Abstract
Adult T-cell leukemia/lymphoma (ATLL) is an incurable disease where most patients succumb within the first year of diagnosis. Both standard chemotherapy regimens and mAbs directed against ATLL tumor markers do not alter this aggressive clinical course. Therapeutic development would be facilitated by the discovery of genes and pathways that drive or initiate ATLL, but so far amenable drug targets have not been forthcoming. Because the IL-2 signaling pathway plays a prominent role in ATLL pathogenesis, mutational analysis of pathway components should yield interesting results. In this study, we focused on JAK3, the nonreceptor tyrosine kinase that signals from the IL-2R, where activating mutations have been found in diverse neoplasms. We screened 36 ATLL patients and 24 ethnically matched controls and found 4 patients with mutations in JAK3. These somatic, missense mutations occurred in the N-terminal FERM (founding members: band 4.1, ezrin, radixin, and moesin) domain and induced gain of function in JAK3. Importantly, we show that these mutant JAK3s are inhibited with a specific kinase inhibitor already in human clinical testing. Our findings underscore the importance of this pathway in ATLL development and offer a therapeutic handle for this incurable cancer.
Introduction
ATLL is an aggressive T-cell neoplasm that is resistant to diverse therapeutic approaches. Because the disease is rare, small case series have been published testing cytotoxic chemotherapy, IFN therapy, nucleoside analogues, stem cell transplantation, and immune therapy, but no specific regimen induces durable remission.1 Cures have been reported after stem cell transplantation but the treatment-related morbidity and mortality are high.2 Unfortunately, newly diagnosed patients can only expect a median survival of < 12 months. Perhaps through better understanding of the unique biology and pathogenesis of ATLL we could generate novel therapeutic targets. Indeed, ATLL is a remarkable example of retrovirally induced cancer in humans.3 ATLL is caused by infection of T cells by human T-lymphotropic virus-1 (HTLV-1) which is endemic in southwestern Japan, the Caribbean islands, West Africa, and South America. HTLV-1 is a complex retrovirus that expresses multiple gene products that contribute to T-cell transformation; among these, the retroviral oncogene tax can induce T-cell immortalization on its own and is thought to be absolutely required for ATLL development.4-6 Interestingly, ATLL arises in only 5% of patients infected with HTLV-1 and disease presents up to 3 decades after initial infection. This suggests that there are probably multiple oncogenic mutations from the initiating hit of HTLV-1 infection to the development of ATLL. In fact, host somatic mutations in specific genes and pathways are probably required for ATLL and could explain the low penetrance and long latency.
One major pathway implicated in ATLL development is the activation of IL-2 (and related cytokines) signaling.3,7,8 In fact, the IL2 and IL2RA (α chain or CD25) genes are trans-activated by tax protein.9 The IL-2Rβ and IL-2Rγ chains are constitutively expressed on T cells but the high-affinity IL-2R requires the IL-2Rα chain.10 Thus, tax-expressing T cells are induced to proliferate because of autocrine signaling from IL-2 to the IL-2R, composed of α, β, γ chains; other HTLV-1 gene products may contribute to this same pathway.11 On ligand binding, the nonreceptor tyrosine kinases, JAK1 and JAK3, initiate phosphorylation of downstream substrates such as the STAT5 transcription factors, A and B, which in turn activate target genes that allow cell-cycle entry (ie, CCND2) and block apoptosis (ie, BCL2L1).12 JAK3 is constitutively bound to the IL-2Rγ (also γc) chain and is used by the receptors for IL-4, IL-7, IL-9, IL-15, and IL-21. Interestingly, IL-9 is up-regulated in some ATLL cells so cytokines other than IL-2 may also use JAK3/γc.13
