Abstract
We sequenced 120 candidate genes in 187 high-risk childhood B-precursor acute lymphoblastic leukemias, the largest pediatric cancer genome sequencing effort reported to date. Integrated analysis of 179 validated somatic sequence mutations with genome-wide copy number alterations and gene expression profiles revealed a high frequency of recurrent somatic alterations in key signaling pathways, including B-cell development/differentiation (68% of cases), the TP53/RB tumor suppressor pathway (54%), Ras signaling (50%), and Janus kinases (11%). Recurrent mutations were also found in ETV6 (6 cases), TBL1XR1 (3), CREBBP (3), MUC4 (2), ASMTL (2), and ADARB2 (2). The frequency of mutations within the 4 major pathways varied markedly across genetic subtypes. Among 23 leukemias expressing a BCR-ABL1-like gene expression profile, 96% had somatic alterations in B-cell development/differentiation, 57% in JAK, and 52% in both pathways, whereas only 9% had Ras pathway mutations. In contrast, 21 cases defined by a distinct gene expression profile coupled with focal ERG deletion rarely had B-cell development/differentiation or JAK kinase alterations but had a high frequency (62%) of Ras signaling pathway mutations. These data extend the range of genes that are recurrently mutated in high-risk childhood B-precursor acute lymphoblastic leukemia and highlight important new therapeutic targets for selected patient subsets.
Introduction
Despite the overall favorable treatment outcome for childhood acute lymphoblastic leukemia (ALL), up to 20% of patients still experience relapse, and survival after relapse is poor.1-3 Accurately identifying those patients with a high probability of relapse at the time of diagnosis and treating them with novel therapies is a rational strategy to improve their outcome, as has been shown for children with Philadelphia chromosome-positive ALL.4 As part of the Children's Oncology Group high-risk B-precursor acute lymphoblastic leukemia (HR B-ALL) TARGET (Therapeutically Applicable Research to Generate Effective Treatments) Project (http://target.cancer.gov), we have used genome-wide profiling of DNA copy number alterations (CNAs), unsupervised analysis of gene-expression profiling data, and candidate gene resequencing of PAX5, IKZF1, and the Janus kinase genes JAK1, JAK2, JAK3, and TYK2 to identify subgroups of childhood ALL with different probabilities of relapse.5-8 Somatic sequence mutations or deletions of IKZF1, which encodes the lymphoid transcription factor IKAROS, are found to be associated with a high risk of relapse in B-cell progenitor ALL.5,9 IKZF1-mutated HR B-ALL cases commonly have a BCR-ABL1-like gene expression signature,5,10 and up to one-half of these cases harbor mutations in Janus kinases (primarily JAK2, but also JAK1 and JAK3) and genomic rearrangements that resulted in high level expression of the CRLF2 cytokine receptor.7,11-13 Collectively, these efforts led to the identification of a new prognostic marker (IKZF1 mutations) and provided a rational therapeutic target (constitutively activated Janus kinases) in a subset of patients.
To examine the genetic basis of HR B-ALL in more detail, we have now sequenced 120 candidate cancer genes in diagnostic leukemia specimens from 187 children and adolescents with HR B-ALL treated with augmented postinduction chemotherapy on the Children's Oncology Group P9906 protocol.14-16 The candidate genes were selected to include genes and pathways targeted by CNAs in ALL,5,17 known targets of sequence mutation in ALL, tyrosine kinase genes, genes commonly mutated in other cancers, and genes exhibiting differential expression across 8 subgroups of HR B-ALL that are defined by unique gene expression signatures.8 Mutations were validated in both tumor and matching remission samples. An integrated analysis of validated somatic sequence mutations and CNAs was then performed to define recurrent alterations in biologic pathways. These analyses revealed a high frequency of recurrent somatic mutations in key signaling pathways, including B-cell development/differentiation, the TP53/RB1 tumor suppressor pathway, Ras signaling, and Janus kinases. Importantly, the frequency of mutations within these 4 pathways varied markedly across the HR-B-ALL subtypes identified by unsupervised analysis of gene expression profiles. These results provide important new insights into the spectrum of genetic alterations in HR B-ALL.
