Abstract
Whereas the final differentiation of conventional dendritic cells (CDCs) from committed precursors occurs locally in secondary lymphoid or peripheral tissues, plasmacytoid dendritic cells (PDCs) are thought to fully develop in the bone marrow from common DC progenitors before migrating to the periphery. In our study, we define, for the first time, a subpopulation of CCR9− major histocompatibility complex class IIlow PDCs in murine bone marrow, which express E2-2 and are immediate precursors of CCR9+ fully differentiated PDCs. However, CCR9− PDCs have the plasticity to acquire the phenotype and function of CD11b+ CD8α− major histocompatibility complex class IIhigh CDC-like cells under the influence of soluble factors produced by intestinal epithelial cells or recombinant GM-CSF. This deviation from the PDC lineage commitment is regulated on the level of transcription factors reflected by down-regulation of E2-2 and up-regulation of ID2, PU.1, and BATF3. Thus, CCR9− PDCs are immediate PDC precursors that can be reprogrammed to differentiate into CDC-like cells with higher antigen-presenting and cytokine-producing capacity under the influence of the local tissue microenvironment.
Introduction
Although plasmacytoid dendritic cells (PDCs) express markers of the “lymphoid” lineage, they can be derived from both common myeloid and common lymphoid progenitors.1 Several elegant studies have convincingly shown that PDCs arise from common dendritic cell (DC) progenitors (or pro-DCs) in the bone marrow (BM),2,3 which have the additional potential to generate precursor cells committed to conventional dendritic cell (CDC) development (pre-CDCs).4,5 In contrast, common DC progenitors derived precursor cells committed exclusively to PDC development have not been described. Generation of PDCs is dependent on transcription factor E2-2, which drives the expression of other key transcription factors involved in PDC development and function (IRF8, Spi-B, IRF7) as well as PDC-specific markers (BST2 and Siglec H in murine PDCs, BDCA2 in human PDCs).6
It has been reported that a population of Siglec-H-negative PDC-like cells in murine BM, which is only present in lymphocytic choriomeningitis virus-infected mice is capable of differentiating into CDCs during viral infection in a type I interferon (IFN)-dependent manner.7,8 The potential of PDCs or committed PDC precursors to differentiate into other DC subpopulations in the absence of infection is unknown.
It has been assumed from the available data that PDCs fully differentiate in the BM, circulate in the blood, and then enter lymphoid organs and peripheral tissues. CDCs, however, are generated from circulating committed precursors (pre-CDCs), which differentiate locally in lymphoid and peripheral tissues under the control of growth factors, such as Fms-like tyrosine kinase 3 ligand (Flt3L).4,9 Thus, the development of CDC subpopulations is shaped by the local microenvironment allowing adaptation to tissue-specific functions. This has been shown recently for DCs in the intestinal lamina propria whose development from precursors is driven by local growth factors and the enteric microbial flora.10-12 It is so far not clear whether PDCs in the intestine or in other peripheral tissues exclusively derive from fully differentiated circulating PDCs13 or whether they can also differentiate locally from committed precursor cells under the influence of the specific tissue microenvironment.
In this study, we identify an immediate PDC precursor in murine BM, which is characterized by expression of the transcription factor E2-2, PDC-specific markers (BST2, Siglec H), and production of type I IFN but lack of CCR9 and low major histocompatibility complex (MHC) class II expression. We show that CCR9− MHCIIlow PDCs spontaneously give rise to fully differentiated CCR9+ MHCII+ PDCs. However, in contrast to CCR9+ PDCs, these cells retain the ability to divert from the PDC lineage and differentiate into MHCIIhigh CD11b+ CD8α− antigen-presenting CDCs under the influence of GM-CSF, which is produced constitutively by intestinal epithelial cells. Down-regulation of the PDC-specific transcription factor E2–2 and concomitant up-regulation of transcription factors involved in CDC development show that this developmental shift is transcriptionally regulated. Our results demonstrate the existence of an immediate PDC precursor whose final differentiation can be shaped by the local tissue microenvironment.
Methods
Mice
Specific pathogen-free, female 6- to 8-week-old C57BL/6 mice were purchased from Harlan Winkelmann. OT II and OT I mice and Cx3cr1-egfp reporter mice (C57BL/6 background)11 were bred under specific pathogen-free conditions. Experiments were performed in accordance with German animal care and ethics legislation and have been approved by the local government authorities.
Generation of Flt3-ligand generated BM-DCs
BM cells were cultured in DC medium containing 20 ng/mL recombinant human (rh) Flt3-L for 7 days as described (FL-DCs).14
Primary cell isolation and sorting
Spleen, Peyer patches, and mesenteric and inguinal lymph nodes were digested with collagenase D and DNase I. Single-cell suspensions from these organs and from BM were stained with the indicated markers and sorted using a FACSAria (BD Biosciences) or MoFlow cell sorter (Beckman Coulter). CD4+ and CD8+ T cells were isolated from splenocytes by negative selection using MACS technology (Milteny Biotec).
Preparation of intestinal epithelial supernatant
The murine colonic epithelial cell line PTK6 was maintained in RPMI 1640/5% fetal bovine serum/1 μg/mL insulin-transferrin-selenium/10 U/mL rmIFN-γ at 33°C as described.15 PTK6 cells were seeded on 0.5 μm pore size transwell filters and cultured at 37°C until formation of polarized monolayers. Monolayers were washed thoroughly with PBS and inserted into fresh DC medium. Supernatants of intestinal epithelial cells (IEC-SN) were harvested from the basolateral side after 24 hours, centrifuged, and sterile-filtered before use.
DC culture and stimulation
FL-DCs and PDC subpopulations sorted from FL-DCs or primary BM cells were cultured with 50% IEC-SN or recombinant GM-CSF or M-CSF (concentrations indicated in figure legends) at a density of 2.5 × 106/mL for 48 hours. Cells were subsequently harvested for RNA isolation, FACS analysis, and cell sorting. Where indicated, CpG 2216 (0.5μM, MWG Biotech) was added after 24 hours for further 24 hours.
Antigen presentation assays
Sorted DC populations were loaded with OVA peptide (MHC class II– specific peptide: ISQAVHAAHAEINEAGR; aa 323-339) or OVA protein (5μM) for 1 hour and cocultured with carboxyfluorescein succinimidyl ester (CFSE)–labeled CD4+ OT II or CD8+ OT I T-cells at a ratio of 1:10 for 4 days. T cells were analyzed by flow cytometry for proliferation (CFSE dilution) and intracellular IFN-γ and IL-2 after restimulation with phorbol myristate acetate (PMA)/ionomycin.