Most remarkably, as ATLL progresses, tax gene expression is lost by methylation or deletion in a majority of patients.3,14 Despite this, many ATLL cell lines remain hypersensitive to γc-using cytokines, such as IL-2, and show constitutive activation of downstream substrates of JAK3 such as STAT5A and PI3K/AKT.7,15,16 Mutations in γc-restricted cytokine signal transduction components (eg, JAK3) could explain the activation of the pathway in the absence of tax transcriptional stimulus. The JAK family kinases are central players in normal and malignant hematopoiesis and are candidates for oncogenic somatic mutations. These nonreceptor tyrosine kinases are recruited for signal transduction by cytokine receptors that do not possess tyrosine kinase activity on their own.12 Activating mutations in JAK2 are clonal in a large number of myeloproliferative disorders (MPDs) and recently mutations in JAK1 and JAK3 have been described in acute myeloid and lymphoblastic leukemias.17-20
The JAK proteins are composed of FERM, SH2, pseudokinase and kinase domains (Figure 1A).12 The FERM (named after founding members band 4.1, ezrin, radixin, and moesin) is predominantly responsible for binding the cytoplasmic tails of cytokine receptor chains.21,22 The pseudokinase domain structurally resembles a kinase domain but does not function as one and is thought to have regulatory function.23 This domain is a hotspot for most of the mutations described in JAK1-3 in MPDs and leukemias. In this study, we analyzed the JAK3 gene in ATLL patients and found 4 ATLL patients with missense mutations in the N-terminal FERM domain. We show that these mutations cause gain of function in the kinase. Most importantly, we found the mutant JAK3s were inhibited by a specific JAK3 inhibitor, tofacitinib, which is already in human clinical testing for autoimmune disorders.24,25 Our studies propose JAK3 kinase as a therapeutic target in ATLL.
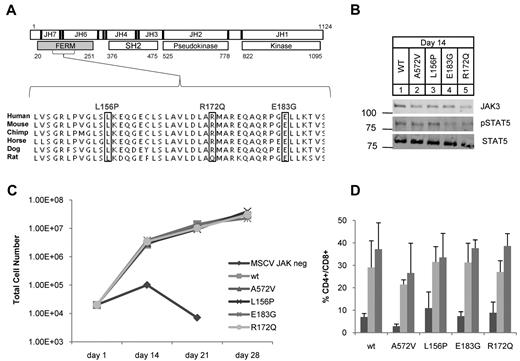
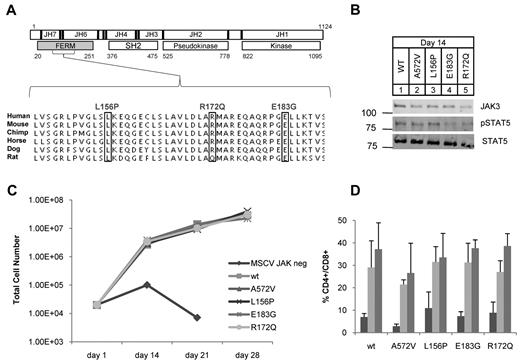
JAK3 FERM mutations identified in ATLL rescue T-cell differentiation in JAK3 knockout hematopoietic progenitors. (A) Schematic shows human JAK3 protein with 7 JAK homology (JH) domains; JH6-7 comprise the FERM (gray). Residue numbers and major structural domains are shown. Sequencing revealed 3 mutations (boxes) in 4 of 36 ATLL patients analyzed. L156P and R172Q were identified in 1 patient each and E183G was found in 2 patients. The mutations occurred in residues highly conserved across mammalian species. (B) Western blot for JAK3 (106 kDa), STAT5A (92 kDa), and phospho-STAT5A (Y694) from T-cell progenitor cell lysates grown on OP9-DL1 at day 14; no lysate was recovered from untransduced cells. (C) Graph shows cell counts over time in OP9-DL1 coculture. JAK3−/− progenitor cells transduced with empty MSCV failed to proliferate and differentiate. Proliferation was rescued for those progenitors transduced with WT or mutant JAK3. (D) Graph shows the percentage of double-positive (CD4+CD8+) cells arising in OP9-DL1 coculture as determined by flow cytometry; bars from left to right show counts at 14, 21, and 28 days. The OP9-DL1 data are representative of 2 independent experiments (mean ± SEM).