Methods
Patients and samples
The cohort was composed of 187 of 267 eligible patients with B-cell progenitor ALL treated on the Children's Oncology Group P9906 study.16 With the exception of presentation white blood cell count, the subset of patients used for genomic profiling studies is representative of the overall cohort, as previously reported.8 This trial included a subset of patients with high-risk ALL by National Cancer Institute/Rome criteria (age at diagnosis at least 10 years or initial white blood cell count at least 50 000/μL) who were at high risk for treatment failure based on the presence of central nervous system or testicular leukemia, MLL gene rearrangement, or the Shuster integrated age, sex, and leukocyte count risk stratification algorithm.15 Patients with BCR-ABL1–positive and hypodiploid ALL, infants less than one year of age, and patients who did not achieve remission after 4-6 weeks of induction chemotherapy were not eligible for this study and were excluded. Patients with favorable cytogenetics (ETV6-RUNX1 or trisomy of chromosomes 4 and 10) were ineligible unless they had central nervous system or testicular leukemia at diagnosis. All patients were treated identically with a modified augmented Berlin-Frankfurt-Münster regimen.16 Outcome data used for these analyses were frozen in September 2009. More detailed information on this patient cohort is available at http://target.cancer.gov. A list of patients, samples, gene expression profile cluster (Recognition of Outliers by Sampling Ends [ROSE]8 ) subgroups, DNA CNAs, and somatic sequence alterations is provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The patients enrolled in the P9906 study and/or their parent(s)/guardian(s) provided informed consent for clinical trial participation, specimen banking, and future research using forms approved by the institutional review boards of Children's Oncology Group member institutions. The current analyses were approved by the institutional review boards of the University of New Mexico and St Jude Children's Research Hospital.
Sequencing and analysis
We selected 120 genes for resequencing of all exons, 1 kb upstream of the translation start site, and flanking splice site junctions by polymerase chain reaction and capillary sequencing using whole genome amplified (RepliG, QIAGEN) DNA from the diagnostic leukemia samples. We have previously observed complete concordance between mutations identified by polymerase chain reaction and Sanger sequencing of WGA material with confirmatory studies performed using unamplified DNA.5,6,17,18 A list of genes sequenced and sequence coverage is provided in supplemental Table 2. Initial sequencing and subsequent validation were performed by Beckman Coulter Genomics. Base calls and quality scores were determined using the program PHRED.19 Sequence variations, including substitutions and insertion/deletions (indel), were analyzed using the SNPdetector20 and the IndelDetector21 software. A useable read was required to have at least one 30-bp window in which 90% of the bases have PHRED quality score of at least 30. Poor quality reads were filtered before variation detection. The minimum threshold of secondary to primary peak ratio for substitution and indel detection was set to be 20% and 10%, respectively. All sequence variations were annotated using a previously developed variation annotation pipeline. Any variation that did not match a known polymorphism (defined as a dbSNP record that does not belong to OMIM SNP or COSMIC22 somatic variation database or germline variations identified by Mullighan et al18 ) and resulted in a nonsilent amino acid change was considered a putative sequence mutation. All putative sequence mutations were confirmed by repeat genomic PCR and sequencing of both tumor and remission DNA, respectively. All traces have been submitted to National Center for Biotechnology Information's trace archive; the linking table is available from the TARGET Data Coordinating Center (http://target.cancer.gov/dataportal). Sequence coverage (supplemental Table 2) refers to the percentage of the targeted bases that are of sufficient quality to be used for single nucleotide polymorphism (SNP) detection by SNPdetector and IndelDetector.20
All data generated by the TARGET project (microarray gene expression, DNA CNA, and sequence data) are made available to the research community through the Website http://target.cancer.gov/dataportal as well as the Cancer Genome Workbench (http://cgwb.nci.nih.gov).
Analysis of BMR
Background mutation rate (BMR) was estimated by calculating the mutation rate of genes that have zero or one nonsynonymous mutation. We took this approach as (1) only nonsynonymous mutations were validated because of cost considerations; and (2) assuming that the majority of singleton mutations discovered in the 187 samples were unlikely to represent driver mutations. Genes that met such criteria were considered unselected genes. The mutation rate was calculated as: (number of mutations in unselected genes)/(sum of the covered bases in the coding region of unselected genes) divided by two-thirds (because the majority of the mutations in the third base of a codon will result in silent mutation).
We reanalyzed TCGA data to evaluate how the results derived from this approach compared with those using silent mutation in the coding regions. Sequence traces generated by the TCGA glioblastoma multiforme sequencing project23 were downloaded from the National Center for Biotechnology Information trace archive and coverage for the 601 target genes across the 91 samples selected for sequencing were computed. Only the validated somatic mutations were used in the calculation.
Statistical analyses
Relapse-free survival was defined as time from achieving complete remission to relapse for those patients with events. Patients who experienced other first events (second malignant neoplasm, remission death) were censored as of the date of event. Patients who did not experience any events were censored as of date of last contact.
