Abstract
The genetics of peripheral T-cell lymphomas are poorly understood. The most well-characterized abnormalities are translocations involving ALK, occurring in approximately half of anaplastic large cell lymphomas (ALCLs). To gain insight into the genetics of ALCLs lacking ALK translocations, we combined mate-pair DNA library construction, massively parallel (“Next Generation”) sequencing, and a novel bioinformatic algorithm. We identified a balanced translocation disrupting the DUSP22 phosphatase gene on 6p25.3 and adjoining the FRA7H fragile site on 7q32.3 in a systemic ALK-negative ALCL. Using fluorescence in situ hybridization, we demonstrated that the t(6;7)(p25.3;q32.3) was recurrent in ALK-negative ALCLs. Furthermore, t(6;7)(p25.3;q32.3) was associated with down-regulation of DUSP22 and up-regulation of MIR29 microRNAs on 7q32.3. These findings represent the first recurrent translocation reported in ALK-negative ALCL and highlight the utility of massively parallel genomic sequencing to discover novel translocations in lymphoma and other cancers.
Introduction
Recurrent chromosomal translocations are common pathogenetic events in hematologic malignancies.1 Among peripheral (post-thymic) T-cell lymphomas, however, the only well-characterized translocations are those involving the anaplastic lymphoma kinase gene ALK.2 ALK is an important prognostic marker and therapeutic target in T-cell anaplastic large cell lymphomas (ALCLs)3,4 ; however, approximately half of ALCLs lack ALK expression, despite nearly identical morphology and phenotype.5 ALK-negative ALCLs can occur either cutaneously or systemically. We previously identified recurrent IRF4 translocations in cutaneous ALCLs,6 but recurrent translocations in the more lethal, systemic form of ALK-negative ALCL have not been reported.
Massively parallel (“Next Generation”) DNA sequencing technology represents a quantum advance in the ability to understand cancer genomes. To identify translocations in ALK-negative ALCL, we performed massively parallel sequencing of a mate-pair DNA library constructed from a systemic ALK-negative ALCL. Using a unique bioinformatic algorithm for translocation discovery, we identified a translocation, t(6;7)(p25.3;q32.3), and demonstrated this translocation in additional ALK-negative ALCLs. This represents the first recurrent translocation reported in systemic ALK-negative ALCL, and demonstrates the utility of mate-pair library sequencing as a tool for translocation discovery.
Methods
Briefly, mate-pair library construction followed the manufacturer's protocol (Illumina) using approximately 5-kb genomic DNA fragments. Sequencing was performed on an Illumina GAIIx, and results were mapped to the genome using a binary indexing algorithm.7 Candidate translocations were validated by polymerase chain reaction (PCR), Sanger sequencing, and fluorescence in situ hybridization (FISH). Gene and microRNA expression levels were assessed using quantitative real-time PCR. The study was approved by the Mayo Clinic Institutional Review Board. Details are included in Supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results and discussion
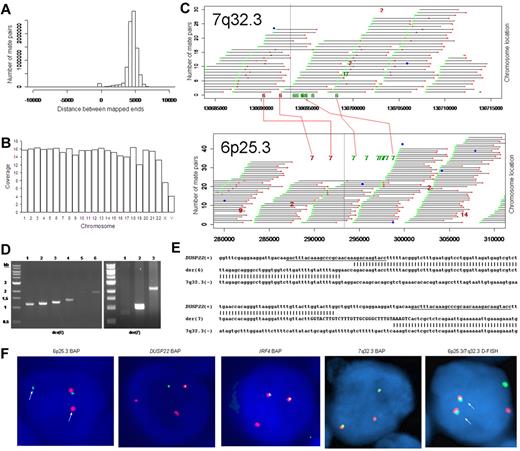
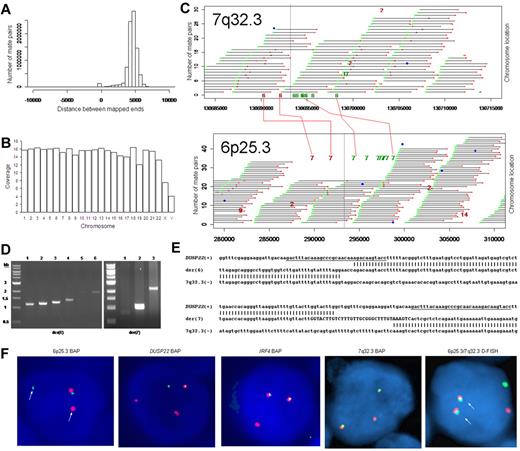
We used massively parallel mate-pair library sequencing to identify the first recurrent translocation reported in ALK-negative ALCL, t(6;7)(p25.3;q32.3). First, we prepared a mate-pair library from approximately 5-kb genomic DNA fragments extracted from a systemic ALK-negative ALCL (Table 1) with known 6p25.3 rearrangement previously shown not to involve IRF4.6 We then sequenced paired ends on one lane of an Illumina flow cell and mapped the results to the human genome using a binary indexing algorithm.7 Of 28.90 × 106 paired sequences, 21.78 × 106 had at least one unique match to the genome. Of these, 4.19 × 106 (29%) were duplicates and were discarded. Of 8.84 × 106 fragments where both ends mapped uniquely, the ends mapped to the same chromosome less than 10 kb apart (peak, ∼ 5 kb) in 8.59 × 106, yielding approximately 7 times the coverage of the genome (Figure 1A-B). There were 249 278 mate pairs in which the 2 ends mapped to distant loci (different