Abstract
B-cell precursor childhood acute lymphoblastic leukemia with ETV6-RUNX1 (TEL-AML1) fusion has an overall good prognosis, but relapses occur, usually after cessation of treatment and occasionally many years later. We have investigated the clonal origins of relapse by comparing the profiles of genomewide copy number alterations at presentation in 21 patients with those in matched relapse (12-119 months). We identified, in total, 159 copy number alterations at presentation and 231 at relapse (excluding Ig/TCR). Deletions of CDKN2A/B or CCNC (6q16.2-3) or both increased from 38% at presentation to 76% in relapse, suggesting that cell-cycle deregulation contributed to emergence of relapse. A novel observation was recurrent gain of chromosome 16 (2 patients at presentation, 4 at relapse) and deletion of plasmocytoma variant translocation 1 in 3 patients. The data indicate that, irrespective of time to relapse, the relapse clone was derived from either a major or minor clone at presentation. Backtracking analysis by FISH identified a minor subclone at diagnosis whose genotype matched that observed in relapse ∼ 10 years later. These data indicate subclonal diversity at diagnosis, providing a variable basis for intraclonal origins of relapse and extended periods (years) of dormancy, possibly by quiescence, for stem cells in ETV6-RUNX1+ acute lymphoblastic leukemia.
Introduction
The translocation t(12;21)(p13;q22) generating ETV6-RUNX1 (TEL-AML1) gene fusion is a consistent genetic abnormality in 25% of childhood B-cell precursor acute lymphoblastic leukemia (ALL).1 It typically occurs in young patients and is only occasionally detected in adolescents and young adults.2 Guthrie card analysis and twin studies have previously shown that the ETV6-RUNX1 fusion gene frequently originates before birth, probably as an initiating lesion.3-5 Preleukemic clones with ETV6-RUNX1 are generated prenatally at a rate considerably in excess of clinical ALL.6 This, plus the modest concordance of ALL in monozygotic twins (10%-15%), suggests that additional, postnatal mutations are necessary for progression to overt malignancy.4 Modeling studies with both murine7 or human8 cells support this contention. Cytogenetic analyses have identified secondary chromosomal abnormalities associated with ETV6-RUNX1+ ALL of which deletion of the non-rearranged ETV6 allele appears the most frequent (∼ 70%).9 This ALL subtype generally has a favorable prognosis.10 Relapses when they do occur are usually in patients off treatment.11 These may occur several years after cessation of treatment12-14 and occasionally after 10-20 years.15 Clonotypic Ig/TCR markers indicate that very late relapsing ALLs are clonally derived from the initially presenting clones.16 A striking feature of late relapsing cases of ALL is that they are chemosensitive with durable term remissions possible.17,18 This implies that their initial survival and reemergence is related to properties other then mutation-based drug resistance. Comparison of the genomic boundaries (by microsatellite markers) of ETV6 deletions at diagnosis and relapse has previously shown that these differ in some but not all cases.19,20 One plausible interpretation of this finding is that the relapse clone can derive from a minor clone present at diagnosis. This view is endorsed by IgH clonotypic markers in which the dominant rearrangement observed at relapse is detectable by PCR as a minor clone at diagnosis.13,19-23 The nature of such minor clones is unclear from such studies, but one suggestion was that they might represent persistent preleukemic stem cells8 with ETV6-RUNX1 but lacking both ETV6 deletion and all other secondary genetic changes.19,20
Recent developments in whole genome scanning provide a greater depth of interrogation of clonal relationships. Genomic analyses with the use of high-density single nucleotide polymorphism (SNP) arrays have recently shown an average of ∼ 6 chromosomal copy number alterations (CNAs) in childhood ALL with ETV6-RUNX1 fusion gene.21 Three recent studies have used high-density CNA analysis to try and elucidate the molecular mechanism of disease recurrence in childhood ALL. These 3 studies have shown an increase in CNAs between presentation and relapse, with B-cell development and cell-cycle regulation pathways most often affected.22,24,25 In most cases the presenting and relapse clones contained some identical CNAs, suggesting a shared clonal origin. This was confirmed by back-tracking PCR some of the genetic lesions originally identified at relapse by SNP analysis. In most cases the relapse “specific” lesion was present at low level at presentation in a minor subclone.22 The investigators of that study equated minor subclones to ancestral clones in a linear clonal succession model.22 If, as speculated previously, some very late relapses in ALL derive from preleukemic clones, then these relapses should have a distinct profile of CNAs absent at diagnosis, and no individual relapse CNAs should be identifiable at diagnosis. We acquired a series of matched presentation/remission/relapse DNA samples from 21 pediatric patients with ETV6-RUNX1+ ALL who had relapses of various time intervals from initial diagnosis to evaluate clonal origins of relapse. The described origin of relapse analysis was performed with the caveat that complete genomic characterization of the cell of origin remains elusive at present. ETV6-RUNX1 itself, we presumed, should provide, as the probable initiating lesion, a stable marker of all descendent clones along with ≥ 1 clonotypic Ig or TCR rearrangements.4,20
