Abstract
Maintaining a steady pool of self-renewing hematopoietic stem cells (HSCs) is critical for sustained production of multiple blood lineages. Many transcription factors and molecules involved in chromatin and epigenetic modifications have been found to be critical for HSC self-renewal and differentiation; however, their interplay is less understood. The transcription factor GA binding protein (GABP), consisting of DNA-binding subunit GABPα and transactivating subunit GABPβ, is essential for lymphopoiesis as shown in our previous studies. Here we demonstrate cell-intrinsic, absolute dependence on GABPα for maintenance and differentiation of hematopoietic stem/progenitor cells. Through genome-wide mapping of GABPα binding and transcriptomic analysis of GABPα-deficient HSCs, we identified Zfx and Etv6 transcription factors and prosurvival Bcl-2 family members including Bcl-2, Bcl-XL, and Mcl-1 as direct GABP target genes, underlying its pivotal role in HSC survival. GABP also directly regulates Foxo3 and Pten and hence sustains HSC quiescence. Furthermore, GABP activates transcription of DNA methyltransferases and histone acetylases including p300, contributing to regulation of HSC self-renewal and differentiation. These systematic analyses revealed a GABP-controlled gene regulatory module that programs multiple aspects of HSC biology. Our studies thus constitute a critical first step in decoding how transcription factors are orchestrated to regulate maintenance and multipotency of HSCs.
Introduction
Multilineage hematopoiesis is maintained by a pool of hematopoietic stem cells (HSCs). To sustain the production of blood cells throughout the lifetime of an individual, HSCs are capable of self-renewal to maintain the HSC pool and have the ability for multilineage differentiation.1,2 Self-renewal relies on a balance between quiescence and cell-cycle progression and a balance between survival and cell death. Recent studies have revealed that these critical processes are under the regulation of a number of transcription factors.3 For example, Gfi-1 and Foxo proteins restrain HSCs from excessive cycling,4-6 and Zfx and Tel/Etv6 are critical in suppressing HSC apoptosis.7,8 Gata2, Fli-1, and Scl/Tal1 act cooperatively in specification of hematopoiesis during embryo development.9 Epigenetic integrity has been demonstrated to be critical for normal HSC activities as well. DNA methyltransferase 1 (Dnmt1)–mediated methylation maintenance10,11 and Dnmt3a/3b-mediated de novo DNA methylation12 are all required for HSC self-renewal. Proteins with histone acetyltransferase activity such as CBP and p300 coactivators were shown to have distinct roles in regulating HSC self-renewal and differentiation.13 The Brg1 ATPase catalytic subunit in the SWI/SNF-related chromatin-remodeling complex was found to be essential for primitive erythropoiesis during embryogenesis.14 Despite increasing numbers of key players that have been identified, their interplay has not been extensively addressed in HSCs.
A functional GA binding protein (GABP) complex is a heterodimer of GABPα and GABPβ subunits. GABPα is one of the Ets family transcription factors and contains a conserved Ets domain responsible for DNA binding. GABPβ is unrelated to Ets factors but heterodimerizes with GABPα and possesses transactivation activity. The GABP complex has been demonstrated to have versatile roles in maintaining basic cellular functions, such as cellular respiration in mitochondria and cell-cycle progression.15 As a result, targeting GABPα in the germline resulted in early embryonic lethality.16,17 Cell type–specific roles of GABP are also well documented. In lymphocytes, we showed previously that GABP critically regulates Pax5 in developing B cells,18 interleukin-7 receptor α chain, and genes involved in T-cell receptor rearrangements in thymocytes.16,19 In this study, we investigated the roles of GABP in HSCs via tissue-specific disruption of GABPα. Recent technological advances in chromatin immunoprecipitation coupled by high-throughput parallel sequencing (ChIP-Seq) allowed genome-wide mapping of transcription factor binding locations.20 Application of this technique in hematopoietic cells, such as ChIP-Seq of GATA1 in erythroleukemia cells21,22 and PU.1 in primary B cells and macrophages,23 has offered comprehensive insights into how these transcription factors operate. Here we report genome-wide chromatin occupancy of GABPα. By combining genetic and bioinformatic approaches with functional assays, our systematic analyses revealed a GABP-controlled gene regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells.
Methods
Mice and pIpC treatment
Generation of GABPαFL/+ and GABPα+/− using 129S6/SvEvTac embryonic stem cells was described previously.24 Mx1Cre transgenic, B6.SJL, and 129/SvEv mice were from The Jackson Laboratory. Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− or bone marrow chimeras derived from these mice were subjected to polyinosinic-polycytidylic acid (pIpC) induction following the treatment schedule detailed in Figure 1A. All mice were maintained at the University of Iowa Animal Facility, and all the mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
Flow cytometry and cellularity
Freshly dissected femurs and tibias were flushed with phosphate-buffered saline containing 2.5% fetal bovine serum, and red blood cells were lysed with ammonium chloride–potassium bicarbonate lysis buffer. For identification of Lin−Sca-1+c-Kit+ cells (LSKs) and long-term HSCs (LT-HSCs), lineage markers (CD4, CD8, B220, CD11c, Gr-1, TER119, NK1.1, and Mac1), Sca-1, c-Kit, Flt3, and CD34, directly conjugated or biotinylated, were used for cell-surface staining (eBioscience or BD Biosciences), and stained cells were analyzed on FACSCalibur or Becton Dickinson LSR II (BD Biosciences). For analysis of bone marrow (BM) chimeras, CD45.1 and CD45.2 antibodies were included to discriminate sources of donor and host cells. Apoptotic state of LSKs was measured using annexin V–PE Apoptosis Detection Kit (BD Biosciences). All the data were analyzed using FlowJo software Version 8.8 (TreeStar Inc). The numbers of LSKs, LT-HSCs, short-term HSCs (ST-HSCs), and multipotent progenitors (MPPs) were calculated from the bone marrow cellularity of femurs and tibias with flow cytometry–detected frequency of each subset.
Antibody conjugation and intracellular staining
Antibodies against GABPα (H-180), Pten (C-20), Brm (N-19), and Brg1 (P-18), along with normal rabbit or goat IgG without carrier proteins, were special ordered from Santa Cruz Biotechnology. The antibodies were labeled with a fluorochrome using the Alexa Fluor 647 monoclonal antibody labeling kit (Invitrogen). Bone marrow cells from pIpC-treated Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were surface stained, then fixed and permeabilized using Cytofix/Cytoperm solutions (BD Biosciences), followed by intracellular staining with self-labeling antibodies or prelabeled Bcl-2 and control antibodies (BD Biosciences).
Generation of BM chimeras and CFU-Spleen12
To generate BM chimeras of Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/−, donor BM cells were stained and analyzed by flow cytometry to determine LSK frequency. BM cells containing 3000 LSKs, alone or in the presence of equivalent numbers of LSKs from B6.SJL mice as protectors/competitors, were intravenously injected into F1 progeny of B6.SJL and 129/SvEv breeding that received 1050 rad whole-body irradiation at least 4 hours before injection. Five weeks later, peripheral blood was sampled and contribution from donor BM cells (CD45.2+CD45.1−) was determined before pIpC treatment. For the colony-formation units in the spleen on day 12 (CFU-Spleen12) assay, Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were treated with pIpC, and on day 9 after last injection, 1 × 105 BM cells were injected into 900 rad–irradiated F1 recipients as described. After another 12 days, spleens from the hosts were harvested and macro-colonies were counted under a dissecting microscope.
Colony-formation assay using methylcellulose
Whole bone marrow cells were harvested from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice on day 4 after the last pIpC treatment. A total of 2 × 104 BM cells were mixed with Methocult M3434 (StemCell Technologies) and plated. Ten days after culture, colony numbers of GEMM (granulocyte, erythroid, macrophage, megakaryocyte), GM (granulocyte macrophage), G (granulocyte), M (macrophage), and BFU-E (burst-forming erythroid-unit) were counted based on their distinct morphology.
Peripheral blood analysis
Peripheral blood was collected from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice on various days after pIpC treatment and analyzed on the Sysmex XT 2000iv automatic hematology analyzer, which is provided on loan to Dr John Widness' laboratory (University of Iowa) from Sysmex Corporation.
BrdU uptake
To determine the proliferation rate of myeloid progenitors and LSK cells, Mx1Cre- GABPαFL/+ and Mx1Cre-GABPαFL/− mice that have been treated with pIpC were given one dose of intraperitoneal 5′-bromo-2′-dexoxy-uridine (BrdU) injection (1 mg). Eighteen hours later, BM cells were isolated and surface-stained, followed by fixation and permeabilization procedures as recommended in the BrdU Flow Kit (BD Biosciences).
ChIP-Seq and data analysis
Mobilized CD34+CD133+ human hematopoietic progenitor cells (HPCs) were purified from peripheral blood lymphocytes of healthy donors using established protocols.25 The chromatin fragments were prepared and immunoprecipitated with an anti-GABPα antibody (H180; Santa Cruz Biotechnology) or a control IgG as described previously.26 The chromatin immunoprecipitation (ChIP) samples were amplified and sequenced using the Solexa 1G Genome Analyzer (Illumina). The resulting 25-bp sequence reads were mapped to the human genome (build hg18) using the Solexa Analysis Pipeline, and only reads that were mapped to unique genomic locations, with mismatches in at most 2 positions, were retained for further analysis. This yielded 4.43 million reads for the GABPα sample and 4.48 million reads for the control IgG sample.