JAK3 FERM mutations identified in ATLL rescue T-cell differentiation in JAK3 knockout hematopoietic progenitors. (A) Schematic shows human JAK3 protein with 7 JAK homology (JH) domains; JH6-7 comprise the FERM (gray). Residue numbers and major structural domains are shown. Sequencing revealed 3 mutations (boxes) in 4 of 36 ATLL patients analyzed. L156P and R172Q were identified in 1 patient each and E183G was found in 2 patients. The mutations occurred in residues highly conserved across mammalian species. (B) Western blot for JAK3 (106 kDa), STAT5A (92 kDa), and phospho-STAT5A (Y694) from T-cell progenitor cell lysates grown on OP9-DL1 at day 14; no lysate was recovered from untransduced cells. (C) Graph shows cell counts over time in OP9-DL1 coculture. JAK3−/− progenitor cells transduced with empty MSCV failed to proliferate and differentiate. Proliferation was rescued for those progenitors transduced with WT or mutant JAK3. (D) Graph shows the percentage of double-positive (CD4+CD8+) cells arising in OP9-DL1 coculture as determined by flow cytometry; bars from left to right show counts at 14, 21, and 28 days. The OP9-DL1 data are representative of 2 independent experiments (mean ± SEM).
Methods
Patient samples, sequencing, and mutational analysis
Genomic DNA was isolated from 36 ATLL patients consented and treated at the NIH Clinical Center under an institutional review board (IRB)–approved protocol. Most of these patients were from Jamaica. Likewise, 24 ethnically matched (Jamaica or Haiti; see supplemental Table 5, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) control patients were consented and had their whole blood drawn for genomic DNA preparation under an IRB-approved protocol. The entire JAK3 gene was analyzed by PCR amplification of specific exons (supplemental Table 2) and analyzed for sequence alterations by SpectruMedix's Reveal Genetic Analysis System on samples where the leukemia cells were purified from the buffy coat. Standard Sanger DNA sequencing (GeneWiz and Vanderbilt Core Sequencing facility) was subsequently used to analyze all potential mutations as determined by Reveal Genetic Analysis System. Multiple independent PCR amplicons cloned into pGEM-TEasy (Promega) were used to analyze samples that did not have pure leukemic samples or in some instances had < 10% leukemic cells. Oligonucleotides used for PCR are available in supplemental Table 2. The proposed JAK3 FERM domain structure was determined by ClustalW alignment with the FAK FERM domain and analyzed with PyMol using FAK FERM coordinates. The crystal structure of the JAK3 kinase domain was available from the RCSB Protein Data Bank.26
Viruses and plasmid constructs
MSCV-ires-GFP (MIG) constructs expressing mutant JAK3 cDNAs were made by introducing mutations into the wild-type (WT) JAK3 using the Stratagene QuikChange kit (supplemental Table 3). The WT JAK3 and A572V JAK3 mutant constructs were provided by Dr Brian Druker (Oregon Health and Science University, Portland, OR) cloned into MIG. Site-directed mutagenesis was verified by full sequencing of the JAK3 cDNA insert. Virus was made by transient transfections of JAK3-MIG constructs into Phoenix cells by the CaPO4 method along with pCL-Eco. Viral titers were determined by flow cytometric analysis of 3T3 cells for GFP expression. pCMVSport6-IL2Rβ and pCMVSport6-STAT5A constructs were obtained from the Mammalian Gene Collection. IL2Rα was cloned into pcDNA 3.1(+; Invitrogen) from pINCY-IL2Rα (Open Biosystems) construct. IL-2Rγ was amplified from HuT-102 cDNA and cloned into pcDNA 3.1(+). All inserts were fully sequenced.