Flow cytometry
For FACS analysis, cells were stained using fluorescently labeled antibodies directed against the indicated cell surface antigens (eBioscience) as described.14 Anti-BST2 (120G8, rat IgG1)16 and anti-Siglec H antibodies (440c, rat IgG2b)17 were conjugated with fluorescein isothiocyanate or biotin. XCR1 staining reagent was kindly provided by R. Kroczek. Propidium iodide was added to exclude dead cells from analysis. T cells were stimulated with PMA/ionomycin (20 ng/mL/1 μg/mL) for 6 hours adding GolgiPlug and GolgiStop (BD Biosciences). Cells were then fixed in 2% paraformaldehyde, permeabilized with 0.5% saponin, and stained with anti–IFN-γ–phycoerythrin and anti–IL-2–allophycocyanin (BD Biosciences). Cells were acquired using a FACSCalibur flow cytometer (BD Biosciences) or a Gallios flow cytometer (Beckman Coulter).
RNA isolation, cDNA preparation, and quantitative real-time PCR
RNA was isolated using the RNeasy mini kit (QIAGEN) and transcribed to cDNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed using TaqMan Gene expression Master Mix together with a TaqMan Step One Plus instrument. TaqMan probes are: HPRT Mm00446968_m1, Tcf4 Mm01262526_g1, Sfpi1 Mm00488142_m1, Batf3 Mm01318274, Irf8 Mm00492567_m1, Csf2ar Mm00438331_g1, Id2 Mm00711781_m1, Cx3cr1 Mm00438354, and Spib Mm01719550_s1 (Invitrogen).
Diagnostic PCR for Ig rearrangement
Genomic DNA was isolated from 5 × 105 FACS-sorted cells using the DNAeasy Blood & Tissue Kit (QIAGEN). PCR primers specific for rearranged and nonrearranged Ig loci were used to diagnose IgH D-J rearrangement as previously described.18
Cytokine measurement by ELISA
Statistical analysis
Paired, 2-tailed Student t test was used to determine statistically significant differences.
Results
CCR9− MHCIIlow PDCs are immediate precursors of CCR9+ MHCII+ PDCs
Compared with spleen and lymph nodes where the majority of CD11c+ DCs are CDCs, murine BM contains a higher percentage of PDCs (39% ± 6.2% of CD11c+ cells, n = 3) accounting for approximately 3% of total BM leukocytes. Among these, we identified a fraction of 24.1% ± 5.3% (n = 4) PDCs lacking expression or expressing very low levels of the chemokine receptor CCR9 (Figure 1A dot plots). This population is designated as CCR9− PDCs in this manuscript. Further characterization of CCR9+ and CCR9− PDCs in the BM revealed that both populations show a phenotype characteristic for PDCs (Figure 1A; supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In contrast to CCR9+ PDCs, however, CCR9− PDCs expressed very low levels of MHC class II and lower levels of costimulatory molecules CD80 and CD86, demonstrating that CCR9− PDCs are less activated than CCR9+ PDCs. In addition, we observed that CD8α, CD4, and Sca-1 were also expressed at lower levels in CCR9− than CCR9+ PDCs. CD115 (macrophage colony-stimulating factor receptor [M-CSFR]) was expressed at similar levels in both populations with a small subpopulation of CD115high CCR9− PDCs. Expression of GM-CSF receptor (csf2ar) on mRNA level was also comparable in both populations (Figure 1B bottom right panel). CX3CR1, which is expressed on common DC progenitors and at variable levels on all splenic DC subpopulations, was expressed on CCR9− as well as CCR9+ BM PDCs with higher expression levels in CCR9− PDCs (Figure 1A-B). Thus, with the exception of very low MHC class II expression and lack of CCR9, CCR9− PDCs phenotypically resemble CCR9+ PDCs, suggesting a close developmental relationship between these subpopulations.
Characterization of CCR9− and CCR9+ PDCs in murine bone marrow. Expression of the indicated markers was analyzed by FACS in primary BM cells. Dot plots show expression of CD11c versus BST2 in propidium iodide-negative cells and expression of BST2 versus CCR9 in BST2+ CD11c+ cells. Expression of the indicated markers in CCR9+ and CCR9− BST2+ CD11c+ cells is shown below (overlay with fluorescence minus one [FMO] control, filled histograms) (A). Relative mRNA expression was quantified in CCR9+/BST2+/CD11c+ and CCR9−/BST2+/CD11c+ PDC populations sorted from primary BM cells by quantitative RT-PCR (B, mean ± SD, n = 3). Immunoglobulin DH-JH rearrangement was detected by PCR in genomic DNA isolated from CCR9+ and CCR9− PDCs (C). CCR9+ and CCR9− PDCs were incubated with medium or CpG 2216 for 24 hours. IFN-α, IL-12p40, and IL-6 were measured in the supernatants by ELISA (D, mean ± SD, n = 3). *P < .05. **P < .01. ***P < .001. n.s. indicates not significant.
Characterization of CCR9− and CCR9+ PDCs in murine bone marrow. Expression of the indicated markers was analyzed by FACS in primary BM cells. Dot plots show expression of CD11c versus BST2 in propidium iodide-negative cells and expression of BST2 versus CCR9 in BST2+ CD11c+ cells. Expression of the indicated markers in CCR9+ and CCR9− BST2+ CD11c+ cells is shown below (overlay with fluorescence minus one [FMO] control, filled histograms) (A). Relative mRNA expression was quantified in CCR9+/BST2+/CD11c+ and CCR9−/BST2+/CD11c+ PDC populations sorted from primary BM cells by quantitative RT-PCR (B, mean ± SD, n = 3). Immunoglobulin DH-JH rearrangement was detected by PCR in genomic DNA isolated from CCR9+ and CCR9− PDCs (C). CCR9+ and CCR9− PDCs were incubated with medium or CpG 2216 for 24 hours. IFN-α, IL-12p40, and IL-6 were measured in the supernatants by ELISA (D, mean ± SD, n = 3). *P < .05. **P < .01. ***P < .001. n.s. indicates not significant.