Methods
Patients
Twenty-one patients with relapsed ETV6-RUNX1–positive childhood ALL were studied (see supplemental Table 1 for patient characteristics, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The samples were obtained from the Academic Department of Paediatric Oncology, The Royal London Hospital, United Kingdom, Great Ormond Street Hospital, United Kingdom, Department of Pediatric Oncology/Hematology, Charité–Medical University, Berlin, Germany, and Childhood Leukemia Investigation Prague, Department of Paediatric Hematology and Oncology, 2nd Faculty of Medicine, Charles University Prague, Czech Republic. The ethics committee or institutional review board of each participating center approved the study. Informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. The diagnosis of ETV6-RUNX1 ALL was established through FISH and PCR analysis as per local protocols. Samples were selected on the basis of availability of DNA or viable cells at initial presentation, remission, and relapse (R) with blast purity of ≥ 75% before extraction of DNA. The median age at diagnosis was 4.5 years (range, 1.93-15.8 years). Time to first relapse from diagnosis was < 2 years (ie, on treatment) to 9 years 11 months. Five of 14 patients with data available had a white cell count > 20 × 109/L. At initial presentation the BM was the only site involved in all patients, whereas at relapse the BM was involved in isolation in 15 patients, combined BM and testis in 3 patients, CNS was involved in 2 relapses, and in 1 patient the data were not available. The patients were treated on a variety of protocols, namely UK ALL X/XI, 97/01, 2003, UKALL R2, BFM 90, 95, 2000, 2002, COALL 92, 97, 2003. At submission of this document 9 patients were in complete remission after first or second relapse. Ten patients died after second relapse. The outcome of 2 patients remains unknown. In 21 patients adequate matched presentation and relapse DNA were available for genomewide CNA and loss of heterozygosity (LOH) investigations. Matched remission DNA was available for all but unique patient number (UPN) 2.
Genome mapping analysis
Mapping analysis was performed with 500 ng of tumor and germline DNA from each patient. DNA was prepared according to the manufacturer's instructions with the use of the GeneChip mapping 500K assay protocol for hybridization to GeneChip Mapping 250K Nsp and Sty arrays (Affymetrix). Briefly, genomic DNA was digested in parallel with restriction endonucleases NspI and StyI, ligated to an adaptor, and subjected to PCR amplification with adaptor-specific primers. The PCR products were digested with DNaseI and labeled with a biotinylated nucleotide analog. The labeled DNA fragments were hybridized to the microarray, stained by streptavidin-phycoerythrin conjugates, washed with the Affymetrix Fluidics Station 450, and then scanned with a GeneChip scanner 3000 7G. The CEL files are available for download on http://www.icr.ac.uk/array/array.html.
Copy number and LOH analysis
SNP genotypes were obtained with the use of Affymetrix GCOS software (Version 1.4) to obtain raw feature intensity and Affymetrix GTYPE software (Version 4.0) with the use of the BRLMM to derive SNP genotypes (see supplemental Table 2 for the 250K Sty/Nsp call rates). Samples were analyzed with CNAG 3.0 (http://plaza.umin.ac.jp/genome/) with the use of paired tumor (test) samples with the self-reference control (reference) samples to determine copy number and LOH caused by imbalance.26 The position of regions of LOH and gain were identified with the University of California Santa Cruz Genome Browser (May 2004 Assembly; http://genome.ucsc.edu/cgi-bin/hgGateway). All samples were also analyzed with dChip (May 2008; www.dchip.org) to verify findings obtained with CNAG.27,28 The remission samples were assigned a copy number of 2 and were used as a reference set to calculate copy number in tumor samples. Median smoothing with a window size of 5 was used to infer copy number along each chromosome. LOH analysis was done with dChip with the use of a Hidden Markov Model to infer the probability of LOH on the basis of the paired control/tumor samples, using an average heterozygosity rate of 0.33.
Quantitative genomic PCR
The genomic copy number of the gene CDKN2A was confirmed with Taqman Real-Time PCR (RQ-PCR). The Taqman probe and primers were designed with Primer Express Version 3.0 (Applied Biosystems). The Taqman probes were labeled with FAM (5′) and BHQ-1 (3′). The PCR reactions were performed in triplicate on the ABI 7900 PCR machine (Applied Biosystems) as per the manufacturer's protocol. The primer sequence used was CDKN2A F, 5′-GACTGCGGAGCAATGAAGACT-3′; CDKN2A R, 5′-GATGCAACTGGCCCTAGTTTG-3′; and probe, CDKN2A 5′-TAGAGGTCTAGTGCCCC-3′. The gene copy number was measured by Relative Quantification assay with the use of RNAseP (Applied Biosystems) as amplification control. The gene copy number was measured in presentation, relapse, and matched remission (germline) DNA. Data evaluation was performed with the ABI 7500 SDS software. A fold change ratio of target gene/housekeeping gene < 0.7 was considered indicative of gene deletion (see supplemental Table 3 for the results of the genomic RQ-PCR).