Flow cytometry and Western blots
Flow cytometry experiments were performed in the Vanderbilt Flow Cytometry Shared Resource core. Flow cytometry data acquisition was performed on an LSRII 3 laser (BD Biosciences) and analyzed with FACSDiva software. BaF3 cells transduced with various MIG-JAK3s were sorted for GFP on a FACS Aria (BD Biosciences). These cells were also stained with Vybrant Dyecycle violet (Invitrogen) and the proportion of cells in each cell cycle stage was determined by flow cytometry and analyzed using FlowJo software. Immunoprecipitations were performed in BaF3 cells transduced with MIG-JAK3 using anti-JAK3 (Santa Cruz Biotechnology Inc) and anti-γc Abs (R&D Biosystems). Detection of total and phosphorylated protein targets was performed using the LI-COR Odyssey Infrared Imaging System. Abs used were anti–phospho-STAT5 (Y694; BD Transduction Laboratories/BD Pharmingen), anti–phospho-AKT (S473), anti-ERK1/2, anti–phospho-ERK1/2 (T202/T204; Cell Signaling Technology), anti-STAT5, anti-IL2Rα, anti-IL2Rβ, anti-γc, anti-JAK3, anti-STAT3, anti–phospho-STAT3 (Y705), and anti–phospho-JAK3 (Y980; Santa Cruz Biotechnology Inc) and 680- and 800-nm infrared dye-conjugated secondary Abs (LI-COR).
Cell lines and inhibitor assays
BaF3 cells were obtained from Dr Elizabeth Yang (Vanderbilt). OP9-DL1 and OP9-GFP cell lines were provided by Dr Juan Carlos Zúñiga-Pflücker (University of Toronto). The following reagents were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health): HuT-102 and MT-2 cell lines. We obtained HEK 293T and Phoenix cell lines through ATCC. All cell lines were maintained in recommended media (supplemental Table 4). The T-cell differentiation assay was performed using JAK3−/− fetal liver cells. JAK−/− mice were acquired from The Jackson Laboratory.27 JAK3−/− fetal liver cells were collected at embryonic day (E) 15.5. Mononuclear cells were isolated using lymphocyte separation medium (MediaTech). Cells were magnetically sorted to enrich for c-Kit+Lin−Sca-1+ (KLS) cells (StemCell Technologies Inc) and verified by flow cytometry. Cells defined as KLS were then transduced with MIG-JAK3, and cocultured with irradiated OP9-DL1 or OP9-GFP cells in the presence of mIL-7 and mFlt3 for 28 days. Lymphocytes were transferred to freshly irradiated OP9-DL1 cells and analyzed by flow cytometry for T- and B-cell differentiation markers every 7 days as described.28 Reconstitution of the IL-2R, JAK3/STAT5 pathway was performed by CaPO4 transient transfection of 2.5 μg of pCMVSport6-STAT5a, pcDNA-3.1-IL2Rα, and pCMVSport6-IL2Rβ; 5 μg of pcDNA3.1-γc and MIG-JAK3 constructs and grown in the presence of hIL-2 for 24 hours. Expression levels of all components were quantified by Western blot or flow cytometry. BaF3 cells were serially transduced 3 times and GFP-positive cells were sorted by flow cytometry after 6 days in culture. Cytokine-independent growth assays were performed as described.29 Once cytokine independence was established, mutant JAK3-expressing cells were maintained in the absence of mIL-3 and used in all subsequent experiments. Protein stability was determined by treating BaF3 cells by serum starvation for 1 hour followed by 100 μg/mL cycloheximide for 0, 0.5, 1, 2, and 3 hours before cells were lysed and JAK3 expression was analyzed by Western blot analysis. The JAK3 inhibition assays were performed on BaF3 cells expressing mutant JAK3s using tofacitinib (formerly tasocitinib or CP-690550; Selleck Chemicals LLC) at concentrations listed at time points 0, 12, and 24 hours. Cells were incubated with anti–annexin V Ab and propidium iodide and analyzed by flow cytometry after 24- and 48-hour incubations with tofacitinib. Protein was obtained at all time points and cells were counted at 0 and 24 hours. Inhibitors BEZ-235 (Selleck), BKM-120 (Active Biochem), and CI-1040 (Dr Carlos L. Arteaga, Vanderbilt) against PI3K/mTOR, PI3K and MEK, respectively, were used in MTT assays as described. For MTT assay, MIG-JAK3-transduced BaF3 cells were plated at 200 000 cells/96 wells at concentrations of 1000 cells/μL in triplicate using media without phenol red at tofacitinib concentrations listed. At 48 hours, 150 μL of media was removed, 20 μL of 5 mg/mL MTT reagent was added and cells were incubated for an additional 4.5 hours. Cells were lysed by adding 100 μL of 0.1N HCl in isopropanol. Plates were scanned on a Molecular Devices Versa Max microplate reader, and data were analyzed using SoftMax Pro software.