The transcription factor E2-2 is critical for development of the PDC lineage.6 We therefore determined relative mRNA expression of E2-2 and other transcription factors involved in DC development in sorted CCR9+ and CCR9− BM PDCs (Figure 1B). E2-2 and IRF8 were expressed in both populations, albeit at slightly lower levels in CCR9− PDCs. No significant difference was observed in the expression of Spi-B, ID2, and BATF3, whereas PU.1 (sfpi1) showed a higher expression in CCR9− PDCs. PCR analysis of genomic DNA obtained from sorted CCR9− and CCR9+ PDCs revealed that only CCR9+ PDCs have undergone DH-JH immunoglobulin gene rearrangement (Figure 1C). High level type I IFN production is a hallmark feature of PDCs, so we stimulated BM PDCs with TLR9 ligand CpG 2216 and were able to show that CCR9− PDCs secreted even higher amounts of IFN-α than the CCR9+ PDCs. In addition, CpG-stimulated CCR9− PDCs produced significant amounts of IL-12 and IL-6, whereas CCR9+ PDCs produced only low levels of IL-12 (1.2 ± 0.3 ng/mL; Figure 1D).
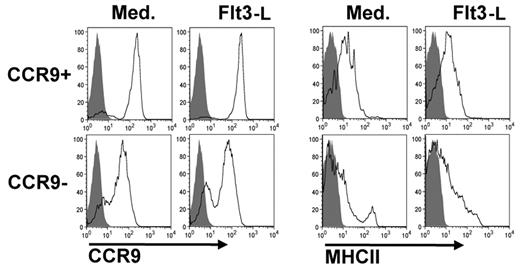
Our analysis shows that CCR9− BM PDCs possess most of the characteristics of CCR9+ differentiated PDCs, with the exception of CCR9 expression and DH-JH rearrangement. We therefore investigated whether CCR9− PDCs can give rise to CCR9+ PDCs by exposing them to culture medium with or without Flt3L for 48 hours. In both conditions, the majority of CCR9− MHCIIlow PDCs differentiated into CCR9+ MHCII+ PDCs (Figure 2). We therefore conclude that CCR9− PDCs are precursors of CCR9+ PDCs.
Differentiation of CCR9− MHCIIlow PDCs into CCR9+ MHCII+ PDCs. CCR9+ and CCR9− PDCs were incubated with or without Flt3-L for 48 hours. Expression of CCR9 and MHC class II was measured by FACS. Filled histograms represent FMO control. Results of one representative experiment are shown.
Differentiation of CCR9− MHCIIlow PDCs into CCR9+ MHCII+ PDCs. CCR9+ and CCR9− PDCs were incubated with or without Flt3-L for 48 hours. Expression of CCR9 and MHC class II was measured by FACS. Filled histograms represent FMO control. Results of one representative experiment are shown.
CCR9− PDCs are also found in spleen and lymph nodes, although the size of this subpopulation is smaller and more variable in these organs than in the BM (11.8% ± 6.4% of splenic PDCs, n = 3, supplemental Figure 1C). CCR9− PDCs were also found in mesenteric lymph nodes and in the colon. In Peyer patches, however, CCR9 expression in PDCs was uniformly lower than in other lymphoid organs, and a distinct population of CCR9− PDCs could not be detected (supplemental Figure 1D). The CCR9− PDC subpopulation in the spleen showed an almost identical phenotype to its counterpart in the BM except for higher expression of MHC class II (supplemental Figure 1B). Thus, CCR9− PDCs exit the BM and could give rise to CCR9+ PDCs in the periphery.
CCR9− MHCIIlow PDC precursors can differentiate to CD11b+ MHCIIhigh CDC-like cells upon exposure to intestinal epithelial cell produced factors
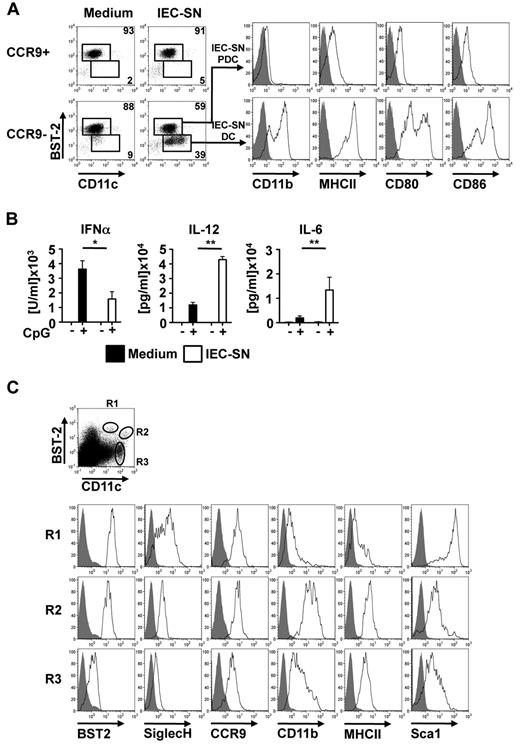
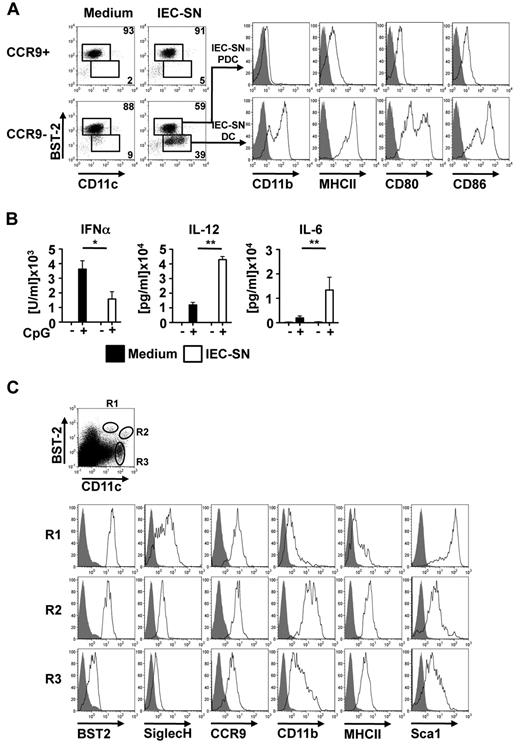
It has been reported that CD11chigh DC subpopulations within the intestinal lamina propria are generated locally from precursors under the influence of the tissue microenvironment. To investigate whether differentiation of PDCs from CCR9− immediate PDC precursors is influenced by the gut microenvironment, sorted CCR9+ and CCR9− BM PDC subpopulations were exposed to 50% IEC-SN or medium alone for 48 hours. As shown in Figure 3A and supplemental Figure 2A, exposure of CCR9−, but not CCR9+, BM PDCs to IEC-SN led to the generation of a substantial population of BST2low/CD11c+ cells (35% ± 7.4%, mean ± SD, n = 5), which were CD11b+ and expressed high levels of MHC class II and costimulatory molecules CD80 and CD86 (IEC-SN DC, Figure 3A histograms). The remaining BST2high PDCs in these cultures (IEC-SN PDCs) maintained their PDC phenotype and further up-regulated CCR9 expression (Figure 3A; supplemental Figure 2B). Extensive phenotypic analysis of BST2low CD11c+ cells newly generated from CCR9− BM PDCs in culture with IEC-SN (IEC-SN-DCs) showed that these cells expressed lower levels of B220, Siglec H, CCR9, CD8α, CD4, and Sca-1, but higher levels of MHC class II, CD80, CD86, CD11b, CD24, CD115, CD117, and CD135 after exposure to IEC-SN than the PDCs present in the same cultures (IEC-SN PDCs), thus resembling CDCs. In contrast, CCR9+ BM PDCs showed a stable PDC phenotype even after exposure to IEC-SN (PDC + IEC-SN, supplemental Figure 2B). Although IEC-SN DCs expressed CD8α at low to intermediate level, they lacked expression of XCR1, a conserved marker of murine CD8α+ splenic CDC equivalents (supplemental Figure 2B).19 In parallel to the observed change in phenotype, CCR9− BM PDCs cultured with IEC-SN produced significantly less IFN-α but more IL-12 and IL-6 in response to CpG 2216 compared with cells cultured with medium alone (Figure 3B).
