Abstract
The human syndrome of dendritic cell, monocyte, B and natural killer lymphoid deficiency presents as a sporadic or autosomal dominant trait causing susceptibility to mycobacterial and other infections, predisposition to myelodysplasia and leukemia, and, in some cases, pulmonary alveolar proteinosis. Seeking a genetic cause, we sequenced the exomes of 4 unrelated persons, 3 with sporadic disease, looking for novel, heterozygous, and probably deleterious variants. A number of genes harbored novel variants in person, but only one gene, GATA2, was mutated in all 4 persons. Each person harbored a different mutation, but all were predicted to be highly deleterious and to cause loss or mutation of the C-terminal zinc finger domain. Because GATA2 is the only common mutated gene in 4 unrelated persons, it is highly probable to be the cause of dendritic cell, monocyte, B, and natural killer lymphoid deficiency. This disorder therefore constitutes a new genetic form of heritable immunodeficiency and leukemic transformation.
Introduction
We recently described an immunodeficiency syndrome that involved loss of dendritic cells (DCs), monocytes, and B and natural killer cells (DCML deficiency).1 A similar disorder of monocytopenia with Mycobacterium avium complex infection has been described as “monoMAC.”2,3 Patients typically present in young adulthood with disseminated mycobacterial and papilloma virus infection occasionally complicated by pulmonary alveolar proteinosis. Left untreated, their lives are shortened by refractory infection, respiratory failure, and leukemic transformation.
Autosomal dominant inheritance among approximately one-half of cases strongly suggests a monoallelic genetic disorder.2 We therefore sequenced exomes to identify novel heterozygous mutations in these patients. Assuming a single gene cause, we reasoned that sequencing 4 unrelated persons would exclude nearly all private but non–disease-associated mutations. We also compared the sequence of constitutional and hematopoietic DNA from 2 persons, to exclude somatic loss of heterozygosity in the hematopoietic stem cell compartment as a potential biallelic mechanism of autosomal dominant inheritance.
Methods
Patients with DCML deficiency were identified as previously described.1 All clinical samples and data were obtained with local ethical permission from the Newcastle and North Tyneside Research Ethics Committee, which approved the study. Genomic DNA was extracted with the QIAamp DNA Mini-Kit (QIAGEN); fragmented to 150-200 bp with the use of the Adaptive Focused Acoustics (Covaris); then end-repaired, adenylated, and ligated to adapters (Illumina Paired-End Sample Preparation kit). Ligated libraries were hybridized with whole-exome baits that covered 27 184 genes (Agilent SureSelect Human All Exon Kit Version 2) with modifications for the SureSelect Target Enrichment System for Illumina Paired-end Sequencing Library (Version 2.0.1). The captured fragments were purified and clonally amplified then sequenced on 2 lanes of an Illumina Genome Analyser IIx with the use of 75 bp paired-end reads.
Sequence was aligned to the human reference genome (UCSC hg19) with the use of Bowtie4 for single-base variants and BWA5 for Indels, then reformatted with the use of SAMtools.6 Eighty-seven percent of exon target sequence was covered by > 10 reads with an average coverage of 90 reads. Single base variants were identified with Varscan,7 and Indels were identified with Dindel.8 Lists of on-target variants were filtered against dbSNP131 (1094 persons) and the exome sequences of 10 unrelated and unaffected persons. Putative disease-causing mutations were identified with MutationTaster.9 GATA2 sequence was covered by 12-96 reads for novel variants and 16-86 reads for reference sequence. Novel GATA2 variants were confirmed by PCR amplification (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and Sanger sequencing and were not found in 11 unaffected first-degree relatives of the patients. An x-ray–derived structure of GATA-binding factor (GATA) zinc finger was used to model the mutations.10 Mutants R398W and T354M were created in Swiss PDB Modeller,11 and the resulting structures were displayed with Pymol (Delano Scientific).