For binding site identification, mapped GABPα reads were processed using the Site Identification from Short Sequence Reads (SISSRs) peak finding tool27 with control IgG reads as a control (see http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/sissrs/SISSRs-Manual.pdf for details). SISSRs 1.4 was run with option “a” (which retains one read per genomic position even if multiple reads were mapped to that position, and thus avoids overrepresentation of one position because of polymerase chain reaction [PCR] amplification–generated bias), fragment length F set to 200, P value set to .01, and the remaining parameters set to their default values. Each identified binding site is associated with a fold-enrichment score, which is the ratio of the normalized number of GABP sequence tags supporting the inferred binding site to the normalized number of control IgG tags supporting the exact same site. Genome-wide distribution of GABPα binding sites was determined with reference to RefSeq genes downloaded from the University of California Santa Cruz genome browser. For Motif analysis, MEME with default parameters was used to identify the consensus binding sequence with identified GABP binding locations. The ChIP-Seq data have been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession number GSE24933.
Validation of direct GABP binding in murine hematopoietic progenitor cells by ChIP
Whole BM cells from C57BL/6 mice were subjected to lineage depletion using biotinylated lineage antibodies coupled with Dynabeads M280 Streptavidin (Invitrogen). The Lin− cells were stained for cell-surface c-Kit, and c-Kit+Lin− BM cells were isolated by either positive selection using CD117 MicroBeads (Miltenyi Biotec) or cell sorting. The chromatin fragments were prepared from Lin− or c-Kit+Lin− BM cells and immunoprecipitated with an anti-GABPα antibody or a control IgG as described previously.19 The enrichment of GABP binding in these murine hematopoietic progenitor cells was assessed by quantitative PCR on ABI 7300 Real Time PCR System (Applied Biosystems). For primer design in a select gene, a cross-species conserved region corresponding to a GABP binding peak based on ChIP-Seq was first identified in the human genome, and the corresponding conserved sequence in the mouse genome was used for quantitative assessment. Each primer set was tested for linear amplification range with input DNA using SYBR Advantage qPCR premix (Clontech). The Rag2 promoter with no GABP binding on ChIP-Seq was used as a negative control. For calculation of enrichment of each selected gene/region, 2−ΔCt was used, where ΔCt is the difference of Ct (crossover threshold) values detected in GABPα antibody- and IgG-precipitated samples. Primer sequences for amplification of each select gene segment are listed in supplemental Table 1A (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Microarray and quantitative PCR
Four days after pIpC treatment, whole BM cells from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were isolated and lineage-depleted, and the Flt3−LSK cells were sorted directly into Trizol LS reagent (Invitrogen). After chloroform extraction, the aqueous phase was mixed with 2 volumes of ethanol and loaded onto a purification column in RNeasy Mini Kit (QIAGEN) for further purification. RNA quality was assessed using the Agilent Model 2100 Bioanalyzer. Total RNAs from 2 wild-type (WT) and 2 GABPα-deficient samples were amplified using the NuGEN WT-Ovation Pico RNA Amplification System (NuGEN). The resulting cDNA probes were hybridized to the GeneChip Mouse GENE 1.0 ST arrays (Affymetrix), scanned with the Affymetrix Model 7G upgraded scanner, and the data were collected using the GeneChip Operating Software. The data were imported into Partek Genomics Suite using Robust Multi-Chip Average normalization. Differential expression and its statistical significance were calculated using linear contrasts with an ANOVA (analysis of variance) model. All microarray data have been deposited at the NCBI Gene Expression Omnibus under accession number GSE23341.
For validation of select gene-expression changes, total RNA was amplified from GABPα-deficient and control Flt3−LSK cells as described and analyzed using quantitative PCR. Relative expression levels of select genes were normalized to that of a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (Gapdh). Primer sequences for each transcript are listed in supplemental Table 1B.
Bioinformatic analysis with GSEA and DAVID
Gene set enrichment analysis (GSEA) software was downloaded and used for analysis according to the instructions.28 The normalized gene-expression profiles of GABPα-deficient Flt3−LSK cells were used as the ranked dataset. The gene sets used in enrichment assessments include C2-curated gene sets from the Broad Institute and the GABP-bound gene set. The latter contained enriched GABP binding within 2 kb of the transcription initiation sites (TISs) in 7782 genes and was constructed in-house based on ChIP-Seq results using human HPCs. For functional annotation of GABP-activated or -repressed direct target genes, the extracted gene list was uploaded onto the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources (http://david.abcc.ncifcrf.gov) and analyzed following reported protocols.29
Results
Ablation of GABPα drastically diminished the HSC and progenitor pool
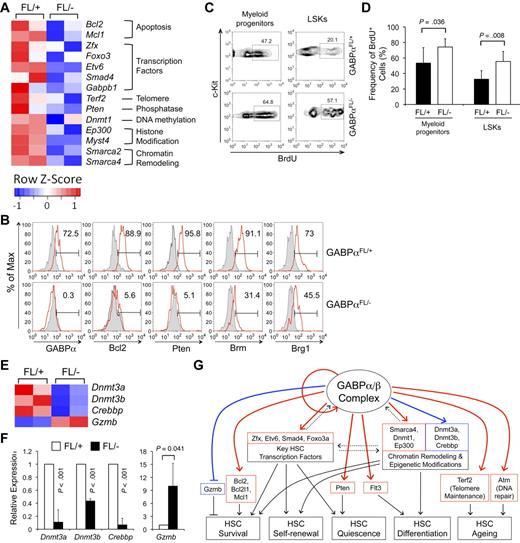
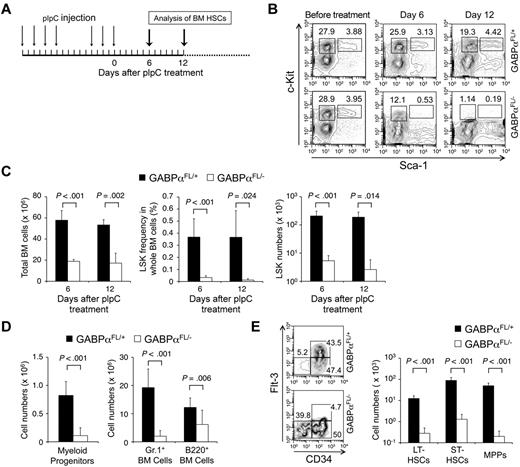
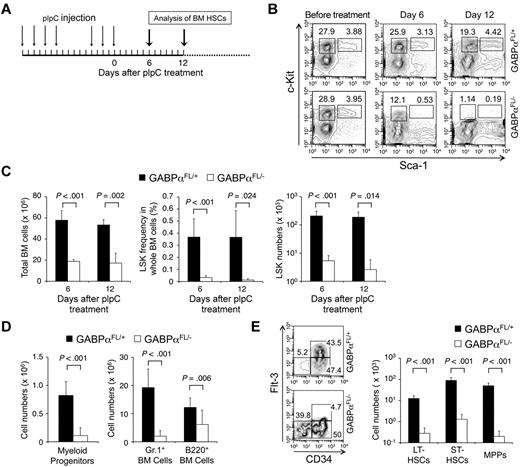
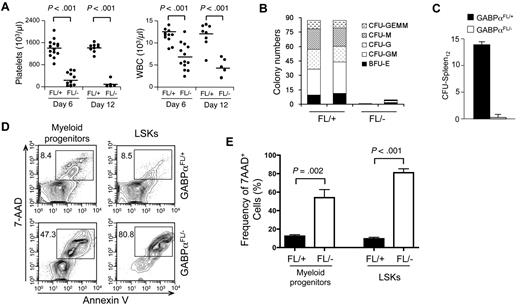
Targeting GABPα in the germline resulted in early embryonic lethality.16,17 To avoid this, GABPα was conditionally targeted by inserting LoxP sites in the Gabpa gene locus to flank exons encoding the DNA binding Ets domain, yielding a Gabpa-floxed allele (GABPαFL/+).24 Crossing GABPαFL/+ mice to an EIIa transgene resulted in conversion of the GABPα-floxed allele to a GABPα-deleted allele in the germline (GABPα+/−).24 To investigate the functional requirements of GABP in HSCs, we used the Mx1Cre transgene to induce specific inactivation of the Gabpa-floxed allele in BM cells through pIpC treatment (Figure 1A). Six to 12 days after the last pIpC treatment, the total BM cells in Mx1Cre-GABPαFL/− mice were reduced to approximately one-third of those in Mx1Cre-GABPαFL/+ controls (Figure 1C). Both lineage-negative, Sca1+, and c-Kithigh (LSK) subset containing HSCs and lineage-negative, Sca1−, and c-Kithigh myeloid progenitors were greatly diminished in both frequency and absolute numbers in Mx1Cre-GABPαFL/− mice (Figure 1B-D). Granulocytes and developing B cells in the BM were also significantly decreased (Figure 1D), indicating an acute failure on induced disruption of GABPα.
GABPα is required for maintaining an HSC and progenitor pool. (A) Induction of GABPα-floxed allele and experimental timeline. Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice at 5 weeks old were injected intraperitoneally with 25 μg/g body weight of pIpC as indicated. The last day of injection was designated day 0 and BM cells were harvested on indicated days for analysis. (B) Flow cytometric analysis of LSKs and c-Kit+ myeloid progenitors in lineage-negative BM cells. BM cells were isolated on indicated days from pIpC-treated Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− mice and surface-stained. Percentages of LSKs and c-Kit+ myeloid progenitors in Lin− BM cells are shown. (C) Total BM cellularity, LSK frequency and numbers. LSK frequency was expressed as percentages of BM nucleated cells. Absolute counts of total BM cells and LSKs were obtained from 2 tibias and 2 femurs in each mouse. (D) Numbers of myeloid progenitors, granulocytes, and developing B cells in the BM. BM cells were isolated from Mx1Cre-GABPαFL/+ or control mice during 6-12 days after pIpC treatment, and stained for Lin−c-Kit+ myeloid progenitors, Gr.1+, and B220+ cells. The absolute counts were from 2 hind limbs as in panel C. (E) Analysis of LT-HSC, ST-HSC, and MPP subsets in LSKs. LSK cells were fractionated based on CD34 and Flt3 expression, and the percentage of each subset was shown in the flow cytometric profile (left panel). Absolute count of each subset was shown in the right panel. All flow cytometric data are representative of at least 3 independent experiments with similar results, and bar graphs are means ± standard deviation (SD) of pooled results (n ≥ 6). Statistical significance was calculated using the Student t test, with P values shown in each relevant panel.
