algorithms provided in GCOS and GTYPE 4.0. Chromosomal aberrations were analyzed using Copy Number Analyzer for Affymetrix GeneChip Mapping arrays (CNAG) 3.0 software employing the AsCNAR algorithm. In 4 patients with 3q gain, the extension of aberration is depicted using red bars and the position coordinates are shown on the right side. Position of EVI1 is indicated. (B-I) qRT-PCR analysis of selected genes as indicated (B: EVI1; C: TERC; D: PAK2; E: SKIL; F: PIK3CA; G: EIF-4G1; H: FXR1; I: DCUN1D1) in FA bone marrow samples with 3q-gains (darker-shaded right 4 columns) and without (brighter-shaded left 2 columns). Transcripts levels are shown as 2−ΔΔCt normalized to normal BM (*P below graph) using β-actin and SDHA as housekeeper genes. Comparison also between mean transcript levels of FA marrows with and without 3q gains (t test).
Aware of the limitations resulting from small sample size, our data imply consistent activation of EVI1 expression with FA characteristic 3q gains, which are uncommon in AML, but prognostically very important.5,6 As EVI1 expression is highest in BM with complex karyotype aberrations, additional genetic changes in FA-associated leukemic transformation might further contribute to EVI1 overexpression, and implies the possibility that EVI1 overexpression might confer clonal advantage specifically in the presence of an FA defect. Altered telomere maintenance, but not low TERC expression, has been described in FA7 and might contribute to bone marrow failure in FA. Our finding of up to 10-fold lower TERC expression in all FA-BM irrespective of chromosomal aberrations or bone marrow morphology implies the possibility of an FA-specific TERC dysregulation.
Authorship
Acknowledgments: We thank Sian Dibben, Dan White, Trevor Carr, and Sophie Gaubert for technical support.
Funding was provided by Cancer Research UK (CRUK) Clinician Scientist Fellowship to S.M. (C1860/A5901), and Charité Universitätsmedizin Berlin. S.M. and A.D.W. are supported by the Leukemia Lymphoma Research Fund (LLR, UK). H.H. is supported by the BMBF network for bone marrow failure syndromes (bmfs) and NIH R01 CA138237-01. We acknowledge the support of the Manchester Biomedical Research Center.
Contribution: S.M. developed the study, designed the research, analyzed microarray data, and wrote the manuscript; C.B. carried out sample preparation and qRT-PCR; S.M., M.W., and S.P. analyzed microarray data; A.D.W. provided scientific input and laboratory facilities and contributed to writing the manuscript; H.H. provided data regarding FA complementation analysis; H.N. and W.E. provided patient material and clinical data and contributed to writing the manuscript; M.W.W. carried out SNP array analysis and contributed to writing the manuscript; and H.T. initiated and developed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.T. is Robert Koch-Institute, Berlin, Germany.
Correspondence: Dr Stefan Meyer, Academic Unit of Paediatric and Adolescent Oncology, University of Manchester, c/o Young Oncology Unit, Christie Hospital,Wilmslow Road Manchester M20 6XB,United Kingdom: e-mail: stefan.meyer@manchester.ac.uk.
References
National Institutes of Health
Supplemental data
Shown are present/absent calls (columns E–L), and normalised 2.0 .cel file data generated using the simpleaffy package in BioConductor (columns Q–X). In columns Z to AK comparisons are shown of mean transcript level comparing i) 6 FA vs 2 control (6vs2, columns Z–AC), ii) normal 3q BM vs 3q gain (4vs 4, columns AD–AG), and iii) FA–BM with 3q gains vs FA–BM with normal 3q (4 vs 2, columns AH–AK), with p-values generated applying non paired t-test, and FDR (false detection rate) values generated using the limma fit model from the limma BioConductor package (www.bioconductor.org).The entire microarray datas et and full details of labelling protocols are available in a Minimum Information About a Microarray Experiment (MIAME) compliant database at http://bioinformatics.picr.man.ac.uk/vice/Welcome.vice.