chromosomes or > 25 kb away on the same chromosome). Only 11 578 of these (< 5%) occurred more than once; the rest probably represented ligation artifact introduced during library construction. We analyzed mate-pairs where: (1) one end mapped to 6p25.3; (2) the other end mapped to a distant locus; and (3) involvement of these loci was supported by more than 4 nonidentical mate-pairs. Only one such instance was found, with 10 mate-pairs mapping to both 6p25.3 and 7q32.3 (Figure 1C). PCR with primers spanning the putative breakpoints (supplemental Table 1) confirmed a balanced t(6;7)(p25.3;q32.3) (Figure 1D). Sanger sequencing showed breakpoints in intron 1 of DUSP22 on 6p25.3 and immediately telomeric to the FRA7H fragile site on 7q32.3 (Figure 1E).
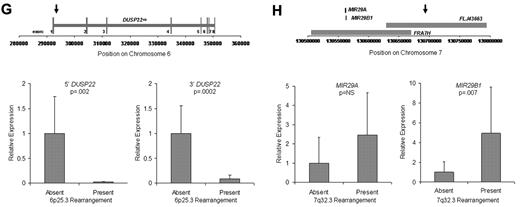
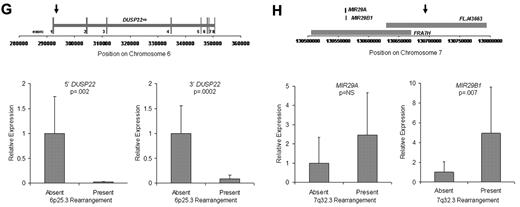
Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative ALCLs using mate-pair library sequencing. (A) Histogram demonstrating the calculated distances between the sequenced ends of mate-pairs in which both ends mapped to the reference genome. Most mate-pairs map approximately 5000 bp apart. (B) Bridged coverage of the genome by chromosome, where “bridged” coverage represents the portion of the genome analyzed for the presence of translocations and includes both the sequenced mate-pairs and the intervening DNA segments (∼ 5000 bp total per mate-pair). Average bridged coverage for the genome was approximately 7x. (C) Schematic representation of mapping of mate-pairs to the reference genome. The region of the known 6p25.3 rearrangement is shown in the bottom panel. The x-axis shows nucleotides according to the February 2009 genome assembly (GRCh37/hg19). Horizontal black bars represent approximately 5000-bp DNA fragments between sequenced mate-pairs, which are colored green and red to represent positive and negative strands, respectively. Blue dots represent mate-pairs in which the 2 ends map to the same chromosome more than 25 000 bp apart. Colored numerals represent mate-pairs in which the 2 ends map to different chromosomes, with the color indicating strand, the number indicating the partner chromosome, and the position along the right-hand y-axis representing the position along the partner chromosome. Ten distinct (ie, nonidentical) mate-pairs map to both 6p25.3 and a narrow region of chromosome 7: 8 (green) on the positive strand and 2 (red) on the negative strand. (Top panel) The corresponding locus on 7q32.3. Because these aberrant mate-pairs have ends that map to distinct regions of the genome, they suggest a possible translocation between loci adjacent to these paired ends. Occasional single numerals (eg, the red “14” in the lower right of the bottom panel) represent sporadic, nonrepeated, aberrant mate-pairs that may be introduced during the ligation step of the mate-pair library preparation. The putative breakpoints lie between the positive-strand mate-pairs (green) and the negative-strand mate-pairs (red). Vertical lines indicate the actual breakpoints confirmed by PCR and sequencing. (D) Gel electrophoreses of PCR reactions amplifying the DNA regions containing the putative breakpoints on der(6) and der(7). Each set of reactions included a single chromosome 6 primer and multiple chromosome 7 primers (supplemental Table 1) at increasing predicted distances from the putative breakpoint. (E) Bands shown in panel D were conventionally sequenced and aligned to the genome. Breakpoints of der(6) and der(7) are shown with their alignment to the positive strand of 6p25.3 (DUSP22) and the negative strand of 7q32.3. Nineteen bases of der(7) do not align but reside within a 33-bp sequence (capitalized) complementary to the positive strand of DUSP22 at the 6p25.3 breakpoint (underlined), suggesting a microinversion associated with the breakpoint on 6p25.3. (F) FISH results in the sequenced case (100×/1.40 NA oil objective). The breakapart (BAP) probe to 6p25.3 shows one normal fusion signal and separation of the red and green components of the other signal (arrows), indicating a translocation. This separation of red and green components also is seen with the DUSP22 BAP probe, confirming the 6p25.3 breakpoint involves DUSP22. The