Generation of CD11b+ MHCIIhigh CDC-like cells from CCR9− MHCIIlow PDCs. CCR9+ and CCR9− PDC populations were sorted from primary BM cells, incubated with medium or IEC-SN for 48 hours, and stained for FACS analysis. Surface expression of BST2 versus CD11c is shown in the dot plots. Expression levels of CD11b, MHC class II, CD80, and CD86 in BST2high IEC-SN PDCs and BST2low IEC-SN DCs generated from CCR9− BM PDCs after treatment with IEC-SN are shown in the histograms (A, filled histograms: FMO control, results of one representative of 5 experiments). CCR9+ and CCR9− PDCs were incubated with medium or 50% IEC-SN. After 24 hours, CpG 2216 (0.5μM) was added and cytokine concentrations were measured in the supernatants by ELISA after further 24 hours (B, mean ± SD, n = 3). Cells were isolated from Peyer patches digested with DNase/collagenase and subsequently analyzed by FACS for surface expression of the depicted markers. Dot blots show expression of BST2 and CD11c in propidium iodide-negative cells. Expression of the indicated markers in BST2high CD11c+ (R1), BST2low CD11chigh (R2) and CD11chigh (R3) is shown below (C, filled histograms: FMO control). *P < .05. **P < .01.
Generation of CD11b+ MHCIIhigh CDC-like cells from CCR9− MHCIIlow PDCs. CCR9+ and CCR9− PDC populations were sorted from primary BM cells, incubated with medium or IEC-SN for 48 hours, and stained for FACS analysis. Surface expression of BST2 versus CD11c is shown in the dot plots. Expression levels of CD11b, MHC class II, CD80, and CD86 in BST2high IEC-SN PDCs and BST2low IEC-SN DCs generated from CCR9− BM PDCs after treatment with IEC-SN are shown in the histograms (A, filled histograms: FMO control, results of one representative of 5 experiments). CCR9+ and CCR9− PDCs were incubated with medium or 50% IEC-SN. After 24 hours, CpG 2216 (0.5μM) was added and cytokine concentrations were measured in the supernatants by ELISA after further 24 hours (B, mean ± SD, n = 3). Cells were isolated from Peyer patches digested with DNase/collagenase and subsequently analyzed by FACS for surface expression of the depicted markers. Dot blots show expression of BST2 and CD11c in propidium iodide-negative cells. Expression of the indicated markers in BST2high CD11c+ (R1), BST2low CD11chigh (R2) and CD11chigh (R3) is shown below (C, filled histograms: FMO control). *P < .05. **P < .01.
Phenotypic analysis of BST2+ SiglecH+ CD11c+ cells in Peyer patches underlying the intestinal epithelium revealed a subpopulation of BST2low CD11chigh cells expressing lower levels of Siglec H and Sca-1 as well as higher levels of MHC class II and CD11b than BST2high CD11clow PDCs (Figure 3C). This DC subpopulation, which was found in Peyer patches but not in spleen or lymph nodes, closely resembles the CDC-like cells generated from CCR9− PDCs in vitro under the influence of IEC-SN, with the exception of CCR9 expression, which may be influenced by local factors. Thus, the observed deviation from the PDC lineage to a CDC-like cell type may also occur in vivo in the intestinal microenvironment of the Peyer patches.
CD11b+ MHCIIhigh CDC-like cells generated from CCR9− PDCs are efficient in presenting antigens to CD4+ and CD8+ T cells
To obtain a larger amount of CCR9− PDCs for functional studies, we turned to CD11c+ BST2+ PDCs from Flt3L-cultured BM cells (FL-DCs), which phenotypically correspond to primary CCR9− BM PDCs and down-regulated BST2 as well as Siglec H while up-regulating expression of CD11b, MHC class II, and CX3CR1 after exposure to IEC-SN (supplemental Figure 3A-B). The differentiation process did not involve proliferation as shown by lack of CFSE dilution during the 48-hour culture period (supplemental Figure 3B histograms). Therefore, with regard to both phenotype and function, CCR9− PDCs from FL-DC cultures are a valid model to study the developmental fate of CCR9− BM PDCs and the functional properties of the resultant IEC-SN DCs.
IEC-SN DCs generated from CCR9− PDCs resembled CDCs in that they expressed high levels of MHC class II and costimulatory molecules and produced high amounts of proinflammatory cytokines. Based on these observations, we hypothesized that these cells would be efficient T-cell activators. To compare their antigen presentation capacity, we sorted BST2low DCs generated by treatment of FL-DCs with IEC-SN (IEC-SN DCs) as well as BST2high PDCs from medium-treated FL-DCs and cocultured them for 4 days with CD4+ OT II T cells or CD8+ OT I T cells after pulsing with OVA peptide or OVA protein, respectively. IEC-SN DCs were significantly more efficient in inducing CD4+ T-cell proliferation and IFN-γ production as well as cross-presenting soluble antigen to CD8+ T cells than PDCs (Figure 4A). IEC-SN DCs were more efficient in inducing proliferation and cytokine production of CD8+ OT I T cells than CD8α− splenic CDCs but less efficient than CD8α+ splenic CDCs (Figure 4B). Thus, CCR9−, but not CCR9+, PDCs maintain the plasticity to divert from the PDC lineage and differentiate into CD11b+ MHCIIhigh CDC-like cells with higher capacity to produce inflammatory cytokines and present as well as cross-present exogenous antigens upon exposure to IEC-derived factors.