The χ2 test was used to analyze the distribution of mutations among different groups. Association of over- or under-representation of mutations in different pathways was determined by comparing each group versus the rest using the Fisher exact test followed by multiple comparison adjustment using the Benjamini-Hochberg method.24 All statistical analyses were performed using R (http://www.R-project.org, Version 2.11.1, with basic and survival packages).
The integrative heatmap view of the somatic mutations and CNAs was generated with the R package heatmap.2. Samples in each ROSE group12 were clustered using the Ward method to illustrate the similarity of mutations and copy number variations across samples.
Results
Frequency of gene mutations in HR B-ALL
For the 120 candidate genes, a total of 2196 amplicons were sequenced bidirectionally generating 50.5 Mb of nonredundant high-quality sequence, covering 92% of the targeted nucleotides (supplemental Figure 1; supplemental Table 2). A total of 147 625 putative variants, including single nucleotide variations and small insertion/deletion variations (indels), were identified by computational analysis of the sequence chromatograms. After removal of noncoding or silent DNA variations, inherited variations present in dbSNP,25 TCGA,23 and inherited variants identified from our recent study sequencing 300 genes in matched diagnosis, relapse, and remission ALL samples,18 680 novel nonsilent variants remained. These variants were resequenced in leukemia and paired remission DNA to verify the alternative alleles and determine whether they were somatic or present in the germline. Of the 680 putative novel variants, 179 were identified to be somatic mutations (supplemental Table 3), 395 were present in the paired germline samples, 64 failed the validation assay (mutant alleles were verified in 53 sites, but the normal samples failed in the assay despite repeated attempts; supplemental Table 4), and 42 were false positives as the alternative allele was not confirmed in the tumor on resequencing.
Recurrent mutations in key signaling pathways
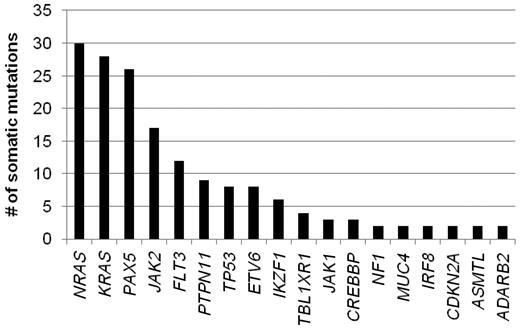
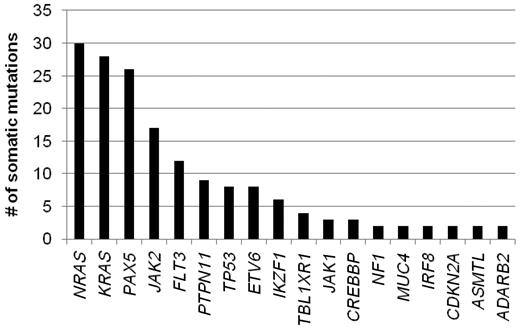
The remainder of our analysis concentrated on the 179 validated somatic nonsynonymous sequence mutations. These mutations occurred in 31 genes, 19 of which were recurrently mutated in this patient cohort (identified in at least 2 patients; Figure 1). The most frequently mutated genes include NRAS, KRAS, PAX5, and JAK2, with each being mutated in at least 10% of the leukemias. The absence of mutation in a number of the sequenced genes is also notable, including 13 of 17 analyzed tyrosine kinase genes known to be expressed in lymphoid cells and PTEN.