GABPα is required for maintaining an HSC and progenitor pool. (A) Induction of GABPα-floxed allele and experimental timeline. Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice at 5 weeks old were injected intraperitoneally with 25 μg/g body weight of pIpC as indicated. The last day of injection was designated day 0 and BM cells were harvested on indicated days for analysis. (B) Flow cytometric analysis of LSKs and c-Kit+ myeloid progenitors in lineage-negative BM cells. BM cells were isolated on indicated days from pIpC-treated Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− mice and surface-stained. Percentages of LSKs and c-Kit+ myeloid progenitors in Lin− BM cells are shown. (C) Total BM cellularity, LSK frequency and numbers. LSK frequency was expressed as percentages of BM nucleated cells. Absolute counts of total BM cells and LSKs were obtained from 2 tibias and 2 femurs in each mouse. (D) Numbers of myeloid progenitors, granulocytes, and developing B cells in the BM. BM cells were isolated from Mx1Cre-GABPαFL/+ or control mice during 6-12 days after pIpC treatment, and stained for Lin−c-Kit+ myeloid progenitors, Gr.1+, and B220+ cells. The absolute counts were from 2 hind limbs as in panel C. (E) Analysis of LT-HSC, ST-HSC, and MPP subsets in LSKs. LSK cells were fractionated based on CD34 and Flt3 expression, and the percentage of each subset was shown in the flow cytometric profile (left panel). Absolute count of each subset was shown in the right panel. All flow cytometric data are representative of at least 3 independent experiments with similar results, and bar graphs are means ± standard deviation (SD) of pooled results (n ≥ 6). Statistical significance was calculated using the Student t test, with P values shown in each relevant panel.
LSK cells are heterogeneous and consist of LT-HSCs, short-term HSCs (ST-HSCs), and MPPs, and these subsets can be distinguished by CD34 and Flt3 expression.30 On day 6 after pIpC treatments when LSKs were still reliably detectable, whereas all 3 subsets were greatly diminished in absolute numbers, the CD34+Flt3+ MPPs were more drastically decreased (Figure 1E). Flt3 marks differentiation of HSCs to more committed pro-genitors,30 and direct binding of GABPα to the FLT3 locus was found in human hematopoietic progenitor cells (HPCs) based on ChIP-Seq data (detailed in supplemental Table 3). The selective disappearance of Flt3-expressing MPPs is thus likely a result of direct regulation of Flt3 by GABP and represents an HSC differentiation defect because of GABPα deficiency. These observations collectively suggest that ablation of GABPα impairs HSC maintenance as well as further differentiation.
GABPα was intrinsically required for HSC activity and survival
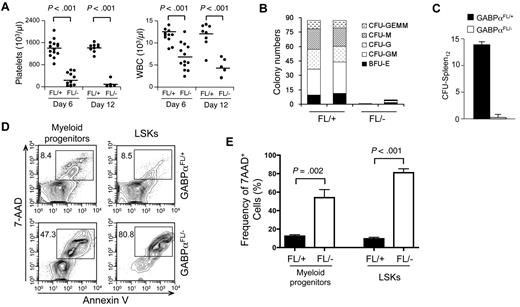
We next determined the impact of GABPα inactivation on HSC differentiation. In peripheral blood, severely diminished platelets and greatly reduced white blood cells were observed in the pIpC-treated Mx1Cre-GABPαFL/− mice (Figure 2A). Using BM cells from pIpC-treated Mx1Cre-GABPαFL/− and control mice in an in vitro methylcellulose colony formation assay, we found substantial and consistently reduced numbers of all different types of colonies as a result of GABPα ablation (Figure 2B). By an in vivo assay of colony-forming units in the spleen on day 12 (CFU-Spleen12), whereas expected numbers of macro-colonies were found in the host spleens when control BM cells were transferred, almost no colonies formed in mice received Mx1Cre-GABPαFL/− BM cells (Figure 2C). These findings indicate that GABPα deletion eliminated HSC activity and resulted in failure to sustain various blood lineages. We next explored the fate of HSCs on induced deletion of GABPα by measuring cell apoptosis. Using annexin-V and 7-AAD staining, both LSKs and Lin−c-Kit+ myeloid progenitors in Mx1Cre-GABPαFL/− mice exhibited greatly increased apoptosis, with LSKs showing more exacerbated cell death (Figure 2D-E). These GABPα deficiency-derived defects are reminiscent of HSCs and progenitors lacking Etv6, or both c-Myc and N-Myc, or both Lyl1 and Scl,8,31,32 indicating the absolute dependence of HSCs and progenitors on GABPα for survival.
GABPα is required for HSC differentiation and survival. (A) Peripheral blood analysis. Blood was collected from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice on indicated days after pIpC treatments and analyzed on Sysmex XT 2000iv automatic hematology analyzer. Numbers of platelets and white blood cells (WBCs) are shown, with horizontal bars denoting mean values in each group. Data are pooled results from 2-3 independent experiments with 5-13 animals analyzed. (B) Colony formation assays using methylcellulose. Total BM cells from Mx1Cre-GABPαFL/+ and Mx1Cre- GABPαFL/− mice were isolated 4 days after the last pIpC injection. A total of 2 × 104 BM cells were mixed and plated with M3434 methylcellulose-based media. Colonies of each type were counted after 10-day culture. GEMM indicates granulocyte, erythroid, macrophage, megakaryocyte; GM, granulocyte macrophage; G, granulocyte; M, macrophage; BFU-E, burst-forming unit-erythroid. Data are representative from 2 independent experiments with 8 animals analyzed. The colony numbers are averages of triplicate measurements of each individual mouse. (C) Ablation of GABPα diminishes CFU-Spleen12 colonies. Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were treated with pIpC, and on day 9 after last injection, 1 × 105 BM cells were injected into 900 rad–irradiated hosts. After another 12 days, spleens from the hosts were harvested and macro-colonies were counted. Shown are representative from 2 independent experiments with similar results (n = 4 in each experiment). (D) Detection of apoptotic cells in LSK and myeloid progenitor subsets. Total BM cells were isolated from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice 2-4 days after pIpC treatment, lineage-depleted, and surface-stained with c-Kit and Sca-1, followed by staining with annexin V and 7-AAD. The percentage of annexin V–7-AAD+ cells in each subset is shown. (E) Enhanced cell death of GABPα-deleted LSKs and myeloid progenitors. Data were pooled results from 2 independent experiments with 6 mice of each genotype analyzed.
GABPα is required for HSC differentiation and survival. (A) Peripheral blood analysis. Blood was collected from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice on indicated days after pIpC treatments and analyzed on Sysmex XT 2000iv automatic hematology analyzer. Numbers of platelets and white blood cells (WBCs) are shown, with horizontal bars denoting mean values in each group. Data are pooled results from 2-3 independent experiments with 5-13 animals analyzed. (B) Colony formation assays using methylcellulose. Total BM cells from Mx1Cre-GABPαFL/+ and Mx1Cre- GABPαFL/− mice were isolated 4 days after the last pIpC injection. A total of 2 × 104 BM cells were mixed and plated with M3434 methylcellulose-based media. Colonies of each type were counted after 10-day culture. GEMM indicates granulocyte, erythroid, macrophage, megakaryocyte; GM, granulocyte macrophage; G, granulocyte; M, macrophage; BFU-E, burst-forming unit-erythroid. Data are representative from 2 independent experiments with 8 animals analyzed. The colony numbers are averages of triplicate measurements of each individual mouse. (C) Ablation of GABPα diminishes CFU-Spleen12 colonies. Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were treated with pIpC, and on day 9 after last injection, 1 × 105 BM cells were injected into 900 rad–irradiated hosts. After another 12 days, spleens from the hosts were harvested and macro-colonies were counted. Shown are representative from 2 independent experiments with similar results (n = 4 in each experiment). (D) Detection of apoptotic cells in LSK and myeloid progenitor subsets. Total BM cells were isolated from Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice 2-4 days after pIpC treatment, lineage-depleted, and surface-stained with c-Kit and Sca-1, followed by staining with annexin V and 7-AAD. The percentage of annexin V–7-AAD+ cells in each subset is shown. (E) Enhanced cell death of GABPα-deleted LSKs and myeloid progenitors. Data were pooled results from 2 independent experiments with 6 mice of each genotype analyzed.