Fluorescence in situ hybridization
Fixed cytospins were prepared from archival viable cells from UPN2 and UPN19. Interphase FISH for the ETV6-RUNX1 fusion gene was performed with a commercial LSI TEL-AML1 extra signal (ES) probe (Vysis; Abbott Laboratories Ltd), according to the manufacturer's instructions. This probe set contains a 350-kb probe for the 5′ end of ETV6 (exons 1-4) and a 500-kb probe covering the entire RUNX1 gene. The FISH signal pattern for ETV6-RUNX1 fusion gene–positive cells with the use of the Vysis probe is 2 red (1 large, 1 small RUNX1 signal), 1 green (ETV6 allele not involved in the translocation), 1 red/green (yellow) fusion signal corresponding to the ETV6-RUNX1 fusion gene. Fosmid probes for PAX5 (Fosmid G248P800846D3) and CDKN2A/CDKN2B (Fosmid G248P882010F5) were obtained from the BACPAC Resource Center, Children's Hospital, Oakland Research Institute (http://bacpac.chori.org). These were labeled by nick translation with biotin-16-dUTP (Roche Diagnostics) as previously described29 and hybridized in combination with the ETV6-RUNX1 ES probe. Hybridization and washes were performed according to the Vysis protocol, with a single layer of cyanine 5–conjugated streptavidin (GE Healthcare) for detection of biotinylated fosmids. Fluorescent signals were viewed with a Zeiss Axioskop fluorescence microscope equipped with filters for DAPI (4′-6′-diamidine-2-phenylindole), FITC/Spectrum Green, Spectrum Orange, and cyanine 5. Images were captured and analyzed with a Hamamatsu ORCA-ER CCD camera and SmartCapture X software (Digital Scientific).
Cloning and backtracking of ETV6-RUNX1 genomic breakpoint fusion sequence
Amplification of the genomic breakpoint fusion sequences for ETV6-RUNX1 at relapse was performed by long-range PCR with the use of conditions and primers previously described.30 The breakpoint sequence was obtained through restriction analysis by primer walking and sequen-cing (supplemental Figure 8). Patient-specific primers for amplification of fusion breakpoints were designed so that PCR yielded a product of 130-300 bp. DNA from presentation material was then amplified with these primers, and resulting products were sequenced to confirm identity with ETV6-RUNX1 intronic fusion in relapse samples (UPN15). In 2 cases (UPN14 and UPN19) backtracking analysis was performed with the sequence of the fusion gene isolated at relapse. For each patient fusion breakpoint specific primers and real-time probe were designed. A dilutional series was made with DNA from relapse diagnosis and DNA pooled from mononuclear cells from 10 healthy volunteers. Genomic real-time quantitative PCR was used to identify and quantify the presence of the fusion sequence at diagnosis (data not shown).
Results
Recurrent CNAs in ETV6-RUNX1–positive ALL
We performed genomewide analysis of CNAs and LOH on matched diagnostic and relapse BM samples from 21 pediatric patients with precursor B-cell ALL carrying an ETV6-RUNX1 fusion gene [t(12;21)(p12;q22)] (see supplemental Table 1 for patient characteristics). DNA copy number and LOH were obtained with the use of the Affymetrix 500K arrays. These arrays interrogate ∼ 500 000 loci. The average distance between SNPs is 5.8 kb.
A total of 159 CNAs at diagnosis and 231 at relapse were identified (excluding physiologic CNAs at the TCR and/or Ig loci) with a mean of 7.57 CNAs per sample (range, 0-17 CNAs per sample) at initial presentation (Table 1; see supplemental Table 4 for a detailed overview of all CNAs detected). Deletions outnumbered gains by ∼ 6.95:1. Eighteen recurrent alterations were found in ≥ 2 diagnostic samples in this series. The most frequently recurring alterations at presentation were deletion of ETV6 (n = 15), CDKN2A (n = 4), CDKN2B (n = 3), 6q16.2-3 (n = 4), FHIT (n = 4), and IL3RA/CSF2RA (n = 5). The most frequently gained regions were 21q22.11-q22.12 (n = 5), Xq26.2-q28 (n = 5), and 12p13.33-p13.31 (n = 2). At diagnosis, 48% of samples showed ≥ 1 deletions in B-cell development regulators; FHIT (n = 4), BTG (n = 3), RAG1/2 (n = 2), CD200/BTLA (n = 2), EBF1 (n = 2), PAX5 (n = 2), IKZF1 (n = 1), and TCF3 (n = 1).
Next we classified gene alterations as probable “driver” or “passenger” mutations. We defined driver mutations as genes that are mutated on a recurrent basis in leukemia or are probably involved in leukemogenesis or both. Passenger mutations are defined as nonrecurrent or have no known or predicted role in leukemogenesis at present or both. The caveat of this approach is the potentially incorrect labeling of drivers as passengers because of the lack of functional studies. Of the 7.36 (mean) CNAs detected at presentation, 3.76 were classified as potential drivers on the basis of recurrence in this or prior series of cases21,22,31-33 (Table 1).