Results and discussion
Exome sequencing identified between 853 and 1968 novel variants in each person (Table 1). Of these, 190-370 variants were predicted to be disease causing by MutationTaster.9 Any 2 persons shared 17-21 genes harboring novel variants and any 3 persons shared 2-6 genes. All shared variant genes contained only heterozygous changes. Only one gene with novel disease-causing variants was found in all 4 patients: GATA2. Identification of a single variant gene in 4 unrelated persons with a common disease phenotype strongly implicates this gene in the pathogenesis of the disorder. Given that 190-370 deleterious variants were detected in 27 184 genes, the chance of observing a novel deleterious variant of any gene is 0.6%-1.3%. The probability of 4 unrelated persons harboring a single variant gene in common is < 1 × 10−3. Each patient had a unique mutation. Neutrophil DNA sequenced in 2 patients also had heterozygous mutation of GATA-2, excluding somatically acquired loss of heterozygosity as a mechanism of pathogenesis.12 These results illustrate the power of exome sequencing to identify disease-causing mutations in a small number of unrelated and sporadic cases of a genetic disorder.
GATA-2 is a transcription factor required for stem cell homeostasis.13 The protein contains 2 highly conserved zinc finger domains that mediate protein-DNA and protein-protein interactions. Examination of the mutations in GATA2 showed a base insertion predicted to cause a frameshift after G200 and subsequent premature stop codon, 2 single nucleotide substitutions causing amino acid changes T354M and R398W, and a splice acceptor site mutation predicted to cause skipping of exon 5 and a 42-aa deletion (Table 2). The frameshift and splice acceptor mutations are expected to cause complete loss of the C terminal zinc finger and are probably null mutations, anticipated to cause DCML deficiency through haploinsufficiency of GATA-2. The amino acid substitutions probably also introduce significant structural variations in the C terminal zinc finger (Figure 1). Substitution of methionine at 354 is predicted to disrupt the conserved threonine string of the zinc finger domain, and replacement of positively charged arginine by tryptophan at the C terminus of the zinc finger probably prevents a critical interaction with the minor groove of DNA. One of these mutations, T354M, has recently been reported as a cause of autosomal-dominant familial myelodysplastic syndrome/acute myeloid leukemia. In this case, in vitro studies indicate dominant-negative function of the mutant protein in certain situations.14 In summary, both haploinsufficiency and dominant-negative loss of GATA-2 function are potential mechanisms of pathogenesis in DCML deficiency.
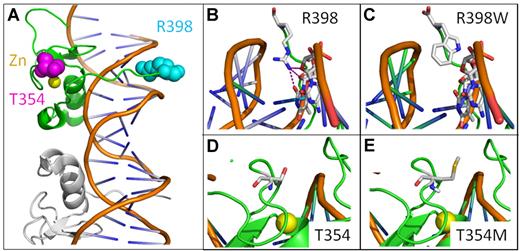
Mutations in GATA zinc finger domain. GATA zinc finger domains bind DNA in 2 conformations the adjacent and opposite forms. Here, we use the adjacent structure (3DFV).10 (A) Illustrated view of single GATA zinc finger (green) bound to DNA with adjacent GATA molecule in gray. R398 (cyan), T354 (magenta), and zinc ion (yellow) shown in space filling representation. (B) Wild-type R398 shown inserting into minor groove and possible H bonds with DNA bases. (C) R398W mutation modeled. (D) Wild-type T354. (E) T354M mutation modeled showing increased side chain size.
Mutations in GATA zinc finger domain. GATA zinc finger domains bind DNA in 2 conformations the adjacent and opposite forms. Here, we use the adjacent structure (3DFV).10 (A) Illustrated view of single GATA zinc finger (green) bound to DNA with adjacent GATA molecule in gray. R398 (cyan), T354 (magenta), and zinc ion (yellow) shown in space filling representation. (B) Wild-type R398 shown inserting into minor groove and possible H bonds with DNA bases. (C) R398W mutation modeled. (D) Wild-type T354. (E) T354M mutation modeled showing increased side chain size.