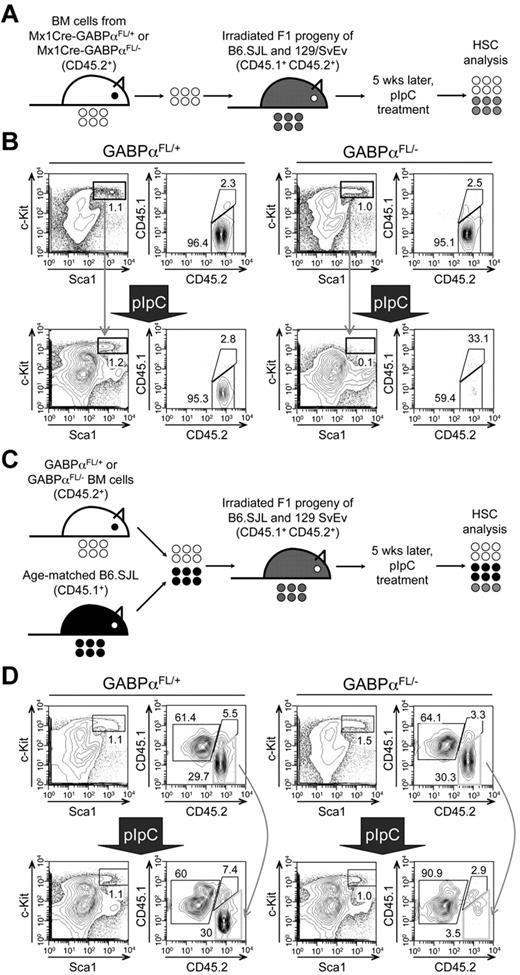
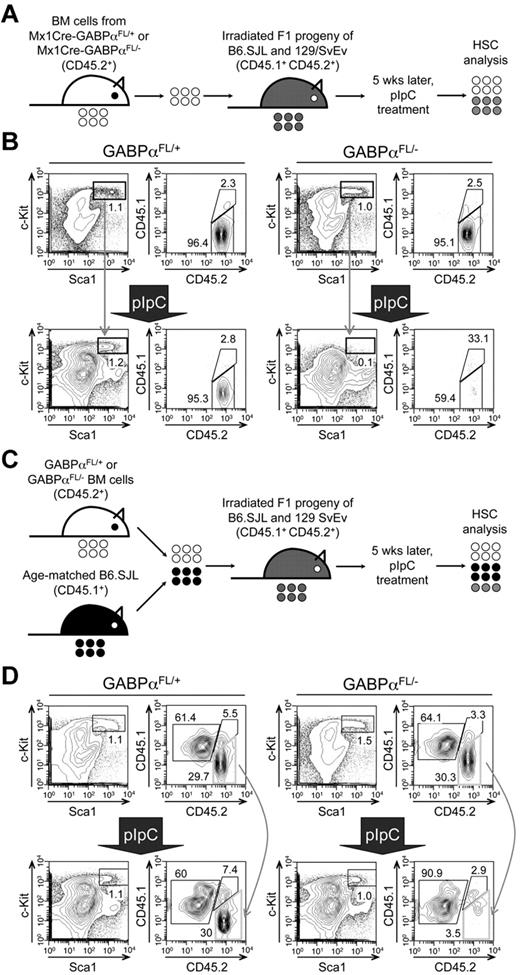
Mx1Cre activity can be induced by pIpC treatment in both nucleated BM cells and other cells in the bones constituting HSC niches. To determine whether the defects in HSC maintenance because of GABPα deficiency are cell intrinsic, we transferred BM cells from Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− mice to irradiated F1 progeny of B6.SJL and 129/SvEv breeding and established BM chimeras (Figure 3A). Treatment with pIpC eliminated GABPαFL/−-derived LSKs but not those from GABPαFL/+ mice (Figure 3B). To further demonstrate the relative maintenance of GABPα-deficient LSKs, we transferred Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− along with B6.SJL BM cells as competitors into irradiated F1 progeny (Figure 3C), and then treated the mixed BM chimeras with pIpC. Whereas the test cell–to–competitor cell ratio was not affected in mice transferred with GABPαFL/+ BM cells after pIpC treatment, GABPαFL/−-derived LSKs were diminished and hence the test to competitor ratio was greatly reduced (Figure 3D). These observations thus indicate that GABPα is intrinsically required for maintaining HSCs.
Cell autonomous requirements for GABPα in maintaining HSCs. (A) Schematic showing the experimental design for induced GABPα inactivation in preestablished BM chimeras. The GABPα-floxed animals were previously generated in 129/SvEv embryonic stem cells and were crossed to C57BL/6 strains (WT or Mx1Cre transgenic) for 4 generations. To avoid potential rejection of BM grafts, the F1 progeny of B6.SJL and 129/SvEv crossing was used as hosts for generation of BM chimeras. The F1 hosts expressed both CD45.1 and CD45.2 congenic markers, whereas the donor-derived cells were positive for CD45.2 only, allowing direct distinguishing of the cell origins in the BM chimeras. (B) BM cells from Mx1Cre-GABPαFL/+ or Mx1Cre- GABPαFL/− were injected into irradiated F1 progeny to establish BM chimeras, and the LSK cells were largely of donor origin (CD45.2+ > 95%, top panels). The BM chimeras were treated with pIpC as in Figure 1A, and 9 days after the last pIpC injection, the LSK population and its origin were analyzed (bottom panels). Note that LSKs from pIpC-treated Mx1Cre-GABPαFL/− BM chimeras were significantly diminished, and a substantial portion of the remaining LSKs was of host origin. (C) Experimental design for induced GABPα inactivation in the presence of WT reference cells. (D) BM cells from Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− were mixed with those from B6.SJL at 1:1 LSK ratio and injected into irradiated F1 progeny to establish mixed BM chimeras (top panels). The hosts were treated with pIpC as in panel B and analyzed for LSK frequency and origin. All data are representative of 3 independent experiments with similar results.
Cell autonomous requirements for GABPα in maintaining HSCs. (A) Schematic showing the experimental design for induced GABPα inactivation in preestablished BM chimeras. The GABPα-floxed animals were previously generated in 129/SvEv embryonic stem cells and were crossed to C57BL/6 strains (WT or Mx1Cre transgenic) for 4 generations. To avoid potential rejection of BM grafts, the F1 progeny of B6.SJL and 129/SvEv crossing was used as hosts for generation of BM chimeras. The F1 hosts expressed both CD45.1 and CD45.2 congenic markers, whereas the donor-derived cells were positive for CD45.2 only, allowing direct distinguishing of the cell origins in the BM chimeras. (B) BM cells from Mx1Cre-GABPαFL/+ or Mx1Cre- GABPαFL/− were injected into irradiated F1 progeny to establish BM chimeras, and the LSK cells were largely of donor origin (CD45.2+ > 95%, top panels). The BM chimeras were treated with pIpC as in Figure 1A, and 9 days after the last pIpC injection, the LSK population and its origin were analyzed (bottom panels). Note that LSKs from pIpC-treated Mx1Cre-GABPαFL/− BM chimeras were significantly diminished, and a substantial portion of the remaining LSKs was of host origin. (C) Experimental design for induced GABPα inactivation in the presence of WT reference cells. (D) BM cells from Mx1Cre-GABPαFL/+ or Mx1Cre-GABPαFL/− were mixed with those from B6.SJL at 1:1 LSK ratio and injected into irradiated F1 progeny to establish mixed BM chimeras (top panels). The hosts were treated with pIpC as in panel B and analyzed for LSK frequency and origin. All data are representative of 3 independent experiments with similar results.
Genome-wide mapping of GABP binding locations in human HPCs
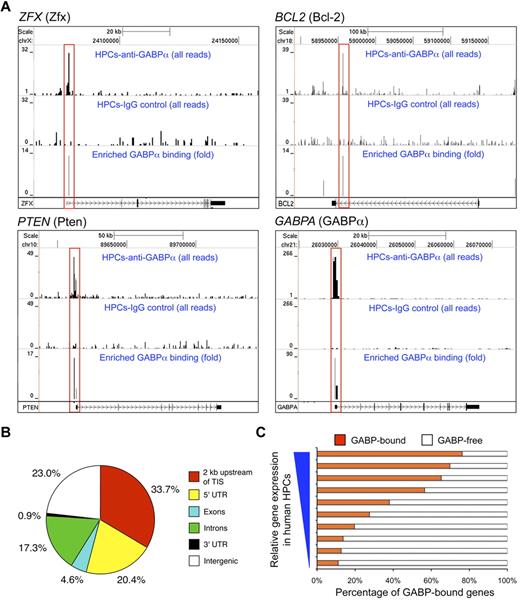
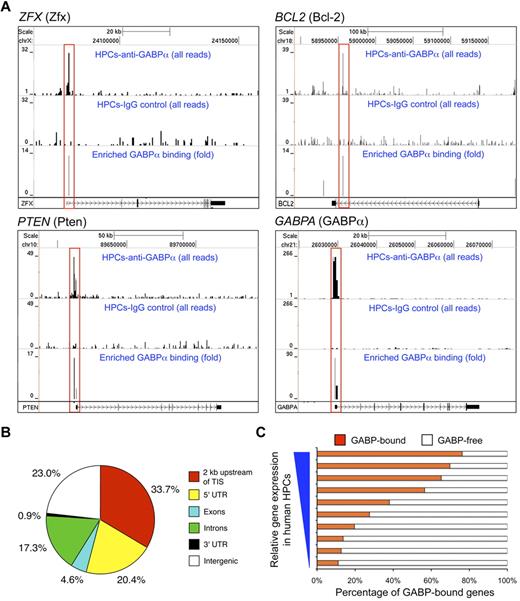
To investigate the molecular basis for regulation of HSC maintenance and biologic activities by GABP, we first used ChIP-Seq to map GABP binding locations across the genome in HSCs.26 Given the paucity of murine LSK cells, we used purified CD34+CD133+c-KitmedThy1med HPCs from human peripheral blood to obtain sufficient genomic DNA for ChIP.26 The sequence tags were processed using SISSRs peak finding tool27 to identify GABP binding sites genome-wide (Figure 4A). By setting of P < .01, a total of 15 767 binding locations were identified and distributed in the promoter, genic, and intergenic regions (Figure 4B). It is of note that more than 50% of GABP binding locations were found to be close to TISs (within 2 kb or in 5′-UTR). This is in contrast to what was found in ChIP-Seq of GATA-1 in mouse erythroleukemia cells, where only 13% of GATA-1 binding was located within 10 kb of TISs.21 De novo motif analysis confirmed the presence of core consensus sequence motif “(a/c)GGAA(g/a)” as the top motif present within more than 80% of the identified GABP binding sites, and further revealed “(c/g)CGGAAGT” as the most preferred GABP binding sequence based on ChIP enrichment ranking (supplemental Figure 1A). The second-ranked sequence motif “GGGA(a/g)(t/a)TGTAGT” was found in approximately 10% of the identified GABP binding sites (supplemental Figure 1B). The secondary motif may represent a site for recruitment of GABP through protein-protein interaction,20 or alternatively a genuine GABP binding consensus sequence, because approximately half of a set of 104 mouse DNA-binding proteins were found to recognize multiple different sequence motifs using a protein-binding microarray technology.33 For the primary GABP binding motif, the ChIP-Seq–predicted GABP binding locations were independently confirmed in select gene loci using in vitro electrophoretic mobility shift assays (supplemental Figure 2). Interestingly, comparative analysis with gene-expression profiles of human HPCs26 revealed that GABP binding was positively correlated with genes expressed at higher levels (Figure 4C). GABP has been known to mediate both transcription activation and repression; however, this observation suggests that GABP appears to be more dominantly associated with activation of gene transcription in HPCs/HSCs.