Eleven of the 19 recurrently mutated genes can be classified into 4 known cancer signaling pathways: B-cell development/differentiation, Ras signaling, JAK/STAT signaling, and the TP53/RB1 tumor suppressor pathway. Combining the identified sequence mutations with CNAs detected using Affymetrix, Version 6.0 SNP arrays5 revealed 68% of the cases to have somatic alterations (mutations and/or CNAs) in genes regulating B-cell development/differentiation, 54% had mutations in the TP53/RB1 tumor suppressor pathway, 54% in Ras signaling, and 11% in Janus kinases (Figure 2). The frequency of TP53 mutations was low (∼ 4%), and the majority of alterations in the TP53/RB1 pathway involved deletions of CDKN2A/CDKN2B. With the exception of the B-cell development/differentiation pathway, the frequency of alterations in the TP53/RB1, Ras, and JAK signaling pathways is much higher in this pediatric HR B-ALL cohort than reported for unselected pediatric B-precursor ALL patients.12,26,27 Nevertheless, the overall frequency of somatic alterations in individual leukemias in this study remains low, with an average of only one sequence mutation (range, 0-4) and 8.36 CNAs per leukemia (range, 0-86).5
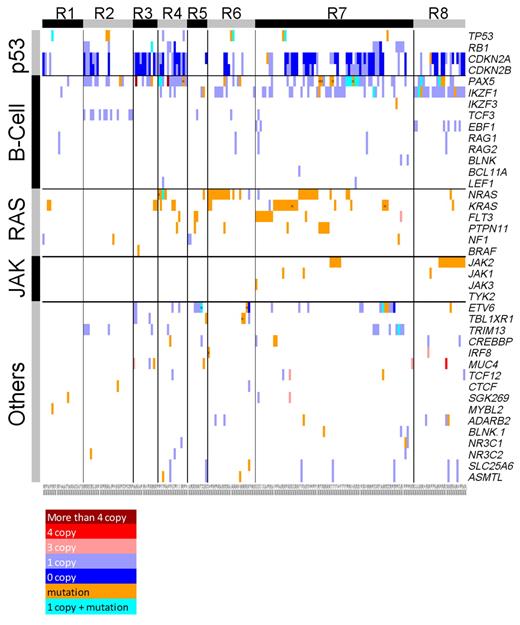
An integrated view of somatic CNA and sequence mutation in genes with recurrent sequence mutations across the 187 patients. Samples are displayed in columns and grouped by their corresponding ROSE clusters.12 Genes are shown in rows grouped by their corresponding pathways. Somatic sequence mutations are marked in orange unless they occur with a somatic deletion (which is shown in cyan). Multiple mutations within a single gene are labeled “x.” Somatic deletions and amplifications are shown in blue and red, and the shades indicate the level of CNAs
An integrated view of somatic CNA and sequence mutation in genes with recurrent sequence mutations across the 187 patients. Samples are displayed in columns and grouped by their corresponding ROSE clusters.12 Genes are shown in rows grouped by their corresponding pathways. Somatic sequence mutations are marked in orange unless they occur with a somatic deletion (which is shown in cyan). Multiple mutations within a single gene are labeled “x.” Somatic deletions and amplifications are shown in blue and red, and the shades indicate the level of CNAs
Although a formal assessment of the intrinsic mutation rate within individual leukemia cases cannot be calculated from our data, an estimate of the average BMR for this cohort can be made. To accomplish this, we limited our analysis to validated mutations (which by design were only nonsynonymous mutations) and removed all genes with recurrent nonsynonymous mutations because these probably represent functionally selected driver mutations. Using the remaining data, we calculated an average mutation rate as the number of identified nonsynonymous mutations divided by the number of sequenced bases within these genes, and then adjusted the resultant number to correct for the degeneracy of the amino acid code in the third codon position. When this approach was applied to a published dataset of 91 adult glioblastoma samples,23 we estimated a BMR of 3.62 × 10−6 per base, which is comparable to the published BMR for this patient cohort of 3.8 × 10−6. Applying this method to our patient cohort resulted in a BMR of 4.6 × 10−7 per base, which is almost 10 times lower than that of adult glioblastoma samples. Interestingly, recent data have shown that the BMR for another childhood tumor, medulloblastoma, is 5-10 times lower than adult solid tumors.28
Despite the low background mutational rate, multiple genes within the 4 individual signaling pathways were mutated in a subset of cases. Within the B-cell development/differentiation pathway, although the majority of the mutations were monoallelic, 26% (33 of 127) of the leukemias with alterations in this pathway had 2 or more pathway genes mutated. Similarly, monoallelic and biallelic mutations in multiple genes within the TP53/RB1 pathway were detected in 16% (16 of 101) of the leukemias containing alterations in this pathway. Even more surprising was the presence of multiple sequence mutations in 2 genes within the Ras signaling pathway in 14% (10 of 73) of the leukemias with alterations in this pathway. This included examples of leukemia with mutation in both NRAS and KRAS, FLT3 and NRAS, FLT3 and PTPN11, and PTPN11 and either NRAS or KRAS (Table 1). In addition, one case (9906_215) contained both KRAS G12D and G13D mutations, whereas another contained both NRAS G12V and G13D mutations (sample 9906_106, supplemental Figure 2), although whether these exist within a single allele or occur on different alleles could not be determined from our data. Collectively, these data suggest a strong selection for mutations within these specific signaling pathways.