additional red signal results from a cross-hybridization to chromosome 16p11.6 No abnormal separation is seen with the IRF4 BAP probe; again, a cross-hybridization (red signal) is seen. A 7q32.3 BAP probe confirms the presence of a translocation; this signal pattern was seen in 45% of ALK-negative ALCLs with 6p25.3 rearrangements. A dual-fusion (D-) FISH probe showed one normal red signal, one normal green signal, and 2 abnormal fusion signals (arrows), confirming a balanced translocation between 6p25.3 and 7q32.3. (G) The 6p25.3 breakpoint (arrow; top panel) lies within intron 1 of DUSP22. (Bottom panels) DUSP22 expression in ALCLs without and with 6p25.3 rearrangements (real-time quantitative PCR, shown as expression relative to the mean value of the nontranslocated cases, mean ± SD: 5′, 1.00 ± 0.74 vs 0.02 ± 0.01; 3′, 1.00 ± 0.55 vs 0.09 ± 0.08). (H) The 7q32.3 breakpoint (arrow; top panel) lies in the noncoding transcript region FLJ43663, immediately telomeric to the fragile site, FRA7H, and the microRNAs, MIR29A and MIR29B1. (Bottom panels) MicroRNA expression in ALCLs without and with7q32.3 rearrangements (real-time quantitative PCR, shown as expression relative to the mean value of the nontranslocated cases, mean ± SD: MIR29A, 1.00 ± 1.34 vs 2.44 ± 2.20; MIR29B1, 1.00 ± 1.05 vs 4.93 ± 4.68).
Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative ALCLs using mate-pair library sequencing. (A) Histogram demonstrating the calculated distances between the sequenced ends of mate-pairs in which both ends mapped to the reference genome. Most mate-pairs map approximately 5000 bp apart. (B) Bridged coverage of the genome by chromosome, where “bridged” coverage represents the portion of the genome analyzed for the presence of translocations and includes both the sequenced mate-pairs and the intervening DNA segments (∼ 5000 bp total per mate-pair). Average bridged coverage for the genome was approximately 7x. (C) Schematic representation of mapping of mate-pairs to the reference genome. The region of the known 6p25.3 rearrangement is shown in the bottom panel. The x-axis shows nucleotides according to the February 2009 genome assembly (GRCh37/hg19). Horizontal black bars represent approximately 5000-bp DNA fragments between sequenced mate-pairs, which are colored green and red to represent positive and negative strands, respectively. Blue dots represent mate-pairs in which the 2 ends map to the same chromosome more than 25 000 bp apart. Colored numerals represent mate-pairs in which the 2 ends map to different chromosomes, with the color indicating strand, the number indicating the partner chromosome, and the position along the right-hand y-axis representing the position along the partner chromosome. Ten distinct (ie, nonidentical) mate-pairs map to both 6p25.3 and a narrow region of chromosome 7: 8 (green) on the positive strand and 2 (red) on the negative strand. (Top panel) The corresponding locus on 7q32.3. Because these aberrant mate-pairs have ends that map to distinct regions of the genome, they suggest a possible translocation between loci adjacent to these paired ends. Occasional single numerals (eg, the red “14” in the lower right of the bottom panel) represent sporadic, nonrepeated, aberrant mate-pairs that may be introduced during the ligation step of the mate-pair library preparation. The putative breakpoints lie between the positive-strand mate-pairs (green) and the negative-strand mate-pairs (red). Vertical lines indicate the actual breakpoints confirmed by PCR and sequencing. (D) Gel electrophoreses of PCR reactions amplifying the DNA regions containing the putative breakpoints on der(6) and der(7). Each set of reactions included a single chromosome 6 primer and multiple chromosome 7 primers (supplemental Table 1) at increasing predicted distances from the putative breakpoint. (E) Bands shown in panel D were conventionally sequenced and aligned to the genome. Breakpoints of der(6) and der(7) are shown with their alignment to the positive strand of 6p25.3 (DUSP22) and the negative strand of 7q32.3. Nineteen bases of der(7) do not align but reside within a 33-bp sequence (capitalized) complementary to the positive strand of DUSP22 