CD4+ and CD8+ T-cell proliferation and cytokine secretion in response to antigen presentation by CD11b+ MHCIIhigh CDC-like cells. BST2high PDCs and BST2low IEC-SN DCs were pulsed with OVA peptide and cocultured with CFSE-labeled CD4+ OT II T cells or were pulsed with OVA protein and cocultured with CD8+ OT I T cells for 4 days. Proliferation was determined by CFSE dilution, and intracellular IFN-γ was measured by FACS after stimulation with PMA/ionomycin (A, mean ± SD, n = 3). *P < .05. PDCs, BST2high IEC-SN PDCs, BSTlow IEC-SN DCs, splenic CD8α+ DCs, and splenic CD8α− DCs were cocultured with OT I T cells in the presence of OVA protein. As control, PDCs were cocultured with OT I T cells in the absence of antigen. After 4 days, proliferation was determined by CFSE dilution, and intracellular IFN-γ and IL-2 were detected by FACS after restimulation with PMA/ionomycin (B, results of 1 representative of 2 experiments are shown).
CD4+ and CD8+ T-cell proliferation and cytokine secretion in response to antigen presentation by CD11b+ MHCIIhigh CDC-like cells. BST2high PDCs and BST2low IEC-SN DCs were pulsed with OVA peptide and cocultured with CFSE-labeled CD4+ OT II T cells or were pulsed with OVA protein and cocultured with CD8+ OT I T cells for 4 days. Proliferation was determined by CFSE dilution, and intracellular IFN-γ was measured by FACS after stimulation with PMA/ionomycin (A, mean ± SD, n = 3). *P < .05. PDCs, BST2high IEC-SN PDCs, BSTlow IEC-SN DCs, splenic CD8α+ DCs, and splenic CD8α− DCs were cocultured with OT I T cells in the presence of OVA protein. As control, PDCs were cocultured with OT I T cells in the absence of antigen. After 4 days, proliferation was determined by CFSE dilution, and intracellular IFN-γ and IL-2 were detected by FACS after restimulation with PMA/ionomycin (B, results of 1 representative of 2 experiments are shown).
Generation of CD11b+ MHCIIhigh CDC-like cells from CCR9− PDC precursors is mediated by GM-CSF
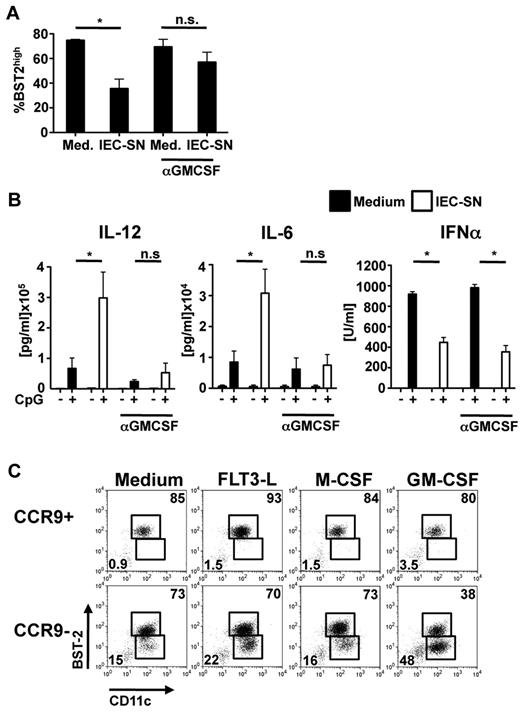
Several soluble factors, which can be produced by IEC and could be involved in inducing differentiation of CCR9− PDCs to CD11b+ MHCIIhigh CDC-like cells were not detectable in IEC-SN (TNF-α, IL-10, TGF-β, IL-6, type I IFN; supplemental Figure 4C). GM-CSF is produced at low levels in various tissues, including the intestine in the steady state, and is further induced during inflammation and infection.20,21 Indeed, IEC-SN obtained from PTK6 cell monolayers contained low concentrations of GM-CSF (56 ± 7.5 pg/mL, n = 3). Pretreatment of IEC-SN with neutralizing antibody against GM-CSF largely abrogated down-regulation of BST2 (Figure 5A) and the enhanced inflammatory cytokine production in FL-DCs exposed to IEC-SN (Figure 5B). The reduction in IFN-α production was unaffected by blocking GM-CSF despite maintenance of the PDC population (Figure 5B). Conversely, addition of recombinant GM-CSF induced down-regulation of BST2, up-regulation of MHC class II, and increased IL-12 and IL-6 production similar to IEC-SN (supplemental Figure 4A-B). In contrast to IEC-SN, however, recombinant GM-CSF did not inhibit but rather enhanced the IFN-α response of FL-DCs to CpG 2216, suggesting that additional IEC-derived factors are involved in down-modulation of the IFN-α response by IEC-SN (supplemental Figure 4B). These results demonstrate that the differentiation of CCR9− MHCIIlow PDCs to CD11b+ MHCIIhigh CDC-like cells with higher inflammatory cytokine-producing ability is mediated by GM-CSF produced by IEC.
Role of GM-CSF for differentiation of CCR9− PDCs to CD11b+ MHCIIhigh CDC-like cells. Neutralizing antibody against GM-CSF (1 μg/mL) was added to IEC-SN or medium 2 hours before addition to FL-DCs for 48 hours (A-B). The percentage of BST2high PDCs of all CD11c+ cells was determined by FACS (A, mean ± SD, n = 3). CpG 2216 (0.5μM) was added after 24 hours, and cytokines were measured in the supernatants after further 24 hours (B, mean ± SD, n = 3). *P < .05. n.s. indicates not significant. CCR9+ and CCR9− PDCs sorted from primary BM cells were cultured with medium alone or medium supplemented with Flt3L (20 ng/mL), M-CSF (1 ng/mL) or GM-CSF (1 ng/mL) for 48 hours. Expression of CD11c and BST2 was measured by FACS. Results of one representative experiment are shown (C).
Role of GM-CSF for differentiation of CCR9− PDCs to CD11b+ MHCIIhigh CDC-like cells. Neutralizing antibody against GM-CSF (1 μg/mL) was added to IEC-SN or medium 2 hours before addition to FL-DCs for 48 hours (A-B). The percentage of BST2high PDCs of all CD11c+ cells was determined by FACS (A, mean ± SD, n = 3). CpG 2216 (0.5μM) was added after 24 hours, and cytokines were measured in the supernatants after further 24 hours (B, mean ± SD, n = 3). *P < .05. n.s. indicates not significant. CCR9+ and CCR9− PDCs sorted from primary BM cells were cultured with medium alone or medium supplemented with Flt3L (20 ng/mL), M-CSF (1 ng/mL) or GM-CSF (1 ng/mL) for 48 hours. Expression of CD11c and BST2 was measured by FACS. Results of one representative experiment are shown (C).