Variation in frequency of pathway-associated gene mutations across known ALL subtypes
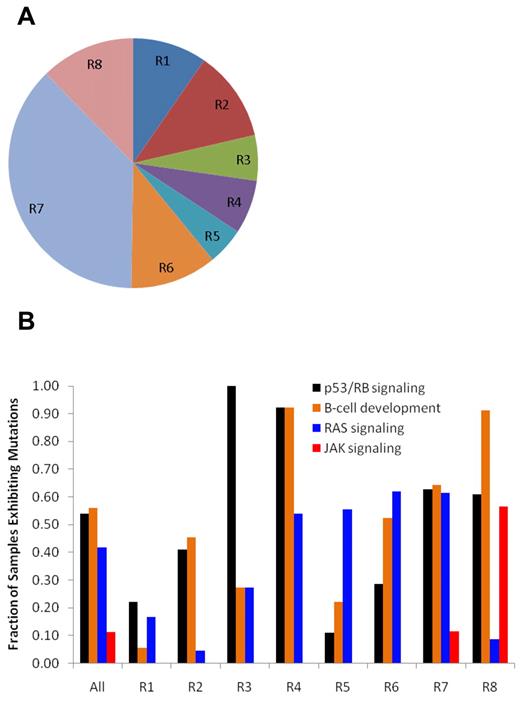
We recently reported 8 distinct subtypes of HR B-ALL based on unsupervised analysis of gene expression profiling data (ROSE clusters R1-R8; Figure 3A).8 A striking finding was a marked difference in the frequency of mutations within the 4 major pathways across these leukemia subgroups (Figure 3B; Table 2). Two of these expression-based leukemia subtypes are characterized by known specific sentinel chromosome translocations: MLL translocations in ROSE cluster 1 (R1) and the t(1;19)(q23;p13.3) that produces TCF3-PBX1 fusion in R2. Similar to the rare occurrence of CNAs in ALL cases with MLL translocations,17 the R1 leukemias have very few identified somatic sequence mutations. The majority of ALL cases in R2 had focal deletions of TCF3 occurring adjacent to the breakpoints in the translocation-encoded TCF3-PBX1 chimeric gene. The R3 and R4 leukemias have a near-obligate mutation in the TP53/RB1 signaling pathway (100% in R3 and 92% in R4) but differ markedly in their frequency of B-cell pathway mutations with 92% of R4 leukemias containing mutations and only 45% of R3 cases. These cases do not have known chromosomal rearrangements. The most notable feature of R5 leukemias is a high frequency of Ras pathway mutations (56%) and/or ETV6 mutations (33%). The R6 subgroup is characterized by a high frequency of RAS mutations (62%) coupled with ERG deletions. The R7 leukemias, which compose the largest and most heterogeneous subgroup, are characterized by a high frequency of mutations in the TP53/RB1 (63%), B-cell development/differentiation (74%), and Ras (61%). Lastly, the R8 subgroup, which is known to be associated with a very poor treatment outcome,8 has a high frequency of mutations in TP53/RB1 (61%), a near-obligate mutation in B-cell development/differentiation (96%), and a remarkably high frequency of JAK pathway mutations (57%) but lower a lower rate of Ras mutations (9%) compared with all other subgroups.
Distribution of cases and mutations according to ROSE clustering subgroups. (A) Distribution of the 8 patient subgroups identified by unsupervised clustering of gene expression profiles using the ROSE method.12 The subgroups, labeled contiguously from R1 to R8, include 2 subgroups associated with known recurrent cytogenetic abnormalities: R1 with translocations of MLL and R2 with TCF3-PBX1 fusion, 1 subgroup that lacks sentinel abnormality (R7), 1 subgroup with good outcome (R6), and 1 subgroup with poor outcome (R8).12 (B) Somatic alteration profiles incorporating both sequence mutations and CNAs in TP53/RB1 signaling, B-cell development, Ras signaling, and JAK signaling pathways. The frequency of patients exhibiting mutations in these 4 pathways among the 8 ROSE gene-expression subgroups differs significantly from the null hypothesis (ie, that the mutations are randomly distributed across all 8 subgroups) with a P value of 9.38 × 10−7, 3.38 × 10−8, 1.53 × 10−7, and 2.16 × 10−10 for TP53/RB1 signaling, B-cell development, Ras signaling, and JAK signaling, respectively by χ2 test.