At relapse the mean number of CNAs per sample increased to 11.0 (range, 0-21), 4.86 being potential drivers (Table 1). No new recurrent genomic alterations were identified compared with diagnosis, but the frequency of particular CNAs changed, as shown in Table 2. At relapse 57% of samples showed deletions in B-cell development regulator genes.
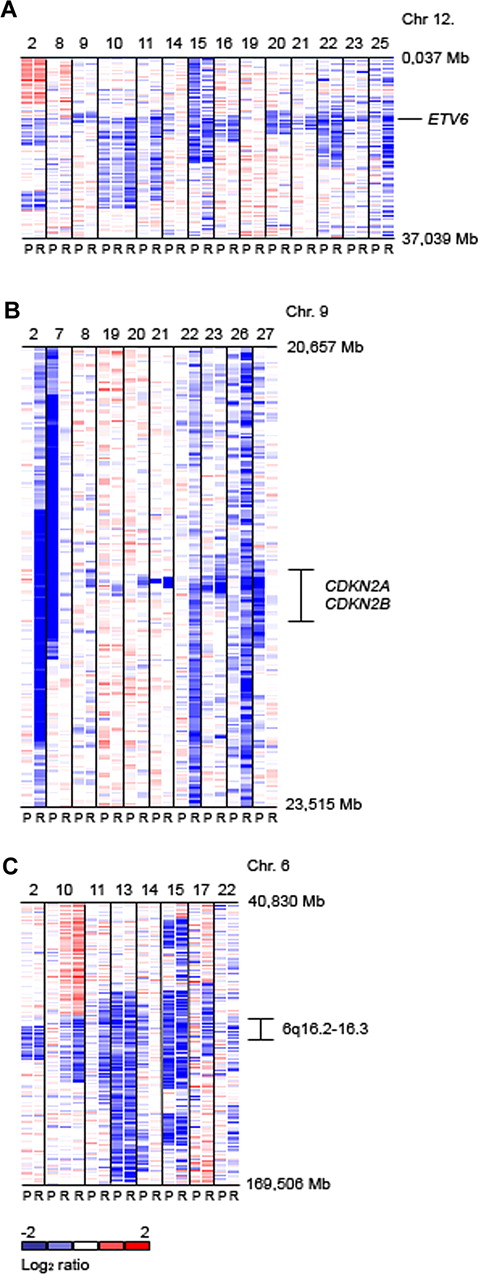
Next, we compared the evolution of individual gene CNAs between presentation and relapse within each patient. Three patterns were established: (1) CNAs at presentation were lost at relapse, (2) new alterations were gained at relapse, or (3) an alteration persisted from presentation to relapse. More detailed analysis of driver mutation evolution identifies a change in the pattern of driver mutations between presentation and relapse in most patients. In the 21 presentation samples a total of 20 copy number gains were detected. Twelve (60%) of these genomic gains persisted in the relapse samples (Table 3). Of the 139 copy number losses at presentation, 98 (71%) remained detectable at relapse. Overall, of 52 driver mutations persisting between presentation and relapse, 49 (94.2%) had identical chromosomal breakpoints at both time points, with the level of resolution determined by 500K SNP array analysis. In 14 patients ≥ 1 identical copy number change in a driver mutation were present at both presentation and relapse (see supplemental Table 5 for evolution of driver mutations present at presentation and relapse). Of the remaining 7 patients, 6 (UPN6, UPN7, UPN11, UPN14, UPN25, and UPN27) shared ≥ 1 TCR or Ig rearrangement or both (confirmed by PCR and sequencing for UPN6 and UPN11) between presentation and relapse. No rearrangement data were available for 1 patient (UPN19) at initial presentation, but for this patient we have demonstrated identical fusion breakpoint sequences at presentation and relapse (see supplemental Table 6 for Ig and TCR rearrangement analyses at presentation and relapse; supplemental Figure 8 for genomic breakpoint sequences). In 16 of 21 relapsed patients a total of 9 new copy number gains and 41 deletions in driver loci appeared at relapse. At relapse the newly acquired gains were distributed on 4 chromosomal regions, namely 21q22.1-22.12 (including RUNX1; n = 3), part or whole of chromosome 16 (n = 3), Xq26.2-28 (n = 2), and 10p (n = 1). The gene loci newly deleted at relapse were CDKN2A/B (n = 7; hemizygous, n = 5; homozygous, n = 2), 6q16.2-3 (n = 4), ETV6 (n = 3), BTG1 (n = 3), DMD (n = 3), EBF1 (n = 2), RB1 (n = 2), 13q14.2-3 (n = 2), MKKS/ORF94 (n = 2), PAX5 (n = 3), LTK (n = 2), DPF3 (n = 2), ELF1 (n = 1), CD200/BTLA (n = 1), PVT1 (n = 1), HIST (n = 1), IL3RA/CSF2RA (n = 1), and TBL1XR1 (n = 1). No new driver mutations were observed at relapse in 5 patients (UPN7, UPN9, UPN13, UPN14, and UPN27). Analysis with dChip for frequent CNAs on chromosome 12p (ETV6), 9p (CDKN2A), and 6q16.2-3 is shown in Figure 1A, B, and C, respectively. The region of deletion on 6q varied in size between the samples. The minimally deleted region was located on 6q16.1-22.1 (95 699 306-115 623 936 bp) containing 72 well-characterized genes and 9 open reading frames, including CCNC, GRIK2, and ARMC2 and 1 microRNA coding region (miR-587).