Characterization of genome-wide GABPα binding locations in human HPCs. (A) GABP binding at selected gene loci inferred from ChIP-Seq. Chromatin fragments prepared from CD34+CD133− human HPCs were subjected to ChIP-Seq analysis. For each gene locus (in individual panels), sequence tags from anti-GABPα (first row) and control IgG (second row) samples were displayed on the University of California Santa Cruz genome browser with their heights (y-axis) denoting the tag numbers. GABP binding locations identified by SISSRs, highlighted in red rectangles, are displayed as enrichment peaks in the third row of each panel, with the y-axis heights corresponding to fold-enrichment of GABP sequence tags over control tags. Gene symbols and corresponding protein names (in parentheses), scale, and relative locations in respective chromosomes are shown on the top, and gene structures and directions of transcription are marked at the bottom of each panel. (B) Genome-wide distribution of GABPα binding locations in human HPCs. TIS indicates transcription initiation site; UTR, untranslated region. (C) Correlation between GABP binding and gene expression in human HPCs. Genes sorted by absolute expression levels in human HPCs (from Cui et al26 ) were binned into 10 groups, and the percentages of genes within each group that bind GABP within 2 kb of TISs are shown.
Characterization of genome-wide GABPα binding locations in human HPCs. (A) GABP binding at selected gene loci inferred from ChIP-Seq. Chromatin fragments prepared from CD34+CD133− human HPCs were subjected to ChIP-Seq analysis. For each gene locus (in individual panels), sequence tags from anti-GABPα (first row) and control IgG (second row) samples were displayed on the University of California Santa Cruz genome browser with their heights (y-axis) denoting the tag numbers. GABP binding locations identified by SISSRs, highlighted in red rectangles, are displayed as enrichment peaks in the third row of each panel, with the y-axis heights corresponding to fold-enrichment of GABP sequence tags over control tags. Gene symbols and corresponding protein names (in parentheses), scale, and relative locations in respective chromosomes are shown on the top, and gene structures and directions of transcription are marked at the bottom of each panel. (B) Genome-wide distribution of GABPα binding locations in human HPCs. TIS indicates transcription initiation site; UTR, untranslated region. (C) Correlation between GABP binding and gene expression in human HPCs. Genes sorted by absolute expression levels in human HPCs (from Cui et al26 ) were binned into 10 groups, and the percentages of genes within each group that bind GABP within 2 kb of TISs are shown.
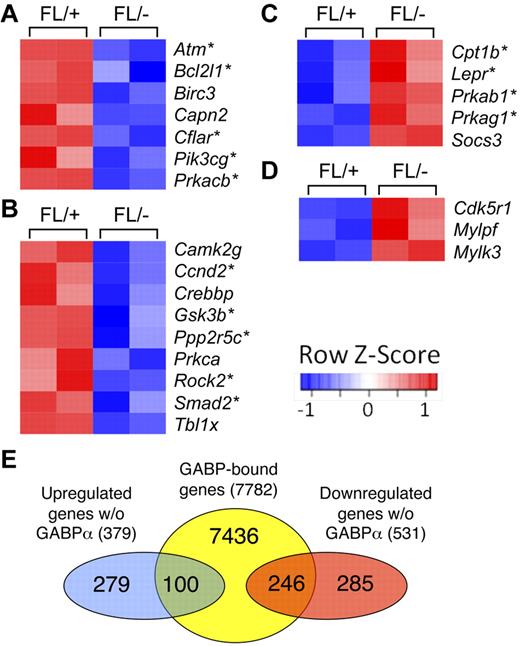
GABPα deficiency–altered transcriptome in HSCs
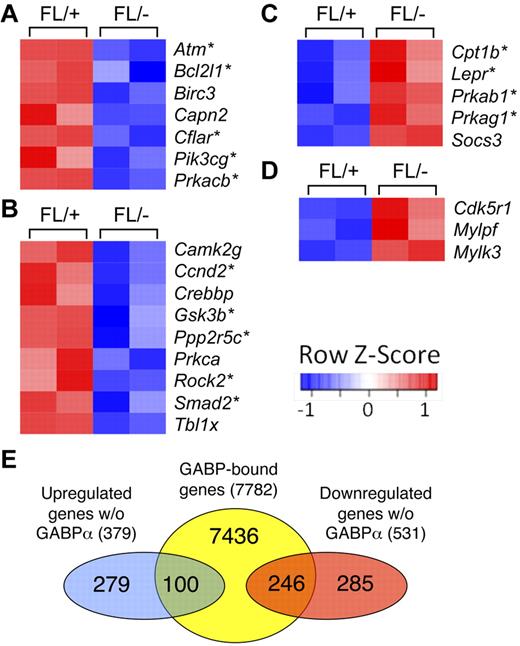
To further investigate the molecular basis of GABPα deficiency–derived HSC defects and functional relevance of GABP occupancy across the HPC/HSC genome, we performed whole-genome transcriptome analysis on GABPα-deficient and control Flt3−LSKs (containing both LT- and ST-HSCs). By the setting of P < .05 for significantly differential expression (based on ANOVA model), 379 genes were up-regulated and 531 genes down-regulated for > 1.5 fold because of GABPα deficiency. Remarkably, among the top 20 down-regulated genes were an Ets family transcription factor Fli-1 and a homeodomain protein Meis1, which have been known to be essential for definitive hematopoiesis. Detailed analysis using functional annotation tools in the DAVID bioinformatic resources revealed that the differentially regulated genes because of GABPα deficiency included transcription factors and genes involved in regulation of apoptosis, cell-cycle progression, DNA repair, and epigenetic modification including DNA methylation and histone acetylation (supplemental Table 2). Further pathway analysis using KEGG (Kyoto Encyclopedia of Genes and Genomes) and BioCarta databases revealed that disruption of GABPα perturbed several pathways that regulate HSC biology. Consistent with increased apoptosis of GABPα-deficient HSCs, we observed diminished expression of several molecules in the apoptosis pathway, including Bcl-XL (Figure 5A). Wnt signaling has been implicated in promoting HSC expansion as well as maintaining HSC quiescence/self-renewal,34 and a number of molecules in this pathway were decreased in expression in GABPα-deficient Flt3−LSK cells (Figure 5B). On the other hand, several players in the adipocytokine signaling pathway and Rac1 cell motility signaling pathway were increased in expression because of loss of GABPα (Figure 5C-D). Bone marrow adipocytes were recently reported to negatively regulate HSC repopulation capacity,35 and alterations in the Rac small GTPases affected mobilization from the bone marrow.36 These analyses suggest that GABP has a multifaceted role in maintaining HSC homeostasis and functionality.
Pathway analysis of differentially expressed genes because of GABPα ablation in HSCs. (A-D) Flt3−LSKs were sorted from bone marrow cells of 2 pairs of Mx1Cre-GABPαFL/− and control mice and subjected to microarray analysis using GeneChip Mouse GENE 1.0 ST arrays. Genes exhibited significant expression changes (> 1.5 fold and P < .05) were analyzed using pathway analysis tools in DAVID bioinformatics resources. Heatmaps of genes in each pathway are shown. Color-coded scale bar indicates Z-score values, denoting number of standard deviations from the mean in each row. Asterisks denote genes containing GABP binding within 2 kb of their TISs as identified by ChIP-Seq. (A) KEGG apoptosis pathway. (B) KEGG Wnt signaling pathway. (C) BioCarta adipocytokine signaling pathway. (D) Rac1 cell mobility signaling pathway. (E) Venn diagram showing the overlap of GABP-bound genes and down-regulated or up-regulated genes because of GABPα deficiency.
Pathway analysis of differentially expressed genes because of GABPα ablation in HSCs. (A-D) Flt3−LSKs were sorted from bone marrow cells of 2 pairs of Mx1Cre-GABPαFL/− and control mice and subjected to microarray analysis using GeneChip Mouse GENE 1.0 ST arrays. Genes exhibited significant expression changes (> 1.5 fold and P < .05) were analyzed using pathway analysis tools in DAVID bioinformatics resources. Heatmaps of genes in each pathway are shown. Color-coded scale bar indicates Z-score values, denoting number of standard deviations from the mean in each row. Asterisks denote genes containing GABP binding within 2 kb of their TISs as identified by ChIP-Seq. (A) KEGG apoptosis pathway. (B) KEGG Wnt signaling pathway. (C) BioCarta adipocytokine signaling pathway. (D) Rac1 cell mobility signaling pathway. (E) Venn diagram showing the overlap of GABP-bound genes and down-regulated or up-regulated genes because of GABPα deficiency.