at the 6p25.3 breakpoint (underlined), suggesting a microinversion associated with the breakpoint on 6p25.3. (F) FISH results in the sequenced case (100×/1.40 NA oil objective). The breakapart (BAP) probe to 6p25.3 shows one normal fusion signal and separation of the red and green components of the other signal (arrows), indicating a translocation. This separation of red and green components also is seen with the DUSP22 BAP probe, confirming the 6p25.3 breakpoint involves DUSP22. The additional red signal results from a cross-hybridization to chromosome 16p11.6 No abnormal separation is seen with the IRF4 BAP probe; again, a cross-hybridization (red signal) is seen. A 7q32.3 BAP probe confirms the presence of a translocation; this signal pattern was seen in 45% of ALK-negative ALCLs with 6p25.3 rearrangements. A dual-fusion (D-) FISH probe showed one normal red signal, one normal green signal, and 2 abnormal fusion signals (arrows), confirming a balanced translocation between 6p25.3 and 7q32.3. (G) The 6p25.3 breakpoint (arrow; top panel) lies within intron 1 of DUSP22. (Bottom panels) DUSP22 expression in ALCLs without and with 6p25.3 rearrangements (real-time quantitative PCR, shown as expression relative to the mean value of the nontranslocated cases, mean ± SD: 5′, 1.00 ± 0.74 vs 0.02 ± 0.01; 3′, 1.00 ± 0.55 vs 0.09 ± 0.08). (H) The 7q32.3 breakpoint (arrow; top panel) lies in the noncoding transcript region FLJ43663, immediately telomeric to the fragile site, FRA7H, and the microRNAs, MIR29A and MIR29B1. (Bottom panels) MicroRNA expression in ALCLs without and with7q32.3 rearrangements (real-time quantitative PCR, shown as expression relative to the mean value of the nontranslocated cases, mean ± SD: MIR29A, 1.00 ± 1.34 vs 2.44 ± 2.20; MIR29B1, 1.00 ± 1.05 vs 4.93 ± 4.68).
FISH (supplemental Figure 1) confirmed t(6;7)(p25.3;q32.3) in the sequenced case (Figure 1F). Among 29 ALK-negative ALCLs with 6p25.3 translocations (Table 1),6,8 13 (45%) had a break at 7q32.3, and all 11 with sufficient material for further testing had t(6;7)(p25.3;q32.3). The translocation involved DUSP22 rather than IRF4 in 9 of 11 cases where the 6p25.3 breakpoint could be characterized. Of 142 peripheral T-cell lymphomas without 6p25.3 rearrangements, 7q32.3 FISH was successful in 108, and none showed a 7q32.3 break (supplemental Table 2). These findings suggest that, among peripheral T-cell lymphomas, the 7q32.3 breakpoint is specifically involved in the t(6;7)(p25.3;q32.3). The t(6;7)(p25.3;q32.3) was seen in both systemic and cutaneous ALK-negative ALCLs. Although recognized as distinct clinicopathologic entities, systemic and cutaneous ALCLs show marked morphologic and phenotypic similarities.5,9 The translocation may contribute to some of these common features; it would not appear to be responsible for the different anatomic distributions and prognoses of the 2 entities.10
Because the 6p25.3 breakpoint in the sequenced case disrupted DUSP22 (Figure 1G), we examined DUSP22 expression in ALK-negative ALCLs with and without 6p25.3 rearrangements and found a significant, up to 50-fold reduction of expression in translocated cases. Consistent with our data that ALCLs typically express IRF4 regardless of translocation status,6,8 no difference in IRF4 gene expression was seen between the 2 groups (supplemental Figure 2). Thus, DUSP22 expression may be affected even when the breakpoint is nearest to IRF4. As DUSP22 and IRF4 are only 40 kb apart, studying additional cases may identify mechanisms for DUSP22 down-regulation besides disruption of DUSP22 itself. DUSP22 is a dual-specificity phosphatase that inhibits T-cell antigen receptor signaling in reactive T cells and Jurkat cells by inactivating the MAPK, ERK2.11 DUSP22-mediated ERK2 inhibition blocks estrogen receptor signaling in breast cancer cells,12 and DUSP22 expression correlates with mutated IGVH status, a favorable prognostic factor, in chronic lymphocytic leukemia.13 Phosphatases have tumor suppressor function in B-cell lymphomas, T-lymphoblastic leukemias, and ALK-positive ALCLs,14-16 but their role in ALK-negative ALCLs is poorly understood. Investigation of DUSP22 as a putative tumor suppressor is warranted.