Log2 ratio SNP 500K copy number data (median smoothed with a window of 5 markers; blue is loss and red is gain). (A) Genomic copy number of chromosome 12p flanking ETV6 for 14 representative cases showing deletions at this locus at presentation (P) and relapse (R). (B) Copy number of chromosome 9p flanking CDKN2A/B. (C) Copy number of chromosome 6q. The chromosomal region of 6q16.2-3 which is recurrently deleted in pediatric ALL is indicated.
Log2 ratio SNP 500K copy number data (median smoothed with a window of 5 markers; blue is loss and red is gain). (A) Genomic copy number of chromosome 12p flanking ETV6 for 14 representative cases showing deletions at this locus at presentation (P) and relapse (R). (B) Copy number of chromosome 9p flanking CDKN2A/B. (C) Copy number of chromosome 6q. The chromosomal region of 6q16.2-3 which is recurrently deleted in pediatric ALL is indicated.
Two patients in this cohort had 2 consecutive relapses each. Patient UPN6 relapsed after 51 (6R1, BM) and 77 (6R2, isolated testes) months, and patient UPN10 after 36 (10R1, BM) and 80 (10R2, BM) months since initial presentation. In the first patient the only driver mutation shared between initial presentation and both relapses was the ETV6-RUNX1 fusion gene. Analysis of TCR and Ig rearrangements by SNP and RQ-PCR/sequencing showed shared clonal origins. Thirteen driver CNAs were shared between R1 and R2. Between R1 and R2 deletion of 6p25.3-25.2 and gain of 16p13.3-12.3 were lost, and deletion of 12p13.2 (ETV6) appeared. In the latter patient, 5 driver mutations were shared between the presenting and relapse clones, and R1 and R2 shared 18 CNAs. At R2 the clone additionally acquired deletion 8q24.21 (PVT1) in comparison with R1. Deletion of 8q12.1 (TOX) was present at presentation and again at second relapse R2. Gain of the short arm of chromosome 16 at R1 was lost at second relapse R2.
Clonal origins of relapse versus time of relapse
Mullighan et al22 and Kuster et al25 used a relapse classification, based on comparative CNA profiles which distinguished clonal origins as either from the “diagnostic” (ie, major) clone or an “ancestral” (ie, precursor and minor) clone present at presentation. Although this is a pragmatic scheme, it makes implicit assumptions about subclonal heterogeneity and ancestral relationships at presentation which are currently uncertain. We therefore selected to use a modified scheme. (Table 4) This classifies relapses into 4 CNA profiles, and for each of these we indicate possible clonal origins with respect to the matched presentation sample. This classification is purely based on driver mutation alterations as detected by the 500K SNP array analysis. In addition, it assumes the persistence of an identical ETV6-RUNX1 fusion gene at presentation and relapse in all cases, as previously shown with matched presentation/relapse samples.20 In our current cohort we confirmed identity of ETV6-RUNX1 genomic sequence in 3 paired diagnostic/relapse samples (UPN14, UPN15, and UPN19; supplemental Figure 8). We distinguished the following 4 profiles: (type 1) the relapse clone resembles the initially presenting leukemia clone, (type 2) the relapse clone has acquired extra driver mutation(s), (type 3) the relapse clone showed losses and gains of driver mutations, and (type 4) the relapse clone has lost all CNAs present initially and acquired a completely new set of alterations. Any samples not fitting these descriptions were defined as unclassified. Of the 21 matched presentation relapse patient samples, 2 patients were classified as type 1 (UPN9 and UPN13), 7 patients were type 2 (UPN2, UPN10, UPN20, UPN21, UPN22, UPN23, and UPN26), 6 as type 3 (UPN8, UPN11, UPN15, UPN16, UPN17, and UPN18), and 4 as type 4 (UPN6, UPN7, UPN14, and UPN19). Two patients were defined as unclassified (UPN25 and UPN 27).