Investigation of the stem cell compartment of patients with DCML deficiency highlighted the absence of multi-lymphoid progenitors and decreased granulocyte-macrophage progenitors in the face of highly elevated flt-3 ligand.1,15 This situation is in marked contrast to all other DC deficiency states, including IRF8 mutation in humans, which are associated with a myeloproliferative response.16 The lack of myeloproliferation in DCML deficiency is in keeping with an intrinsic stem cell defect, as might be predicted to occur with GATA2 mutation. Further work will be required to elucidate the preferential drop-out of mononuclear cell precursors that gives rise to immunodeficiency early in the disease.1,2 In mice with haploinsufficiency of Gata-2 because of heterozygous truncation of the C-terminal zinc finger domain, the region primarily affected in the persons described, specific defects of the granulocyte/macrophage progenitor pool have been reported.17 Intriguingly, there is also selective loss of monocytes and lymphoid cells but preservation of granulopoiesis when Gata2 heterozygous stem cells are compared with wild-type stem cells in competitive transplantation assays.13 These results indicate that haploinsufficiency of GATA-2 modifies hematopoietic differentiation and could be sufficient to cause autosomal-dominant DCML deficiency in humans. Numerous downstream targets of GATA-2 are involved in terminal hematopoietic differentiation. Of note, GATA-2 interacts with the myeloid transcription factor PU.1,18 which is required to drive monocyte colony-stimulating factor receptor and Flt-3 expression, both essential growth factor receptors in the development of DCs and monocytes.19 We previously found a marked reduction in Flt-3 expression by CD34+ cells in DCML deficiency.1 The regulation of phagocytosis by GATA-2 may also explain loss of function in alveolar macrophages, leading to pulmonary alveolar proteinosis in some patients.20
Mutations of GATA-2 also cause familial myelodysplastic syndrome/acute myeloid leukemia.14 Although DCML deficiency can exist for many years without tri-lineage dysplasia, the evolution of progressive pancytopenia, and acquired cytogenetic abnormalities, including monosomy 7 and trisomy 8, have been highlighted in a recent study.3 This provides a unified view of GATA-2 mutation as a cause of both immunodeficiency and myeloid malignancy through its unique function in maintaining both mononuclear cell differentiation and stem cell homeostasis.
Note added in proof: Coincident with submission of our paper, Holland's group published a paper reporting the association of GATA2 mutations with the MonoMac syndrome.21
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Steven Holland for giving access to unpublished data relating to his laboratory's study on GATA-2 mutation,21 which was received while this manuscript was in preparation.
This work was supported by Leukaemia and Lymphoma Research, UK (R.E.D. and M.C.); Medical Research Council, United Kingdom (V.B., P.F.C., and S.H.); the Wellcome Trust (M.H. and P.F.C.); British Heart Foundation (B.K.); and by the National Institute of Health Research Biomedical Research Center at Newcastle Hospitals NHS Foundation Trust, The Newcastle Health Care Charity, and the Newcastle on Tyne Hospitals NHS Charity.
Wellcome Trust
Authorship
Contribution: R.E.D. performed sequencing, analyzed data, and wrote the manuscript; H.G. designed and performed bioinformatic analysis and wrote the manuscript; V.B. identified the phenotype of subjects, designed the study, and wrote the manuscript; L.N.R. processed samples and commented on the manuscript; R.H. performed sequencing and commented on the manuscript; M.H. identified the phenotype of patients and commented on the manuscript; J.H.L. performed structural modeling; T.R. performed sequencing and wrote the manuscript; X.-N.W., N.Mc.G., and D.Mc.D. processed samples; S.P. and S.C. provided technical assistance; I.C., J.W., A.C., and S.H. identified patients; M.W., B.K., P.F.C., J.L., and S.H. analyzed data and commented on manuscript; M.S.-K. performed bioinformatic analysis and commented on manuscript; and M.C. designed the study, analyzed data, and wrote the manuscript
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew Collin, Institute of Cellular Medicine, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, United Kingdom; e-mail: matthew.collin@ncl.ac.uk.