Systematic identification of GABP-activated and -repressed direct target genes
Using the genome-wide GABP occupancy data in human HPCs, we wished to determine which genes are direct targets of GABP. Focusing on genes that harbor GABP binding within 2 kb of their TISs, we constructed an HSC/HPC GABP-bound gene set containing 7782 unique genes (hereafter referred to as GABP-bound genes). By direct comparison with differentially regulated genes described in the previous section, 246 of 531 down-regulated genes and 100 of 379 up-regulated genes were GABP-bound genes (Figure 5E). These genes constitute only 3.3% for down-regulated and 1.3% for up-regulated among all the GABP-bound genes. So as not to overlook other GABP direct targets that mediate regulation of HSC biologic activities, we used GSEA, which employs the Kolmogorov-Smirnov statistics to assess the degree of enrichment of a given gene set in the ranked gene-expression profiling of GABPα deficiency, independent of fold change–based preselection/cutoff.28 This approach is powerful in identifying relevant genes that are associated with one particular biologic process, which may be hard to distinguish at the level of individual genes. Comparison of the GABP-bound gene set with gene-expression profiles using GSEA revealed that 2130 genes (27.4%) are at the leading edge of positively correlated genes (hereby defined as GABP-activated direct targets; Figure 6A). In contrast, only 294 genes (approximately 3.8%) were negatively correlated (defined as GABP-repressed direct targets; Figure 6A). This analysis substantially expanded GABP direct targets and made it more evident that GABP acts more dominantly as a transcriptional activator as observed in Figure 4C. Functional annotation of the expanded GABP direct target genes using DAVID was summarized in supplemental Table 3, revealing that GABP directly regulates genes involved in telomere maintenance and chromatin remodeling, in addition to functional categories in supplemental Table 2. A recent study mapping binding locations of Scl/Tal1 in a hematopoietic precursor cell line identified and validated that SCL directly regulates 16 other critical transcription factors.37 By stringent fold-expression change and P value cutoff, only Fli-1 and Hhex out of the 16 factors were found to be GABP direct targets. However, GSEA identified 4 additional factors including Runx1, Runx2, Myb, and Lyl1 as GABP-activated genes (Figure 6B). The conventional analysis and GSEA thus provided complementary information that allows us to systematically map GABP target genes.
Comparative analysis of genome-wide GABPα occupancy and expression profiles by GSEA broadens GABP direct target genes/pathways. (A) GSEA profile of correlation between GABP-regulated genes and GABP-bound gene set. GABP-activated direct targets are at the leading edge of positive correlation (in red box on the left), and GABP-repressed direct targets are at the leading edge of negative correlation (in blue box on the right). Functional annotation of these 2 subsets is summarized in supplemental Table 3. (B) Downstream transcription factors that are regulated by both GABP and Scl/Tal1. Shown is the heatmap with color-coded scale bars showing Z-scores. (C) GSEA profiles of correlation between genes positively regulated by GABP and gene sets containing stem cell signatures. (D) GSEA profile of correlation between genes negatively regulated by GABP and a gene set containing genes enriched in mature blood cells compared with HSCs (shared between fetal liver and adult BM HSCs). Other negatively correlated gene sets include HSC_MATURE_FETAL (P = 0, false discovery rate [FDR] q = 0.009) and HSC_MATURE_ADULT (P = 0, FDR q = 0.069) in GSEA, containing genes up-regulated in mature blood cells compared with fetal liver and BM HSCs, respectively (both from Ivanova et al38 ). (E) GSEA profiles of correlation between genes positively regulated by GABP and gene sets of select pathways. For all the GSEA in panels A and C-E, enrichment plots are shown, with gene set names, nominal P values, and FDR q values marked on top of the plots. More detailed description of all the gene sets in panels C through E, genes at the leading edge, and heatmaps are given in supplemental Figure 3. Note that a reported P = 0 indicates an actual P < 1/number of permutations. In our analysis the number of permutations was set at 1000, and thus the reported “P = 0” is equivalent to P < .001.
Comparative analysis of genome-wide GABPα occupancy and expression profiles by GSEA broadens GABP direct target genes/pathways. (A) GSEA profile of correlation between GABP-regulated genes and GABP-bound gene set. GABP-activated direct targets are at the leading edge of positive correlation (in red box on the left), and GABP-repressed direct targets are at the leading edge of negative correlation (in blue box on the right). Functional annotation of these 2 subsets is summarized in supplemental Table 3. (B) Downstream transcription factors that are regulated by both GABP and Scl/Tal1. Shown is the heatmap with color-coded scale bars showing Z-scores. (C) GSEA profiles of correlation between genes positively regulated by GABP and gene sets containing stem cell signatures. (D) GSEA profile of correlation between genes negatively regulated by GABP and a gene set containing genes enriched in mature blood cells compared with HSCs (shared between fetal liver and adult BM HSCs). Other negatively correlated gene sets include HSC_MATURE_FETAL (P = 0, false discovery rate [FDR] q = 0.009) and HSC_MATURE_ADULT (P = 0, FDR q = 0.069) in GSEA, containing genes up-regulated in mature blood cells compared with fetal liver and BM HSCs, respectively (both from Ivanova et al38 ). (E) GSEA profiles of correlation between genes positively regulated by GABP and gene sets of select pathways. For all the GSEA in panels A and C-E, enrichment plots are shown, with gene set names, nominal P values, and FDR q values marked on top of the plots. More detailed description of all the gene sets in panels C through E, genes at the leading edge, and heatmaps are given in supplemental Figure 3. Note that a reported P = 0 indicates an actual P < 1/number of permutations. In our analysis the number of permutations was set at 1000, and thus the reported “P = 0” is equivalent to P < .001.
We next performed GSEA using C2 curated gene sets, which revealed strikingly strong positive correlation of GABP-regulated genes with those previously defined HSC-related datasets38 (Figure 6C top panels; supplemental Figure 3), supporting direct association of GABP with regulation of stem cell signature genes. Interestingly, among negatively correlated genes (ie, up-regulated expression because of loss of GABPα), highly enriched are gene sets with up-regulated expression in mature blood cells compared with BM or fetal liver HSCs38 (Figure 6D; supplemental Figure 3). This observation implies that GABP might be actively involved in repression of genes expressed in more differentiated blood cells, contributing to sustaining multipotency of HSCs.
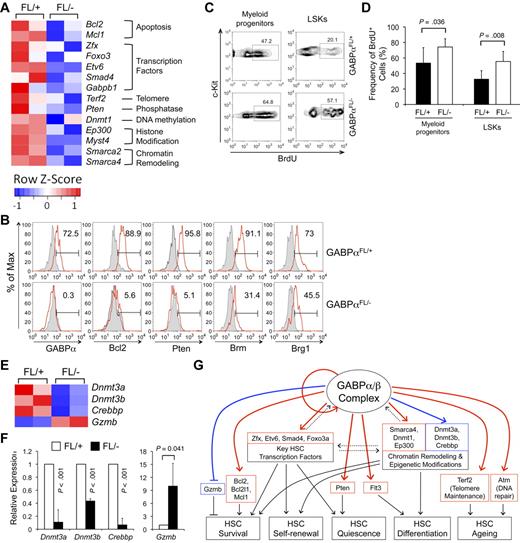
Consistent with massive apoptosis of HSCs on loss of GABPα (Figure 2D-E), an apoptosis-related gene set (containing Bcl-2 and Bcl-XL) was enriched in genes positively regulated by GABPα (Figure 6E). Recent studies demonstrated critical requirements of Pten,39,40 p53,41 and TGF-β42 pathways in restraining HSCs from hyperproliferation and thus maintaining their relative quiescence. Additionally, DNA damage repair43 and telomere maintenance44 are critical for maintaining HSC functionality during the process of aging. Further detailed GSEA analysis revealed that a substantial portion of genes involved in each of these pathways was positively regulated by GABP (Figure 6E; supplemental Figure 3). These analyses suggest that GABP is controlling a wide spectrum of HSC activities, including maintaining HSC quiescence, self-renewal, and multipotency, in addition to survival.
Based on analysis with GSEA and DAVID and relevance to HSC biology, we validated GABP occupancy in select gene loci in both Lin− and c-Kit+Lin− murine hematopoietic precursors and their expression changes because of induced GABPα inactivation in Flt3−LSK cells (including both LT- and ST-HSCs). As summarized in Figure 5A, Figure 7A, and Table 1, GABP directly regulated the expression of prosurvival Bcl-2 family members including Bcl-2, Bcl-XL, and Mcl-1,45 and Zfx and Etv6 transcription factors known to be critical for maintaining the HSC pool.7,8 These data thus provide a molecular explanation for the indispensable role of GABP in HSC survival. Also validated to be GABP-activated direct target genes are Smad4 transcription factors in the TGF-β signaling pathway,46 Atm in the DNA repair pathway,43 Terf2 in telomere maintenance,44 Foxo3a transcription factor,5,6 and Pten itself39,40 in Pten signaling pathway. To further substantiate these findings, we examined protein expression of select GABP target genes by intracellular staining. Induced disruption of the Gabpa alleles eliminated GABPα expression in LSK cells from the Mx1Cre-GABPαFL/− mice (Figure 7B). Consistent with their greatly diminished transcript levels (by approximately 10-fold or more), Bcl-2 and Pten proteins were substantially decreased in GABPα-deficient LSKs (Figure 7B). Reduction of the Bcl-2 and Pten proteins was also confirmed in myeloid progenitor cells from the Mx1Cre-GABPαFL/− mice (supplemental Figure 4). For other genes such as Atm and Zfx that were dramatically down-regulated on induced ablation of GABPα but were not validated on protein levels because of lack of intracellular staining antibodies, we further validated their reduction on transcript levels using a second independent primer set (supplemental Figure 5). These observations thus lend additional support of our high-throughput approaches in identifying GABP targets in HSCs and progenitors.