Next, we considered the duration of remission in each of the classified patients and sought a potential relationship between remission duration and the clonal origin of relapse, with the realization that our patients received different therapies and that outcome for ALL is protocol driven, potentially clouding interpretation of data. We used the following time cutoffs: very early relapse < 2 years, early relapse 2-5 years, and late relapse > 5 years since initial presentation. A random distribution of relapse types 1-4 was observed in the very early, early, and late relapse categories, as shown in Table 4, without a clear increase in type 3 or 4 in case the remission duration was ≥ 5 years. We also observed no correlation between total number of driver mutations at relapse and the length of remission. When we compared the number of newly acquired driver mutations at relapse in function of the length of remission duration, we observed an increase in the average number of new drivers with an increase in remission duration (< 2 years: average of 0.75 new drivers; 2-5 years: average of 2.5 new drivers; > 5 years: average of 3.0 new drivers); however, this difference was not significant. The total number of driver mutations or newly acquired driver mutations at relapse did not correlate with survival. In our cohort, the type of relapse did correlate with survival. All patients with type 4 relapses (4 of 4) remain in complete continuous remission (median follow-up, 8.9 years; range, 4.7-10.2 years), whereas only 5 of 15 patients with a type 1-3 relapse are alive and in continuous remission (median follow-up, 6.1 years; range, 3.2-10.2 years; supplemental Table 1).
Backtracking of relapse clone to original presentation
In our series, patient UPN2 experienced the longest remission duration, namely 9 years 11 months. To establish whether in this case the relapse clone was present as a minor subclone at initial presentation, we used FISH to detect relapse clone specific CNAs in the presentation sample. The SNP array data suggested duplication of the ETV6-RUNX1 fusion gene and loss of wild-type ETV6 at presentation and relapse and hemizygous loss of 9p (including CDKN2A and PAX5) in addition to loss of the other CDKN2A allele (homozygous loss of CDKN2A) at relapse (supplemental Table 4). FISH analysis of the presentation and relapse samples was performed in 702 and 582 cells, respectively (supplemental Table 7). The FISH analysis of the presentation sample confirmed the presence of a dominant leukemic clone with duplication of the fusion gene and loss of the nontranslocated ETV6 in 72.4% of cells. These cells did not show deletion of CDKN2A. Hemizygous loss of CDKN2A occurred in 21.3% of cells. A small clone (0.4%) was detected with a genotype identical to the relapse clone, with duplication of the fusion gene, loss of wild-type ETV6, and homozygous loss of CDKN2A (Figure 2). At relapse, most cells showed duplication of the fusion gene, loss of the nontranslocated ETV6, and homozygous loss of CDKN2A (79.8%). Leukemic cells with no or hemizygous loss of CDKN2A were visualized in 11.5% and 1.4%, respectively (supplemental Figures 4,6). CDKN2A deletion-specific backtracking PCR was unsuccessful in this patient. Parallel FISH analysis at presentation and relapse with probes for ETV6, RUNX1, and PAX5 was performed and confirmed the genotype of the dominant clones at both time points, that is, duplication of the fusion gene and 2 copies of PAX5 in 89.14% of cells at presentation and duplication of the fusion gene and 1 copy of PAX5 in 60.4% of cells (supplemental Table 7; supplemental Figures 5-6).
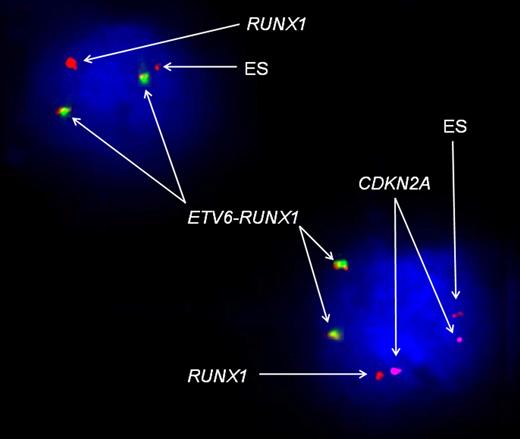
FISH analysis of UPN2 at presentation, using the LSI TEL/AML1 ES Dual-Color translocation probe (Vysis) and CDKN2A probe. The red signal represents the RUNX1 probe; green, the ETV6 probe; yellow, the ETV6-RUNX1 fusion gene; and purple, the CDKN2A probe. This image represents 2 different leukemic cells at presentation. The cell in the right bottom corner represents the dominant leukemic clone (according to the SNP array data analysis), with duplicated fusion gene, loss of wild-type ETV6, 2 copies of CDKN2A, 1 normal RUNX1, and 1 ES (72% of total cells analyzed). In the left top corner FISH analysis identified a leukemic cell with a constellation of signals corresponding to the dominant relapse clone, that is, a duplicated fusion gene, loss of wild-type ETV6, 1 normal RUNX1, 1 ES, and complete loss of CDKN2A (0.4% of total cells analyzed). In control experiments with normal blood lymphocytes from 5 donors, using the same probe combination, this CNA pattern was not observed in any cells (< 0.2%).