Distribution of cases and mutations according to ROSE clustering subgroups. (A) Distribution of the 8 patient subgroups identified by unsupervised clustering of gene expression profiles using the ROSE method.12 The subgroups, labeled contiguously from R1 to R8, include 2 subgroups associated with known recurrent cytogenetic abnormalities: R1 with translocations of MLL and R2 with TCF3-PBX1 fusion, 1 subgroup that lacks sentinel abnormality (R7), 1 subgroup with good outcome (R6), and 1 subgroup with poor outcome (R8).12 (B) Somatic alteration profiles incorporating both sequence mutations and CNAs in TP53/RB1 signaling, B-cell development, Ras signaling, and JAK signaling pathways. The frequency of patients exhibiting mutations in these 4 pathways among the 8 ROSE gene-expression subgroups differs significantly from the null hypothesis (ie, that the mutations are randomly distributed across all 8 subgroups) with a P value of 9.38 × 10−7, 3.38 × 10−8, 1.53 × 10−7, and 2.16 × 10−10 for TP53/RB1 signaling, B-cell development, Ras signaling, and JAK signaling, respectively by χ2 test.
Frequent inactivating mutations in other noncanonical pathway genes
In addition to mutations in genes involved in the 4 major pathways, recurrent point mutations were also detected in ETV6 (n = 6), TBL1XR1 (n = 3), CREBBP (n = 3), MUC4 (n = 2), ASMTL (n = 2), and ADARB2 (n = 2). A total of 8 ETV6 sequence mutations were detected in 6 cases, 7 of which were frameshift mutations predicted to generate a truncated protein (Figure 4A). When ETV6 sequence mutations were combined with somatic copy number deletions, 12% of the leukemias had loss-of-function mutations in this gene, with 18 of 23 being monoallelic and 5 of 23 biallelic (Figure 4B). Loss-of-function mutations of the nonrearranged ETV6 allele are a frequent secondary event in ALLs that contain a t(12;21)–encoded ETV6-RUNX1 chimeric gene.17 The high frequency of ETV6 inactivating mutations in our cohort was surprising in that only one ETV6-RUNX1 containing ALL was present in the analyzed cohort, and it lacked an alteration in the nonrearranged ETV6 allele.
Somatic mutations in ETV6. (A) Distribution of somatic sequence mutations in ETV6. The N-terminal pointed (PNT) is involved in protein-protein interactions with itself and other proteins, whereas the C-terminal ETS domain (red) is involved in DNA binding. (B) Single and double mutations in ETV6. The genomic region in display is chromosome 12:11 619 584–12 112 117 (genome build hg18). Each row represents a sample with somatic alterations. Gray lines indicate deletions; and black lines, homozygous deletions. Sequence mutations are shown as vertical black boxes.
Somatic mutations in ETV6. (A) Distribution of somatic sequence mutations in ETV6. The N-terminal pointed (PNT) is involved in protein-protein interactions with itself and other proteins, whereas the C-terminal ETS domain (red) is involved in DNA binding. (B) Single and double mutations in ETV6. The genomic region in display is chromosome 12:11 619 584–12 112 117 (genome build hg18). Each row represents a sample with somatic alterations. Gray lines indicate deletions; and black lines, homozygous deletions. Sequence mutations are shown as vertical black boxes.
Transducin β-like protein 1 (TBL1)–related protein TBL1XR1 serves as a specific adaptor to mediate an exchange of the nuclear receptor corepressors, SMRT–N-CoR, for coactivators on ligand binding. This activity is mediated through an E3 ubiquitin ligase activity of TBL1XR1 that recruits the ubiquitin/proteasome machinery to the nuclear receptor to degrade the corepressors. More recently, TBL1-TBL1XR1 was also identified as a key player in WNT signaling.29 Interestingly, the TBL1XR1 mutations detected in our patient cohort are predicted to be monoallelic loss-of-function mutations, suggesting that reduction in TBL1XL1 activity is probably mechanistically involved in leukemogenesis. CREBBP is a transcriptional coactivator and acetylator of histone and nonhistone targets that is frequently mutated in relapsed ALL and non-Hodgkin lymphoma.18,30 Of the 3 missense mutations discovered in this study, one (R1446C) is at the critical histone acetyltransferase domain and is located at a residue previously identified as a target of mutation in lymphoid malignancies.18,30
Additional novel mutations in B-ALL
A number of novel mutations were identified in genes with putative roles in leukemogenesis and tumor formation, epigenetic modification, and kinase signaling. These included mutations in genes involved in lymphoid development and signaling, including BLNK (encoding B-cell linker, an adapter protein required for B-cell signaling); IKZF3 (encoding the lymphoid transcription factor AIOLOS); JAK3 (Janus kinase 3), which is known to be mutated in megakaryoblastic leukemia,31 but not B-lineage ALL; and mutations in the glucocorticoid and mineralocorticoid receptor genes NR3C1 and NR3C2 (Table 3).