Chromosomal fragile sites and subtelomeric regions are targets for translocations17,18 ; thus, the proximity of the 7q32.3 breakpoint to FRA7H and the subtelomeric location of DUSP22 and IRF4 probably contribute to the formation of t(6;7)(p25.3;q32.3). The 7q32.3 breakpoint also coincides with FLJ43663, a putative noncoding transcript (Figure 1H). A B-cell lymphoma with an FLJ43663 breakpoint showed down-regulation of the microRNA, MIR29B1,19 which, along with MIR29A, resides within FRA7H. Unexpectedly, we found both miRNAs (particularly MIR29B1) overexpressed in ALCLs with 7q32.3 rearrangements. MIR29 has tumor suppressor activity in B-cell lymphomas by targeting the TCL1 oncogene,20 which is not expressed in ALCLs.21 Thus, MIR29 may have oncogenic function in t(6;7)(p25.3;q32.3)-positive ALCL, as has been proposed in acute myeloid leukemia and breast cancer.22,23
Whole-genome sequencing currently demands resources beyond the reach of most cancer investigators. Transcriptome sequencing has been used to identify novel fusion genes derived from chromosomal translocations in solid tumors.24 In lymphomas, however, translocations often do not produce a fusion gene1 and, in some cases, disrupt a putative tumor suppressor gene.25 Transcriptome sequencing is unlikely to detect such translocations. Therefore, we used a mate-pair genomic DNA sequencing approach for lymphoma translocation discovery. In this resource-efficient approach, only the ends of approximately 5-kb DNA fragments are sequenced, such that we obtained approximately 7 times the bridged coverage of the genome despite base coverage of only approximately 0.35 times, requiring only one lane of an Illumina flow cell. Our binary indexing algorithm maximizes computer memory and speed while allowing the reference genome and its reverse compliment to be stored in RAM. This allows mapping approximately 2 × 108 sequences to a 3 × 109-nucleotide genome in one day. This approach is much faster and less labor-intensive than approaches such as long-distance inverse PCR. In addition, the whole-genome coverage and 5-kb resolution are more informative than karyotyping or FISH. Because the World Health Organization emphasizes the incorporation of genetic data into lymphoma diagnosis,2 our approach may represent a robust clinical platform for translocation detection in tissue samples from patients with lymphomas and other cancers.
In conclusion, we used a novel approach to identify lymphoma translocations, combining mate-pair library construction, massively parallel sequencing, and a unique bioinformatic algorithm. We discovered the first recurrent translocation in ALK-negative ALCL, t(6;7)(p25.3;q32.3), which was associated with down-regulation of DUSP22 and up-regulation of MIR29A. As our approach was guided by knowledge of one of the involved loci, we currently are expanding our algorithm with additional filters to discover unsuspected translocations across the entire genome in cancer tissues.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Waterman Biomarker Discovery grant from the Mayo Clinic Center for Individualized Medicine. A.L.F. is a Damon Runyon Clinical Investigator supported by the Damon Runyon Cancer Research Foundation (CI-48-09). This work also was supported in part by a Career Development Award from the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence (Public Health Service grant P50 CA097274) (A.L.F.) and the National Cancer Institute.
National Institutes of Health
Authorship
Contribution: A.L.F. designed and performed research, analyzed data, and wrote the paper; A.D. and D.I.S. designed research and analyzed data; M.E.L. performed research and analyzed data; S.M.A. and S.H.J. analyzed data; J.C.P., N.O., and B.W.E. performed research; E.D.W. designed research; and G.V. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The Mayo Clinic and A.L.F., A.D., D.I.S., M.E.L., S.H.J., J.C.P., and G.V. have a potential financial interest in technology associated with this research. The Mayo Clinic has filed a nonprovisional patent application for that technology. The remaining authors declare no competing financial interests.
Correspondence: George Vasmatzis, Department of Molecular Medicine and Center for Individualized Medicine, Mayo Clinic, 200 1st St SW, Rochester, MN 55905; e-mail: vasmatzis.george@mayo.edu.