FISH analysis of UPN2 at presentation, using the LSI TEL/AML1 ES Dual-Color translocation probe (Vysis) and CDKN2A probe. The red signal represents the RUNX1 probe; green, the ETV6 probe; yellow, the ETV6-RUNX1 fusion gene; and purple, the CDKN2A probe. This image represents 2 different leukemic cells at presentation. The cell in the right bottom corner represents the dominant leukemic clone (according to the SNP array data analysis), with duplicated fusion gene, loss of wild-type ETV6, 2 copies of CDKN2A, 1 normal RUNX1, and 1 ES (72% of total cells analyzed). In the left top corner FISH analysis identified a leukemic cell with a constellation of signals corresponding to the dominant relapse clone, that is, a duplicated fusion gene, loss of wild-type ETV6, 1 normal RUNX1, 1 ES, and complete loss of CDKN2A (0.4% of total cells analyzed). In control experiments with normal blood lymphocytes from 5 donors, using the same probe combination, this CNA pattern was not observed in any cells (< 0.2%).
The SNP array analysis has identified duplication of the ETV6-RUNX1 fusion gene and loss of wild-type ETV6 at presentation and relapse and hemizygous loss of 9p (including CDKN2A and PAX5) in addition to loss of the other CDKN2A allele (homozygous loss of CDKN2A) at relapse (supplemental Table 4). The homozygous loss of CDKN2A and hemizygous loss of PAX5 in the relapse clone could have arisen in several different ways. The FISH analysis at presentation and relapse with the use of using ETV6, RUNX1, CDKN2A or ETV6, RUNX1, and PAX5 probes suggested focal loss of CDKN2A before loss of the unaffected chromosome 9p containing CDKN2A and PAX5 (supplemental Table 7; supplemental Figure 7).
Discussion
The patterns of CNAs we observed at diagnosis and relapse of ETV6-RUNX1–positive ALL parallel those reported in previous studies of unselected ALL. Overall, there was an increase in the number of CNAs per case in relapses.22,24,25,31 The genes or regions predominantly involved were also as previously described with the 2 most frequently implicated pathways being cell-cycle regulation and B-cell development.21,22,24,31-33 In agreement with a recent study,34 we identified frequent gain of part or whole of chromosome 16 (supplemental Figure 1). This finding has occasionally been described in hematologic malignancy but its significance is unclear. Another novel observation was the deletion of plasmacytoma variant translocation 1 (PVT1) at 8q24 in 3 cases (supplemental Figure 2). The significance of deletion of PVT1 in our cohort remains unclear. Pvt1 is a nonprotein coding gene located 3′ of myc, and this locus harbors several miRNAs. Hsa-miR-1204 is often fused to the immunoglobulin light chain in variant Burkitt lymphoma and present in MYC/PVT1 coamplified tumors.35 Coamplification of MYC and PVT1 has also been identified in human astrocytomas,36 in breast and ovarian cancers,37 and in acute myeloid leukemias and myelodysplastic syndromes.38
In 14 of 21 cases, ≥ 1 driver CNAs were shared between diagnosis and relapse. Ig/TCR clonality data when available (see supplemental Table 6, data available on 15 patients) and conservation of ETV6-RUNX1 fusion (as shown for UPN14, UPN15, and UPN19; supplemental Figure 8) indicated that relapses originated from within the clones present at diagnosis.
A principal and novel objective of this study was to compare SNP array–defined CNAs in relapse versus matched diagnosis samples to gain some insight into the clonal origins of late and very late relapse in ETV6-RUNX1+ ALL. Other studies have looked at this issue in childhood ALL in general or ETV6-RUNX1+ ALL in particular25 and have drawn the conclusion that relapses (undefined by time from diagnosis) can emerge from either major or minor subclones present at diagnosis.22,23,39 Distinct from previous studies we focused entirely on the subset of ALL with ETV6-RUNX1 to ascertain whether the apparent origin from major or minor subclones was associated with time to relapse. In particular, we were interested in the biologically interesting phenomenon of very late relapse in childhood ALL. These relapses can occur intraclonally ≤ 20 years after initial diagnosis.15,16,40 These have included ETV6-RUNX1+ cases. A striking feature is that they are often clinically responsive rather than drug resistant, and durable second remissions and cure are possible. One possibility we and others have previously suggested was that persistent preleukemic clones harboring ETV6-RUNX1, the initiating lesion but no other genetic changes, might be the reservoir for such late relapses.7,8,13,19,20,41
Others have interpreted clonal origins of relapse in ALL in the context of a linear model of clonal evolution in ALL with minor subclones considered as ancestral to major subclones.22,25 Recent single-cell analysis with multiple FISH probes for detecting CNAs indicates rather a complex, nonlinear, branching clonal structure.42,43 In concordance with our previous analyses, the FISH analysis performed in patient UPN2 showed genetic diversity at both presentation and relapse. At presentation we observed a dominant clone with duplicated fusion gene and 2 copies of CDKN2A, alongside a minor clone with duplicated fusion and 1 copy of CDKN2A. At relapse this clone with loss of 1 copy of CDKN2A is not detectable, but “presentation cells” (duplication of fusion gene and 2 copies of CDKN2A) represent a minor clone next to the dominant relapse clone (duplication of fusion gene and complete loss of CDKN2A). The use of single-cell FISH underlines the limitations of using SNP arrays to study the genetics underlying the biology of relapse. Deductions on clonal origins of relapse on the basis of SNP array data should therefore, we suggest, be limited to the likelihood that they are derived from either the dominant or major subclone at diagnosis or from a minor subclone, irrespective of subclonal evolutionary relationships. We devised a scheme to accommodate these possibilities, defining 4 types of relapses in total (Table 4). A clear conclusion from this analysis is that, in accord with other studies, relapses can originate from either major or minor subclones present at diagnosis.