Kinase assay
Kinase assays were performed using LANCEUltra time-resolved fluorescence resonance energy transfer (TR-FRET) from PerkinElmer. Assays use a europium chelate donor fluorophore-conjugated Ab, PT66 (Eu), against the phosphotyrosine of substrate ULight-JAK-1 (Y1023) which is a short peptide sequence from JAK1 that is phosphorylated by JAK3. The ULight substrate has a small-molecular-weight acceptor dye with red-shifted fluorescent emission. This assay was optimized with recombinant human JAK3 (Signal Chem Inc; amino acids 781-end) containing only the kinase domain. WT or mutant JAK3 enzymes were purified from BaF3 cells by immunoprecipitation. Enzymes were incubated in Lance JAK3 assay buffer with 10nM ULight-JAK-1 (Y1023) and 20μM ATP. Kinase reactions were terminated after 1 hour by the addition of 2 times detection mixture containing PT66 Eu (8μM final) and EDTA (6mM final). After 30-minute incubation in the detection mixture, FRET was detected at 665 nm after excitation at 340 nm.
Results
JAK3 FERM domain mutations in ATLL patients do not show loss of function
We analyzed 36 ATLL patients for mutations in the JAK3 gene by heteroduplex analysis, PCR cloning, and direct sequencing.19,30 In 4 patients (11%), we found sequence variants that were not present in single nucleotide polymorphism (SNP) databases or in 24 ethnically matched control individuals31 (Figure 1A), supplemental Figure 1, supplemental Tables 1 and 5). Some of our samples were purified leukemic blasts from apheresed patients and some were admixed with normal cells of the buffy coat. The frequency of one of these mutations based on cloned PCR products (10%) correlated with the frequency of leukemic cells in genomic DNA samples as estimated by quantitative PCR for tax (supplemental Figure 2). Missense mutations L156P, R172Q, and E183G occurred in highly conserved residues and were somatically acquired (Figure 1A). The E183G mutation was recurrent because it was found in 2 patients. All mutations clustered within the FERM domain of JAK3 whose major function is to bind the cytoplasmic tail of γc.21,32-35 In humans, autosomal-recessive SCID is caused by loss of function of JAK3.36 Some of the germline SCID mutations described occur in the FERM domain of JAK3 and abolish binding to γc.35 Therefore, we sought to rule out loss of function of JAK3 because of these newly identified mutations in ATLL. We isolated JAK3−/− fetal liver (Lin−Sca-1+c-Kit+) hematopoietic stem and progenitor cells (HSPCs) and cocultured them with OP9-GFP or OP9-DL1 stromal cells as described.28 JAK3 knockout cells failed to proliferate or differentiate into CD4+CD8+ double-positive (DP) T cells but JAK3−/− HSPCs transduced with WT JAK3 were rescued (Figure 1C). A previously described activating mutant JAK3 A572V and the ATLL mutations also rescued proliferative and differentiation defects in the knockouts (Figure 1B-D).19,27,28 Interestingly, T-cell differentiation and proliferation did not occur in the transduced cells if IL-7 was omitted from culture. We evaluated our mutant JAK3s for γc binding by coimmunoprecipitation assays and found that mutant JAK3s bound γc to the same extent as WT JAK3 (supplemental Figure 3). These experiments showed that the ATLL JAK3 mutations in the FERM domain did not cause loss of function but also did not show gain of function in this assay.