Discussion
A total of 179 nonsynonymous validated somatic mutations were identified by sequencing 120 candidate genes in 187 high-risk ALL patients, 81% of which occur in genes involved in TP53/RB1 signaling pathway, B-cell development pathway, Ras signaling, and JAK/STAT signaling pathways. Both DNA CNA5 and sequence variant data were used to identify patients with somatic alterations in these pathways. In addition, we evaluated somatic alteration profiles among the 8 patient subgroups identified by unsupervised analysis of gene expression profiles in this patient cohort.8 The somatic alteration profiles of the 4 pathways are distinct among the 8 gene-expression subgroups, indicating differential gene expression patterns may be driven by underlying differences in the spectrum of mutations that occur in a given case of leukemia. Specifically, cases in the R8 cluster, which commonly have a gene-expression profile similar to that of BCR-ABL1 ALL and poor outcome, have a high frequency of mutations targeting B-cell development and JAK signaling. These cases have a high frequency of IKZF1 alteration (supplemental Figure 3), Janus kinase mutations, and CRLF2 rearrangement resulting in overexpression of this cytokine receptor as previously described.6,7,12 Interestingly, this subgroup also has a low frequency of Ras pathway mutation. Four additional subgroups have significant under- or over-representation of mutations in at least one pathway. Because only 187 cases were analyzed in this study, it is possible that some of the associations that are not statistically significant might be real and clinically significant if a larger number of cases were analyzed. Because we did not sequence every gene involved in the 4 pathways, these results provide a minimum estimate of the pathway mutations rates in HR B-ALL. It is possible that some somatic mutations were missed, as there were an additional 53 sequence variations that could not be resolved in that the mutant alleles were verified in the diagnostic leukemia specimen, but sequence of the normal sample could not be determined definitively despite multiple attempts (supplemental Table 3).
Somatic sequence mutations are rare in leukemias with MLL translocations (R1) or TCF3-PBX1 fusion (R2), with only 5 mutations identified in 17 cases, and 5 mutations in 22 cases, respectively. Mutations activating FLT3 and Ras signaling have been previously described in MLL-rearranged leukemia,32,33 but the paucity of sequence mutations and somatic CNAs targeting other genes in MLL-rearranged ALL suggests that few additional mutations are needed to cause leukemia, or that other mechanisms of change may be present in these cases, such as changes in gene expression driven by epigenetic changes.34 In contrast, 50% of the cases in the R2 have somatic CNAs in TP53/RB1 pathway and B-cell development pathway.
One surprising finding in this study is the lack of mutual exclusivity in sequence mutations of the Ras signaling pathway: Five samples have multiple mutations at NRAS or KRAS G12/G13 (Table 1). Given that the estimated BMR in this cohort is almost 10 times lower than that of the adult glioblastoma in which mutual exclusivity of mutations in signaling pathways was reported, we speculate that clonal heterogeneity could be a plausible cause for this observation, as has been recently described in ALL.18,35-38 The presence or number of mutations in the Ras signaling pathway were not associated with relapse-free survival (supplemental Figure 4). By contrast, in B-cell development pathway, the number of somatic alterations was associated with progressively worse outcome.5 Ras pathway mutations have been previously described in ALL,39-43 but the observed 50% mutation frequency in this pathway is higher than in prior reports and contrasts sharply with known genetic alterations in T-cell ALL where Ras pathway alterations are relatively uncommon (< 10% of cases).44 In addition, PTEN somatic mutations and deletions occur in ∼ 25% to 35% of cases of T-cell ALL,45 but no PTEN alterations were found in this cohort, suggesting differential activation of Ras and PI3K signaling pathways in T-cell and HR B-precursor ALL.
Because patients with ETV6-RUNX1 have a favorable outcome, they were excluded from the P9906 clinical trial unless they had central nervous system or testicular leukemia. Hence, only 3 patients with ETV6-RUNX1 fusion were included in P9906, only one of which was included among the 187 patients in this cohort. We identified 21 cases with ETV6 somatic alteration, including 5 cases with double-hits. Almost all sequence mutations and > 50% of the somatic CNAs are truncation mutations expected to cause premature stop of the ETV6 protein. The ETV6 mutation pattern and frequency resemble that previously described for AML.46 In addition, the same study also reported absence of ETV6 protein in AML, which is consistent with the double-hit mutations found in our data. It is possible that, similar to AML, ETV6 may function as a tumor suppressor gene in ETV6-RUNX1-negative ALL.