The observed effect of GM-CSF on the differentiation of CCR9− PDCs was confirmed in CCR9− PDCs freshly isolated from primary BM cells. After exposure to GM-CSF (even at very low doses) but not Flt3L or M-CSF CCR9−, but not CCR9+, PDCs down-regulated BST2 expression (Figure 5C; supplemental Figure 4E) and up-regulated CD11b and MHC class II expression (supplemental Figure 4D). Whereas the majority of CCR9− PDCs further differentiated to CCR9+ PDCs in the absence of growth factors or in the presence of Flt3L or M-CSF, a substantial fraction of CCR9− PDCs differentiated into CD11b+ MHCIIhigh CDC-like cells on exposure to IEC-SN containing GM-CSF or low concentrations of recombinant GM-CSF. Thus, CCR9− immediate PDC precursors constitute an alternative source for the generation of CD11b+ MHCIIhigh CDC-like cells under conditions where GM-CSF is produced.
Diversion of CCR9− PDC precursors to CD11b+ MHCIIhigh CDC-like cells is regulated by transcription factors
We hypothesized that the observed differentiation of CCR9− PDCs to CD11b+ MHCIIhigh CDC-like cells is not merely a transient shift in phenotype but rather a true diversion of lineage commitment that should be reflected in altered expression of lineage-specific transcription factors. We therefore sorted BST2high PDCs (IEC-SN PDCs) and BST2low CDCs (IEC-SN DCs) generated in cultures of primary CCR9− BM PDCs with IEC-SN as well as BST2high PDCs, which had been cultured with medium alone. For comparison, CD8α+ and CD8α− CDCs were sorted from splenocytes. Expression levels of transcription factors involved in PDC and CDC development were quantified by quantitative RT-PCR.
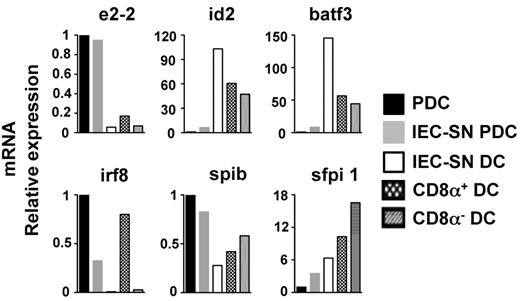
Expression of E2-2 was greatly reduced in IEC-SN DCs compared with the remaining IEC-SN PDCs or PDCs treated with medium alone (Figure 6). Expression of E2-2 regulated transcription factors IRF8 and SpiB was also reduced in IEC-SN DCs compared with PDCs. IEC-SN DCs expressed similarly low levels of IRF8 as CD8α− splenic CDCs distinct from CD8α+ splenic CDCs. Expression of transcription factors regulating CDC development (ID2, BATF3, and PU.1),22-24 was greatly increased in IEC-SN DCs compared with PDCs, thus resembling the expression pattern found in splenic CDCs. Thus, loss of PDC-specific phenotype and function directly correlates with down-regulation of E2–2 and concomitant up-regulation of transcription factors involved in CDC development. Upon exposure to GM-CSF–containing IEC-SN CCR9− immediate PDC precursors undergo a developmental shift from the PDC lineage to the CDC lineage, which is regulated on the level of transcription factors.
Transcription factor expression pattern in CD11b+ MHCIIhigh CDC-like cells generated from CCR9− PDCs. CCR9+ and CCR9− PDCs sorted from primary BM cells were incubated with medium alone or 50% IEC-SN for 48 hours. Relative mRNA expression of transcription factors was measured by quantitative RT-PCR in sorted BST2low IEC-SN DCs and BST2high IEC-SN PDCs generated from CCR9− PDCs cultured with IEC-SN as well as in sorted BST2high PDCs generated from CCR9+ PDCs cultured with medium alone. The expression patterns of transcription factors in splenic CD8α+ and CD8α− DCs are shown for comparison. Results of one representative of 2 experiments are shown.
Transcription factor expression pattern in CD11b+ MHCIIhigh CDC-like cells generated from CCR9− PDCs. CCR9+ and CCR9− PDCs sorted from primary BM cells were incubated with medium alone or 50% IEC-SN for 48 hours. Relative mRNA expression of transcription factors was measured by quantitative RT-PCR in sorted BST2low IEC-SN DCs and BST2high IEC-SN PDCs generated from CCR9− PDCs cultured with IEC-SN as well as in sorted BST2high PDCs generated from CCR9+ PDCs cultured with medium alone. The expression patterns of transcription factors in splenic CD8α+ and CD8α− DCs are shown for comparison. Results of one representative of 2 experiments are shown.
Discussion
In this study, we identify an immediate CCR9− MHCIIlow precursor of PDCs imprinted with a PDC developmental program, which leads to spontaneous differentiation into CCR9+ MHCII+ PDCs. This population can be reprogrammed to differentiate into a CD11b+ MHCIIhigh CDC-like population upon exposure to supernatants of IECs with low concentrations of GM-CSF as an essential factor. This developmental shift correlates with down-regulation of E2-2 and up-regulation of transcription factors promoting CDC development. Thus, immediately before the final differentiation step, exposure to GM-CSF forces CCR9− PDCs to branch off their developmental course and contribute to the generation of CDCs. This alternative differentiation is likely to be relevant in peripheral tissues, such as the intestine or at sites of inflammation, where GM-CSF is produced.
We show in our study that CCR9− MHCIIlow BM PDCs are closely related to CCR9+ MHCII+ PDCs: Their phenotype largely overlaps with the characteristic phenotype of CCR9+ PDCs in BM as well as secondary lymphoid organs with few notable exceptions: Lack of CCR9 expression, very low expression of MHC class II, and lower expression of CD4, CD8α, CD80, and CD86 demonstrating the lower differentiation and activation status of CCR9− versus CCR9+ PDCs. Higher levels of Sca-1 expression may reflect that CCR9+ BM PDCs are developmentally closer to splenic CCR9+ PDCs, which are Sca-1high (supplemental Figure 1). The CCR9− BM PDC population secreted high amounts of IFN-α upon stimulation and expressed E2-2, the essential transcription factor driving PDC development. Thus, CCR9− BM PDCs already possess most of the characteristic features of differentiated PDCs. Interestingly, DH-JH immunoglobulin rearrangement was not detected in CCR9− PDCs in contrast to CCR9+ PDCs, yet it is already known that expression of recombination activation gene 1 (RAG1) and subsequent DH-JH immunoglobulin rearrangement occurs only in a fraction of PDCs (∼ 50%) during later steps of development after the common DC progenitor stage.25,26 The Rag1−/− BM PDC population characterized by Pelayo et al25 responded to CpG stimulation with higher IFN-α and inflammatory cytokine production, similar to our results obtained with CCR9− BM PDCs.