Validation of GABP target genes and predicted role of GABP in maintaining HSC quiescence. (A) Heatmaps of select GABP-activated direct target genes from the transcriptomic analysis. Color-coded scale bars denote Z-scores. Validation of GABP binding and transcript changes of all these genes is summarized in Table 1. (B) Validation of expression changes of select genes on protein levels. BM cells from pIpC-treated mice were sequentially surface-stained and intracellularly stained with fluorochrome-conjugated antibodies against GABPα, Bcl-2, Pten, Brm, or Brg1. The expression of each protein in LSKs is shown in red lines, with shaded histograms denoting isotype controls. For GABPα and Pten staining, self-conjugated normal rabbit IgG was used as an isotype control, and for Brm and Brg1 staining, self-conjugated normal goat IgG was as an isotype control. The values in histograms indicate the percentages of LSKs expressing indicated proteins. Data are representative from 3 independent experiments analyzing 3-6 pairs of Mx1Cre-GABPαFL/− and control mice. (C) Proliferation status in myeloid progenitors and LSK cells. Four to 6 days after pIpC treatment, Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were pulsed with BrdU via intraperitoneal injection for 18 hours. BM cells were surface-stained followed by intracellular detection of BrdU uptake. The percentage of BrdU+ cells in each subset is shown. (D) Increased proliferation of GABPα-deleted LSKs and myeloid progenitors. Data were pooled results from 3 independent experiments with 5 mice of each genotype analyzed. (E) Heatmaps of select genes that do not have GABPα binding within 2 kb of TISs but are affected in expression by GABPα deficiency. Color-coded scale bars showing Z-scores are the same as in panel A. The gene-expression changes were validated by quantitative RT-PCR in panel F. (G) Proposed model for the roles of GABP in regulating HSC activity. A GABP-controlled gene regulatory module in HSCs is illustrated, showing GABP auto-regulation, potential interregulation with other key transcription factors and epigenetic modification molecules, and coregulation of downstream effector genes involved in HSC survival, self-renewal, quiescence, and differentiation. Solid red and blue lines denote direct and indirect regulatory connection confirmed in this study, respectively. Dashed lines denote possible interaction in the regulatory module. Solid black lines are regulatory roles based on literature. Arrows indicate positive regulation, and lines ending in bars indicate negative regulation.
Validation of GABP target genes and predicted role of GABP in maintaining HSC quiescence. (A) Heatmaps of select GABP-activated direct target genes from the transcriptomic analysis. Color-coded scale bars denote Z-scores. Validation of GABP binding and transcript changes of all these genes is summarized in Table 1. (B) Validation of expression changes of select genes on protein levels. BM cells from pIpC-treated mice were sequentially surface-stained and intracellularly stained with fluorochrome-conjugated antibodies against GABPα, Bcl-2, Pten, Brm, or Brg1. The expression of each protein in LSKs is shown in red lines, with shaded histograms denoting isotype controls. For GABPα and Pten staining, self-conjugated normal rabbit IgG was used as an isotype control, and for Brm and Brg1 staining, self-conjugated normal goat IgG was as an isotype control. The values in histograms indicate the percentages of LSKs expressing indicated proteins. Data are representative from 3 independent experiments analyzing 3-6 pairs of Mx1Cre-GABPαFL/− and control mice. (C) Proliferation status in myeloid progenitors and LSK cells. Four to 6 days after pIpC treatment, Mx1Cre-GABPαFL/+ and Mx1Cre-GABPαFL/− mice were pulsed with BrdU via intraperitoneal injection for 18 hours. BM cells were surface-stained followed by intracellular detection of BrdU uptake. The percentage of BrdU+ cells in each subset is shown. (D) Increased proliferation of GABPα-deleted LSKs and myeloid progenitors. Data were pooled results from 3 independent experiments with 5 mice of each genotype analyzed. (E) Heatmaps of select genes that do not have GABPα binding within 2 kb of TISs but are affected in expression by GABPα deficiency. Color-coded scale bars showing Z-scores are the same as in panel A. The gene-expression changes were validated by quantitative RT-PCR in panel F. (G) Proposed model for the roles of GABP in regulating HSC activity. A GABP-controlled gene regulatory module in HSCs is illustrated, showing GABP auto-regulation, potential interregulation with other key transcription factors and epigenetic modification molecules, and coregulation of downstream effector genes involved in HSC survival, self-renewal, quiescence, and differentiation. Solid red and blue lines denote direct and indirect regulatory connection confirmed in this study, respectively. Dashed lines denote possible interaction in the regulatory module. Solid black lines are regulatory roles based on literature. Arrows indicate positive regulation, and lines ending in bars indicate negative regulation.
Given the known role of Foxo3a and Pten in restraining HSC proliferation,5,6,39,40 one would predict that their decreased expression in GABPα-deficient HSCs may result in HSC hyperproliferation. After pulsing the mice with 5′-bromo-2′-dexoxy-uridine (BrdU) for 18 hours, we indeed found increased BrdU incorporation in myeloid progenitors and more evidently in LSK cells from pIpC-treated Mx1Cre-GABPαFL/− mice (Figure 7C-D). Thus, molecular analysis of GABP target genes is instructive in identifying a novel role of GABP in sustaining relative quiescence of HSCs in addition to their survival. It is noteworthy that hyperproliferation and apoptosis have been known to be interconnected in both biologic and pathologic conditions.47
Chromatin remodeling as well as epigenetic modifications including DNA methylation and histone acetylation are essential to multiple aspects of HSC activities. We found that GABP directly regulates the expression of Brg1 (encoded by Smarca4) and Brm (Smarca2) subunits in SWI/SNF chromatin remodeling complex, DNA methyltransferase 1 (Dnmt1), and p300 (Ep300) and Myst4 histone acetyltransferases in HSCs (Figure 7A and Table 1). Reduction of Brm and Brg1 was validated on protein levels in GABPα-deficient LSKs as well as myeloid progenitors (Figure 7B and supplemental Figures 4-5), albeit the Brg1 protein was decreased to a lesser extent, proportional to its smaller reduction in transcript (Table 1). Whereas no direct binding of GABP was found within 2 kb of TISs of the Dnmt3a, Dnmt3b, and Crebbp gene loci (encoding DNA methyltransferases 3a, 3b, and CBP coactivator/histone acetyltransferase, respectively), all these genes showed diminished expression in GABPα-deficient Flt3−LSK cells (on microarray as in Figure 7E and validated by quantitative RT-PCR in Figure 7F), suggesting that they are either nondirect targets for GABP or being regulated through distal GABP-bound cis-elements. Collectively, these findings suggest that GABP can regulate gene expression by directly or indirectly affecting epigenetic modifications in HSCs. Interestingly, among nondirect GABP-repressed genes, granzyme B (Gzmb) was substantially up-regulated in GABPα-deficient Flt3−LSK cells (Figure 7E-F). This is reminiscent of c-Myc and N-Myc double deficiency, where increased granzyme B expression impaired HSC survival.31
Discussion
Through systematic genetic, bioinformatic, and functional analyses, our data reveal that GABP controls a key gene-regulatory module and has multifaceted biologic roles in HSCs (Figure 7G). The GABP-controlled module contains several layers of regulation that connect with HSC biologic activities. First, GABP directly regulates the prosurvival Bcl-2 family members that are critical for HSC survival, Flt3 for HSC differentiation, Pten for HSC quiescence, and genes involved in DNA damage repair and telomere maintenance that control HSC aging. The next level of regulation comes from direct regulation of other key transcription factors with known roles in HSC activities. These include Zfx and Etv6 that are essential for HSC survival and Foxo3a that has pivotal roles in maintaining HSC quiescence and self-renewal. Interestingly, GABPα directly binds to its own gene locus and regulates the expression of its interacting partner, GABPβ1 (Table 1), suggesting that GABP forms a positive feedback loop on its own expression through self-enforcing auto-regulation. The third regulatory effect by GABP lies in direct or indirect regulation of genes involved in chromatin remodeling and epigenetic modifications, which may globally impact HSC gene expression, including other GABP direct targets described above. Collectively, our systematic approaches elucidate the breadth of molecular events controlled by GABP in HSCs.
The genome-wide profiling of transcription factor binding locations in a given cell type has allowed an unprecedented opportunity to systematically examine global activity of this factor. Application of this powerful approach to murine embryonic stem cells (ESCs) has led to elucidation of an interactive gene-regulatory network,48 which facilitated molecular understanding of pluripotency and self-renewal of ESCs and efficient generation of induced pluripotent stem cells. As increasing numbers of transcription factors and chromatin/epigenetic modifiers have been and continue to be identified to critically regulate HSCs, elucidation of their interplay using such global approaches seems both imperative and revealing. Wilson et al identified 228 high-confidence binding sites for Scl/Tal1 using a hematopoietic precursor cell line and by transgenic models they validated direct regulatory connections between Scl/Tal1 and 16 other key transcription factors in hematopoiesis.37 Of the 228 sites/gene loci occupied by Scl/Tal1, 42 are GABP-activated and 1 is GABP-repressed, and among the 16 validated transcription factors, 6 are GABP direct targets (Figure 6B). Such comparative analysis of high-throughput data suggests that these common downstream targets may serve as an interface for crosstalk between GABP- and Scl/Tal1-controlled transcription modules. The study by Wilson et al and our study thus constitute a starting point, and a similar approach can be extended to other critical transcription factors with well-defined roles in HSCs, aiming to determine how individual modules interact with each other to form a transcriptional network in the adult stem cells.