The size of the subclone at diagnosis might be anticipated to influence time to relapse, as reported in some previous studies.39,44 However, in this series, there was no discernable relationship between early or late relapses and origins from minor or major clones at diagnosis. The basis for this discrepancy with prior data are unclear, but it might reflect particular biologic properties of the ETV6-RUNX1+ subset of ALL or the fact that prior studies included more on-treatment, early relapses. The latter might be more likely to be drug-resistant subclones selected by treatment as evidenced by the low frequency of sustained second remissions in such cases. Off-treatment (> 3 years after diagnosis) relapses of ETV6-RUNX1+ ALL are usually responsive to second remission inductions, suggesting that their protracted survival after initial diagnosis and therapy was based on properties other than mutation-based drug resistance. There is some evidence that such subclones (identified by clonotypic IgH sequences) may respond slowly to induction chemotherapy.39 One possibility is that this reflects relative quiescence and associated drug insensitivity rather than more classic, mutation-based resistance. Sustained quiescence would explain the extraordinary length of clinical remission in some of these cases. In the absence of a clear relationship between size of relapse clone at initial presentation and time to relapse in our series, we sought potential relationships between our newly devised relapse classification, number of relapse driver mutations, and overall outcome. No correlation was observed between our relapse classification, relapse drivers, and remission duration. As a consequence we cannot recommend a monitoring scheme to identify low- or high-risk relapses. We did however observe that all patients with type 4 relapses (4 of 4) remain in complete continuous remission, whereas only 5 of 15 patients with a type 1-3 relapse are alive and in continuous remission (supplemental Table 1). One possible explanation for this observation is that premalignant clones without CNAs have a more abbreviated proliferative history and, if so, will be statistically less likely to harbor drug-resistance mutations.45
Preleukemic clones in ALL have been detected as harboring ETV6-RUNX1 gene fusions but no other discernable genetic alterations.4,8 Studies with monozygotic twins with discordant ALL, neonatal blood spot scrutiny, and cord blood screening all suggest that these clones exist at a low level, 10−3 to 10−4, of B lineage cells and can persist throughout childhood.3,5,8,46-48 We previously suggested that such preleukemic clones, perhaps as a consequence of slow growth or quiescence, might be relatively resistant to ablation by chemotherapy, would persist during and after maintenance chemotherapy, and occasionally give rise to another de novo ALL masquerading as a conventional although late off-treatment relapse.19,20 Evidence that appeared to support this view was obtained by comparing genomic boundaries of ETV6 deletion at relapse versus diagnosis whereby they frequently were found to be different. This indicated that the relapse emerged from a minor subclone that did not harbor the ETV6 deletion that was present in most cells at diagnosis.19,20 The comprehensive genetic profiles made available by high-resolution SNP arrays provide a more stringent assessment of this hypothesis. If persistent preleukemic cells (defined as ETV6-RUNX1 fusion-only clones) do provide the source of off-treatment late or very late relapses, then clearly not only ETV6 but all the specific CNAs observed at relapse should be absent at diagnosis, either at the major or minor subclone level. This would correspond to our type 4 relapses (Table 2). As described, we observed 4 such cases. Three cases relapsed between 3 and 5 years from diagnosis, and 1 case presented as a very late relapse. Of the very late relapses (5-9 years 11 months) only 1 (UPN19; supplemental Figure 3) was classified as a type 4 relapse, possibly of preleukemic clonal origin. Overall our data indicate variable but often prolonged quiescence for ALL stem cells with multiple genetic abnormalities.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank patients, parents, and hospital staff for their valuable collaboration.
This work was supported by a Leukemia Research Fund (LRF) grant (M.G.), Kay Kendall Intermediate Fellowship (F.W.v.D.) and (grants MSM0021620813 and NS/1000-4; J.Z.).
Authorship
Contribution: F.W.v.D., L.K., and M.G. designed the research; F.W.v.D. and S.H. performed the research; F.W.v.D. and M.G. analyzed the data and wrote the paper; S.C. and K.A. performed confirmatory FISH; C.B. provided helpful discussion; H.K., J.Z., C.E., and V.S. contributed patient samples; A.F. provided research supervision; and C.E. and A.F. cloned the ETV6-RUNX1 genomic fusion gene breakpoints.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mel Greaves, Section of Haemato-Oncology, Institute of Cancer Research, Brookes Lawley Bldg, 15 Cotswold Rd, Sutton, SM2 5NG, United Kingdom; e-mail: mel.greaves@icr.ac.uk.