CCR9− MHCIIlow PDCs are immediate PDC precursors that retain the ability to divert from commitment to the PDC lineage and differentiate into CD11b+ MHCIIhigh CDC-like cells in the presence of IEC-SN or GM-CSF. This population, however, is distinct from previously described common DC progenitors or pro-DCs (Lin− c-kitint Flt3+ M-CSFR+) because of expression of lineage markers B220, CD11c, CD4, CD8α, and CD86, heterogeneous expression of M-CSFR (CD115), and lack of proliferation and CDC differentiation in response to Flt3L.2,3 CCR9− MHCIIlow PDC precursors may be contained within CD11c+ Ly6C+ CD31+ B220+ “preimmunocytes” or CD11c+ MHCII− B220+ precursors in the BM.27,28 In contrast to these studies, which were performed before the identification of PDC-specific markers, we clearly show that CCR9− MHClow PDC precursors are a well-defined mostly homogeneous population, which is far advanced in their differentiation to CCR9+ MHCII+ PDCs and committed to PDCs under steady-state conditions (culture with medium or Flt3L). Because of the expression of the PDC-specific markers BST2 and Siglec H, this population does not overlap with the CD11c+ B220+ BST2− CCR9− DCs in the spleen, which give rise to splenic CDC subsets.29
Despite their close relationship to finally differentiated CCR9+ PDCs, the CCR9− MHCIIlow PDC precursors identified in our study retain the potential to significantly contribute to the generation of CD11b+ MHCIIhigh CDC-like cells under conditions where GM-CSF is produced. Outside of the BM, CCR9− PDC precursors, which have the same phenotype as their BM counterparts but express MHC class II at higher levels, were also found in spleen and lymph nodes, suggesting that these immediate PDC precursors leave the BM. In CCR9-deficient mice, the PDC population is greatly diminished in the small intestine but only partially in Peyer patches13 and not reduced in the colon.30 Therefore, PDC precursors may reach Peyer patches and colonic lamina propria despite the lack of or low expression of CCR9. We found a subpopulation of BST2low CD11chigh cells in Peyer patches, which phenotypically closely resembles the CDC-like cells generated from CCR9− PDCs in vitro. The PDC-specific marker Siglec H was still expressed on this subpopulation, although at lower levels than in PDCs, suggesting a close developmental relationship to PDCs. Thus, differentiation of immediate PDC precursors into CDC-like cells may also occur in vivo at this site. The final differentiation of PDC precursors to either CCR9+ PDCs or CD11b+ MHCIIhigh CDC-like cells is therefore subjected to conditions of the local tissue microenvironment, such as the intestine, allowing adaptation to local requirements in the steady state as well as during inflammation or infection.
Careful phenotypic analysis of the MHCIIhigh CDC-like cells, which were generated from CCR9− PDC precursors under the influence of GM-CSF containing IEC-SN (supplemental Figures 2, 3), revealed that these cells expressed CD11b and CX3CR1, low levels of CD8α and CD4, but lacked significant XCR1 and CD103 expression, markers of spleen CD8α+ DC equivalents.19 This phenotype therefore resembles that of CD8α− CD11b+ splenic CDCs or related DCs in peripheral tissues.
Very low IRF8 expression combined with increased expression of BATF3, ID2, and PU.1 is also congruent with this phenotype. The CD11b+ MHCIIhigh CDC-like cells, which were generated from CCR9− PDC precursors might also be related to the CX3CR1+ CD8α+ DC population recently identified in the spleen whose gene expression profile partially overlaps with that of PDCs.31 Functional analysis showed that the CD11b+ MHCIIhigh CDC-like cells generated from CCR9− PDC precursors produce higher levels of inflammatory cytokines and present exogenous antigens to CD4+ T cells and CD8+ T cells more efficiently than PDCs. The cross-presenting capacity of this CDC-like population generated from CCR9− PDCs in vitro was higher than that of splenic CD8α− CDCs, but lower than that of splenic CD8α+ CDCs. We conclude from these functional data that diversion from the PDC commitment at this late stage in development allows the generation of CDC-like cells with higher potential for efficient induction of adaptive immune responses, which may be beneficial in specific organs or during infections.
We found that GM-CSF is the major factor produced by IEC, which was responsible for the observed differentiation of CCR9− PDC precursors to CD11b+ MHCIIhigh CDC-like cells in response to IEC-SN. Type I IFN, which has been shown to drive the generation of BST2+ Siglec H− PDC-like cells in the BM, which convert to CDCs during lymphocytic choriomeningitis virus infection, was not produced by IEC and could thus be excluded as a relevant factor.7,8 GM-CSF is not detectable in the serum in the steady state but increases during inflammation.20 It has been assumed that GM-CSF is dispensable for DC development in the steady state but influences DC differentiation during inflammation by inducing the generation of “inflammatory” DCs from precursors. However, it has been shown recently that GM-CSF controls the development of subpopulations of lamina propria CDCs and dermal DCs from precursor cells.10,12,32 Thus, the observed differentiation of CCR9− PDC precursors to CDCs could contribute to the generation of CDCs in peripheral tissues in the steady state. However, GM-CSF is strongly induced and critical for host defense during intestinal infection with the mouse enteric pathogen Citrobacter rodentium.21 GM-CSF is also induced and plays an essential role in autoimmune inflammatory diseases, such as experimental autoimmune encephalomyelitis, collagen-induced arthritis, and myocarditis.20,33 Thus, differentiation of CCR9− PDC precursors to CDC-like cells is probably relevant during infection and inflammation, when GM-CSF expression is induced.