Dissection of GABP-regulated genes provided molecular explanation of biologic defects in the GABPα-targeted animals. The direct regulation of prosurvival members of Bcl-2 family, Zfx and Tel/Etv6 transcription factors, and indirect regulation of granzyme B by GABP explain the rapidly vanishing HSC pool on loss of GABPα. Identification of Pten and Foxo3a as GABP direct targets assisted revealing a novel role of GABP in maintaining HSC quiescence. Other GABP-regulated genes, including CBP and p300 coactivators, DNA methyltransferases, Atm, and Terf2 further predict that GABP deficiency may result in impaired self-renewal during serial transplantation and/or accelerated ageing. The critical requirement of GABPα for HSC survival has limited us to the use of type I interferon–mediated acute disruption of GABPα, because of the strong selection against GABPα-null cells when a constitutively expressed lineage-specific Cre recombinase was used.19 Recent studies have demonstrated that HSCs respond to interferon-α stimulation, showing increased phosphorylation of STAT1 and Akt.49 It is thus possible that a portion of the differentially regulated genes in GABPα-deficient Flt3− LSKs may represent differences of these cells in responding to interferon-α. The global GABPα occupancy data may have helped minimize this side effect, albeit not completely. In addition, the dominant impact of GABPα ablation on HSC survival makes it unfeasible to experimentally demonstrate all possible functional alterations inferred from bioinformatic analyses in GABPα-null HSCs. These caveats might be overcome through systematic structure-function analysis using mixed chimeric mice generated from WT blastocysts and embryonic stem cells expressing mutant GABPα proteins, as has been done for the p300 coactivator.50 The GABPβ subunit in the complex is known to exist in multiple isoforms, including GABPβ1L and GABPβ1S (both encoded by Gabpb1 as splice variants) and GABPβ2 (encoded by Gabpb2). In contrast to early embryonic lethality caused by GABPα deficiency, mice lacking GABPβ1L or GABPβ2 were viable.51,52 We recently found that mice lacking both GABPβ1L and GABPβ2 remained viable and exhibited age-dependent loss of HSCs (S.Y. and H.-H.X., unpublished observations, October 2010). Further analyses along these lines will help dissect distinct roles of GABPα structural domains and GABPβ isoforms in regulating HSC activities.
It is of note that not all GABP-bound genes are altered in expression on GABPα inactivation and that among all the validated GABP-activated direct target genes, some including Atm, Pten, and Brm are more strictly dependent on GABPα expression whereas others are not. One plausible explanation might be that loss of GABPα was compensated, to different degrees, by other Ets family transcription factors because multiple Ets proteins such as Ets1, Elf-1, Elk-1, and GABPα can co-occupy the same cis-regulatory elements as revealed by genome-wide mapping of their binding locations.53,54 It is also proposed that a transcription factor can function as an individual at a subset of its binding sites and have different “community” function at other sties.20 The “community” roles may be mediated through interaction with other factors and/or recruitment of chromatin remodeling or histone modification molecules. As a result, loss of a single factor may be less consequential to transcription of the nearby genes. Detailed analysis of such sites awaits accumulation of more global data on transcription factor binding and chromatin/histone modification states in the same cell type. Our studies, along with those of Wilson et al,37 constitute initial steps toward a comprehensive understanding of the gene-regulatory network in HSCs. Decoding how key transcription factors are wired together in HSCs will guide successful manipulation of the outcomes of HSC homeostasis and differentiation for therapeutic purposes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Steven Burden (Skirball Institute, NYU Medical School) for providing GABPα-targeted mice. We thank Earl Gingerich and Robert Schmidt for kind assistance with operating the Sysmex XT 2000iv hematology analyzer; Garry Hauser and Tom Bair (DNA Facility) for performing microarray analysis and assistance with the data analysis, respectively; Dr Justin Fishbaugh for assistance with the use of the LSR II flow cytometer and cell sorting; Amanda Kalen at the University of Iowa Radiation Core Facility for mouse irradiation; Dr Dustin Schones for the operation of the Illumina Solexa 1G Genome Analyzer; and Baishu Sheng for preparing the tables. We thank Drs Gail Bishop, John Colgan, John Harty, Dana Levasseur, Thomas Waldschmidt, and Nicolas Zavazava for critical reading of the manuscript.
This study is supported by NIH grants AI042767 and HL095540 (H-H.X.), the intramural research program of the National Heart, Lung and Blood Institute, NIH (K.Z.), and the National Institute of Environmental Health Sciences (1ZIAES102625-01; R.J.).
National Institutes of Health
Authorship
Contribution: S.Y. performed all the experiments unless indicated otherwise and analyzed the data; K.C. performed ChIP-Seq; R.J. analyzed the ChIP-Seq data; D.-M.Z. contributed to flow cytometric experiments and data analysis; X.J. contributed to EMSA experiments; K.Z. directed the ChIP-Seq analysis; H.-H.X. conceived the research, analyzed the data, and wrote the manuscript; and all authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for X.J. is Indiana University School of Medicine, Northwest Campus, Gary, IN.
Correspondence: Hai-Hui Xue, 51 Newton Rd, BSB 3-710, Iowa City, IA 52246; e-mail: hai-hui-xue@uiowa.edu.
References
Author notes
S.Y., K.C., and R.J. contributed equally to this work.

![Figure 6. Comparative analysis of genome-wide GABPα occupancy and expression profiles by GSEA broadens GABP direct target genes/pathways. (A) GSEA profile of correlation between GABP-regulated genes and GABP-bound gene set. GABP-activated direct targets are at the leading edge of positive correlation (in red box on the left), and GABP-repressed direct targets are at the leading edge of negative correlation (in blue box on the right). Functional annotation of these 2 subsets is summarized in supplemental Table 3. (B) Downstream transcription factors that are regulated by both GABP and Scl/Tal1. Shown is the heatmap with color-coded scale bars showing Z-scores. (C) GSEA profiles of correlation between genes positively regulated by GABP and gene sets containing stem cell signatures. (D) GSEA profile of correlation between genes negatively regulated by GABP and a gene set containing genes enriched in mature blood cells compared with HSCs (shared between fetal liver and adult BM HSCs). Other negatively correlated gene sets include HSC_MATURE_FETAL (P = 0, false discovery rate [FDR] q = 0.009) and HSC_MATURE_ADULT (P = 0, FDR q = 0.069) in GSEA, containing genes up-regulated in mature blood cells compared with fetal liver and BM HSCs, respectively (both from Ivanova et al38). (E) GSEA profiles of correlation between genes positively regulated by GABP and gene sets of select pathways. For all the GSEA in panels A and C-E, enrichment plots are shown, with gene set names, nominal P values, and FDR q values marked on top of the plots. More detailed description of all the gene sets in panels C through E, genes at the leading edge, and heatmaps are given in supplemental Figure 3. Note that a reported P = 0 indicates an actual P < 1/number of permutations. In our analysis the number of permutations was set at 1000, and thus the reported “P = 0” is equivalent to P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/7/10.1182_blood-2010-09-306563/5/m_zh89991066200006.jpeg?Expires=1766338599&Signature=X~zRk1mF1RwNukJzCkk1tzrJhdQbj2FodMpo463L4bmtIGpZxEwTDr072NLAEtALZrMqMdJlEikqgvXKc6ArerajHHItuuyMl8KpuAONtE0nvJReTiMTtaKbaOB4NsmRBoDit97fAqda6CUM6wORjSqOrd0mcbaIMvYVwjnHgpDbk400pvAy8Q0tidKndM~1Q0jdoOIkET6l2BAfxImJU8JAZNvx1Pgm~sZDjdAW6RivJ0r8nQWdf9K-Cwowav7gafz2frk9AulGB8BYyVSZWvyI6pdsQmNDnJd6-pj1I~CcK5ozsH4OrJ1VLyoT96cT3UqQMBA2QEp~WV16U9CoVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Comparative analysis of genome-wide GABPα occupancy and expression profiles by GSEA broadens GABP direct target genes/pathways. (A) GSEA profile of correlation between GABP-regulated genes and GABP-bound gene set. GABP-activated direct targets are at the leading edge of positive correlation (in red box on the left), and GABP-repressed direct targets are at the leading edge of negative correlation (in blue box on the right). Functional annotation of these 2 subsets is summarized in supplemental Table 3. (B) Downstream transcription factors that are regulated by both GABP and Scl/Tal1. Shown is the heatmap with color-coded scale bars showing Z-scores. (C) GSEA profiles of correlation between genes positively regulated by GABP and gene sets containing stem cell signatures. (D) GSEA profile of correlation between genes negatively regulated by GABP and a gene set containing genes enriched in mature blood cells compared with HSCs (shared between fetal liver and adult BM HSCs). Other negatively correlated gene sets include HSC_MATURE_FETAL (P = 0, false discovery rate [FDR] q = 0.009) and HSC_MATURE_ADULT (P = 0, FDR q = 0.069) in GSEA, containing genes up-regulated in mature blood cells compared with fetal liver and BM HSCs, respectively (both from Ivanova et al38). (E) GSEA profiles of correlation between genes positively regulated by GABP and gene sets of select pathways. For all the GSEA in panels A and C-E, enrichment plots are shown, with gene set names, nominal P values, and FDR q values marked on top of the plots. More detailed description of all the gene sets in panels C through E, genes at the leading edge, and heatmaps are given in supplemental Figure 3. Note that a reported P = 0 indicates an actual P < 1/number of permutations. In our analysis the number of permutations was set at 1000, and thus the reported “P = 0” is equivalent to P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/7/10.1182_blood-2010-09-306563/5/m_zh89991066200006.jpeg?Expires=1766566777&Signature=MohqPEHR5I7ZGyWaFBOLxyHiWVYpkx-Kmq4DHQX2lPV-veDxsstHHYVRMsV9uW78hwAW0LOyQRFlZe5LDnseShR3jPRfyLnu0GVmQmINxtigeAQthOCly94eW45Xjpz0QtzThOxRgpLUlrIqR45bUSmNtmlWILLwI0me8uKxCPIzeZiLYqNnsHs237iC5PoLzYUWmf~0FUClRqGOZkZ90fdKV6G4nCAe8JPuzIkVXHxJZLyfhKnOAJ6c8vEZh9gx8~MQMJc7M8sjtZSLeavv9w3T~0nIyYDDadqnVfEVHAaTgJimPSNHgxqeMVb4wBguQ81CxiUVLT6q0WWKUmXTow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)