TBL1XR1 is involved in transcription regulation through the recruitment of the ubiquitin/19S proteasome complex to nuclear receptor-regulated transcription units. TBL1XR1 has been noted to be focally deleted in pediatric ALL,17 with deletions being most common in ETV6-RUNX1 ALL.17,47 However, this is the first report of sequence mutations in TBL1XR1 in leukemia patients. The histone acetyltransferase CREBBP is involved in acute myeloid leukemia through translocations involving multiple fusion partners (eg, MLL, MOZ, and MORF).48,49 Most recently, recurrent mutations in CREBBP have been discovered in 21% of the relapsed ALL cases with a hotspot in the histone acetyltransferase domain.18 In this study, 3 somatic mutations were found in CREBBP, one of which (R1446C) changes the residue R1446 in the histone acetyltransferase domain. The same amino acid has also been somatically altered (R1446H) in relapsed ALL.
This is the first reported large-scale sequence analysis of somatic sequence mutations in newly diagnosed high-risk pediatric ALL. The availability of somatic CNAs and gene expression data from previous studies allows us to assemble a comprehensive view of the somatic alteration profiles to identify pathways with recurrent mutations and to assess genetic heterogeneity in this cohort. Because we did not sequence every gene involved in the 4 pathways, and as some mutations could not be confirmed as somatic due failure of validation assays in matched normal material l (supplemental Table 4), these results provide a minimum estimate of the pathway mutations rates in HR B-ALL. Moreover, as we sequenced only a small fraction of the coding genome, it is almost certain that there are other genes and pathways that are commonly mutated in pediatric ALL. These limitations notwithstanding, our data extend prior knowledge of the genetic basis of HR B-ALL and suggest potential new therapeutic approaches, such as targeting of the Ras/MAPK signaling pathway.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Jim Jacobson for his role in the genesis of this and other cancer genomics projects. Dr Jacobson passed away from complications of acute leukemia during the course of this project.
This work was supported by the Children's Oncology Group (Chair's Award CA098543, G.H.R.; and a supplement to that grant to support the TARGET initiative); National Cancer Institute Strategic Partnering to Evaluate Cancer Signatures (Program Award CA114762, W.L.C., I.-M.C., R.C.H., and C.L.W.); National Institutes of Health Cancer Center (Support Grant CA21765, J.R.D. and C.G.M.; and Support Grant CA118100, R.C.H., I.-M.C., and C.L.W.), which also provided support for critical Shared Resources at St Jude Children's Research Hospital and University of New Mexico Cancer Center; National Cancer Institute (grant U10 CA98413, supporting the Statistical Center, G.H.R.; grant U24 CA114766, Human Specimen Banking in CNI Supported Cancer Trials); Leukemia & Lymphoma Society Specialized Center of Research (grant 7388-06, C.L.W.); CureSearch; St Baldrick's Foundation (M.L.L.); National Health and Medical Research Council (Australia; C.J. Martin Traveling Fellowship, C.G.M.); and the American Lebanese Syrian Associated Charities of St Jude Children's Research Hospital. C.G.M. is a Pew Scholar in the Biomedical Sciences. S.P.H. is the Ergen Family Chair in Pediatric Cancer. The sequencing was funded with federal funds from the National Cancer Institute, National Institutes of Health (contract N01-C0-12400).
National Institutes of Health
Authorship
Contribution: J.Z., C.G.M., G.W., X.C., M.E., and K.H.B. analyzed data; R.C.H. and I.-M.C. performed laboratory assays; M.D. performed statistical analyses; D.S.G. coordinated genomic profiling; W.L.C., M.L.L., G.H.R., M.V.R., B.M.C., W.P.B., and C.L.W. provided clinical samples and data; J.Z., C.G.M., R.C.H., W.L.C., D.S.G., M.L.L., G.H.R., M.V.R., M.A.S., C.L.W., J.R.D., and S.P.H. selected genes for sequencing; J.Z., C.G.M., R.C.H., D.S.G., M.L.L., C.L.W., J.R.D., and S.P.H. designed the study; and J.Z., J.R.D., and C.G.M. wrote the manuscript, with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James R. Downing, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN, 38105; e-mail: james.downing@stjude.org; Stephen P. Hunger, Center for Cancer and Blood Disorders, Children's Hospital Colorado, 13123 E 16th Ave B115, Aurora, CO 80045; e-mail: stephen.hunger@childrenscolorado.org; and Cheryl L. Willman, University of New Mexico Cancer Center, 1201 Camino de Salud NE, Rm 4630, MSC07-4025, 1 University of New Mexico, Albuquerque, NM 87131-0001; e-mail: cwillman@salud.unm.edu.
References
Author notes
J.Z. and C.G.M. contributed equally to this study.
C.L.W., J.R.D., and S.P.H. contributed equally to this study.