E2-2 is the critical transcription factor for PDC development and lineage stability.6 CCR9− PDCs differentiating to CD11b+ MHCIIhigh CDC-like cells in the presence of IEC-SN down-regulate expression of E2-2 and E2-2–regulated Spi-B and IRF8, whereas CCR9+ PDCs maintain their high level expression of E2-2. DC development is regulated by transcription factors in a dose-dependent manner,6,22 and differentiation to DC subpopulations appears to depend on the ratio of the different transcription factors. ID2-mediated inhibition of E protein activity by formation of inactive ID2/E2-2 heterodimers prevents PDC development34 and allows the generation of CDCs.35 Thus, down-regulation of E2-2 and up-regulation of ID2 in CCR9− PDCs exposed to IEC-SN is expected to prevent the activation of E2-2 target genes required for full differentiation and stability of PDCs. The decrease in the E2-2/ID2 ratio found in CCR9− PDCs on treatment with IEC-SN therefore provides a good explanation for the observed diversion from the PDC lineage commitment and differentiation into CDC-like cells. PU.1 dose-dependently regulates DC development by inducing and maintaining Flt3 and GM-CSF–receptor expression, which is more critical for CDC than for PDC development. Divergence of the CDC and PDC lineages was shown to be paralleled by high versus low PU.1 expression.22 This is confirmed in our study also for the reprogramming of PDC precursors to CDC-like cells.
In conclusion, we identify CCR9− PDCs, which are found in BM and secondary lymphoid organs as immediate PDC precursors whose final differentiation can be shaped by the local tissue microenvironment. In the presence of GM-CSF, which is produced constitutively in peripheral tissues or is induced at sites of inflammation, these immediate PDC precursors undergo profound changes in transcription factor expression pattern leading to a developmental shift toward CD11b+ MHCIIhigh CDC-like cells with higher cytokine-producing and antigen-presenting capacity. This plasticity at a late stage in the PDC developmental pathway may promote inflammation but may also be beneficial for immune defense against pathogens.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Giorgio Trinchieri, Marco Colonna, and Richard Kroczek for providing reagents, Viktoria Doll for cooperation, Annika Bosch and Julia Geitner for excellent technical assistance, and the Faculty Graduate Center Weihenstephan of TUM Graduate School at Technical University Munich, Germany for support.
A. K., A. S., J. L., and W. R. were supported by the German Research Foundation (grants KR2199/1-4, KR2199/3-1, SFB 455, SFB 571, and GRK 1482). J.-H.N. is supported by the German Research Foundation (grants Ni575/6-2 and Ni575/7-1).
This work is submitted in partial fulfillment of the PhD thesis of A.S.
Authorship
Contribution: A.S. designed research, performed experiments, analyzed data, and wrote the paper; J.L., K.M., and L.H. performed experiments; R.V., M.S., and J.-H.N. contributed reagents and mice; H.E. analyzed data; and W.R. and A.K. designed the study, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anne Krug, II Medizinische Klinik, Klinikum Rechts der Isar, Technical University Munich, Trogerstrasse 32/1.04, D-81675 Munich, Germany; e-mail: anne.krug@lrz.tum.de.

![Figure 1. Characterization of CCR9− and CCR9+ PDCs in murine bone marrow. Expression of the indicated markers was analyzed by FACS in primary BM cells. Dot plots show expression of CD11c versus BST2 in propidium iodide-negative cells and expression of BST2 versus CCR9 in BST2+ CD11c+ cells. Expression of the indicated markers in CCR9+ and CCR9− BST2+ CD11c+ cells is shown below (overlay with fluorescence minus one [FMO] control, filled histograms) (A). Relative mRNA expression was quantified in CCR9+/BST2+/CD11c+ and CCR9−/BST2+/CD11c+ PDC populations sorted from primary BM cells by quantitative RT-PCR (B, mean ± SD, n = 3). Immunoglobulin DH-JH rearrangement was detected by PCR in genomic DNA isolated from CCR9+ and CCR9− PDCs (C). CCR9+ and CCR9− PDCs were incubated with medium or CpG 2216 for 24 hours. IFN-α, IL-12p40, and IL-6 were measured in the supernatants by ELISA (D, mean ± SD, n = 3). *P < .05. **P < .01. ***P < .001. n.s. indicates not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/24/10.1182_blood-2010-12-326678/4/m_zh89991172800001.jpeg?Expires=1766068178&Signature=xVKTpTanyIFEhBtV835f7Vxcn1Sonky4BCV9xn7B7ZmCrKZ0Xm81qEyqekuoo7D6gLHTx3y0vDfnUiyxdBrEBBpQOhORGvJhi3-sazyqynUF1x2xhmqQmd1yw7GjPPWVna6RomredmlPO9R-9tS5tS9V5s6Hygo7UeHlNATp-jxABgOJtHEDJOeCANr4jDAsA1XY5ubdlnP2XoDBIpLhIuXuud-ppPumNjHRwfUPr1jtv1Y9pLsaz0WWUexeAmvuJhKuI8RTvQ1M9lkZYjwNMjCBSMAW3In5sr0mESqBphpsp9jz~Ox-jENzU-ZaufuU~-fIHEByg5s2Q~LKYJPXKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 1. Characterization of CCR9− and CCR9+ PDCs in murine bone marrow. Expression of the indicated markers was analyzed by FACS in primary BM cells. Dot plots show expression of CD11c versus BST2 in propidium iodide-negative cells and expression of BST2 versus CCR9 in BST2+ CD11c+ cells. Expression of the indicated markers in CCR9+ and CCR9− BST2+ CD11c+ cells is shown below (overlay with fluorescence minus one [FMO] control, filled histograms) (A). Relative mRNA expression was quantified in CCR9+/BST2+/CD11c+ and CCR9−/BST2+/CD11c+ PDC populations sorted from primary BM cells by quantitative RT-PCR (B, mean ± SD, n = 3). Immunoglobulin DH-JH rearrangement was detected by PCR in genomic DNA isolated from CCR9+ and CCR9− PDCs (C). CCR9+ and CCR9− PDCs were incubated with medium or CpG 2216 for 24 hours. IFN-α, IL-12p40, and IL-6 were measured in the supernatants by ELISA (D, mean ± SD, n = 3). *P < .05. **P < .01. ***P < .001. n.s. indicates not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/24/10.1182_blood-2010-12-326678/4/m_zh89991172800001.jpeg?Expires=1766068179&Signature=vcIJjIfsp2Bz~S8I3MnGtUkim3~tXg5Ho3U7A0oxtLnc6FhI0t-VZUR29cM4~HxyngfHgy29uFJrDX-ib0abfrf1zo0V55yt1Pt33Hn1jzfuaYTSwULAtyMkW0YgrxDLG799SAiIF2HAUryFksLRx~-e5OFs5eSPJdhBA0vVGV7YjlQEH8uqQd-iG5CgRzlIwynSORWqMKn15U~~u-KyvrtqNXqof9W1Xi36UAR6ggMxLitclMi47-hKMG9i8vXm4~3ypnXCdeRdSb3WQRXLurWtFGQ1rs9eHPiD5uqeOo84WIjH4pYXhB7Fhh5ulllsH1WMrzmrOv3dUX2lbktsYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)