ATLL mutations show increased phosphorylation of STAT5A
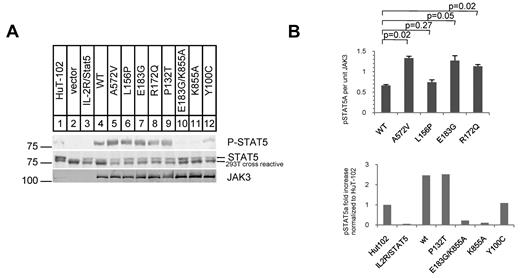
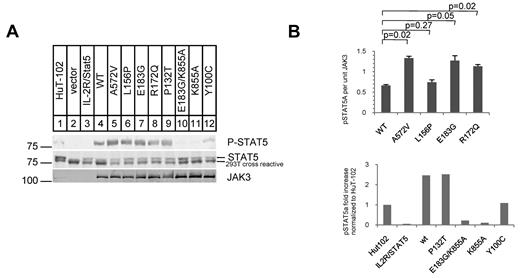
Next, we reconstituted IL-2 signaling in HEK 293T cells, which lack IL-2R proteins, by cotransfecting plasmids expressing the heterotrimeric IL-2Rα, β, γc, WT or mutant JAK3, and the JAK3 substrate STAT5A. Equivalent amounts of protein were expressed in each transfection as quantified by Western blotting with infrared dye-labeled secondary Abs. We quantified phosphorylated STAT5A using a primary Ab specific to phospho-Y694 of STAT5A (Figure 2), a residue uniquely phosphorylated by JAK3.37 Cotransfection of the IL-2R components with STAT5A showed no phospho-STAT5A in the absence of JAK3 (lane 3 of Figure 2A). Cotransfection of IL-Rα, β, γc, STAT5A, and WT JAK3 induced phospho-STAT5A only in the presence of exogenous IL-2 (100 U/mL; Figure 2A lane 4). We consistently found increased phospho-STAT5A with mutant JAK3 cotransfection in comparison to WT JAK3 cotransfection (quantification in Figure 2B). Equivalent expression of JAK3 was seen in each transfection and phospho-STAT5A levels were normalized to JAK3 protein expression (Figure 2B). The L156P JAK3 mutant did not show statistically significant increase in phospho-STAT5A levels in this assay but was biologically active in BaF3 cells (Figure 3A).
JAK3 FERM mutations cause increased phosphorylation of STAT5A in reconstituted IL-2 signaling in 293T cells. (A) Western blots show phospho-(P-)STAT5A (92 kDa), STAT5A, and JAK3. Lane 1 shows whole-cell lysate of the HuT-102 cell line. STAT5A phosphorylation requires JAK3 and IL-2 (100 U/mL). (B) Graph shows quantification of Western blots; P-STAT5A protein per unit of expressed JAK3 protein. Mutant JAK3s were compared with WT JAK3 in 4 independent cotransfection experiments and showed consistently higher P-STAT5A levels (mean ± SEM; top panel). P values were generated from the Student t test (2-tailed). The bottom panel shows P-STAT5A protein levels in cotransfection experiments with various other JAK3 mutants normalized to endogenous levels in HuT-102. The K855A mutant causes loss of kinase activity in JAK3. The Y100C mutation was found in a SCID patient and P132T is a SNP.
JAK3 FERM mutations cause increased phosphorylation of STAT5A in reconstituted IL-2 signaling in 293T cells. (A) Western blots show phospho-(P-)STAT5A (92 kDa), STAT5A, and JAK3. Lane 1 shows whole-cell lysate of the HuT-102 cell line. STAT5A phosphorylation requires JAK3 and IL-2 (100 U/mL). (B) Graph shows quantification of Western blots; P-STAT5A protein per unit of expressed JAK3 protein. Mutant JAK3s were compared with WT JAK3 in 4 independent cotransfection experiments and showed consistently higher P-STAT5A levels (mean ± SEM; top panel). P values were generated from the Student t test (2-tailed). The bottom panel shows P-STAT5A protein levels in cotransfection experiments with various other JAK3 mutants normalized to endogenous levels in HuT-102. The K855A mutant causes loss of kinase activity in JAK3. The Y100C mutation was found in a SCID patient and P132T is a SNP.
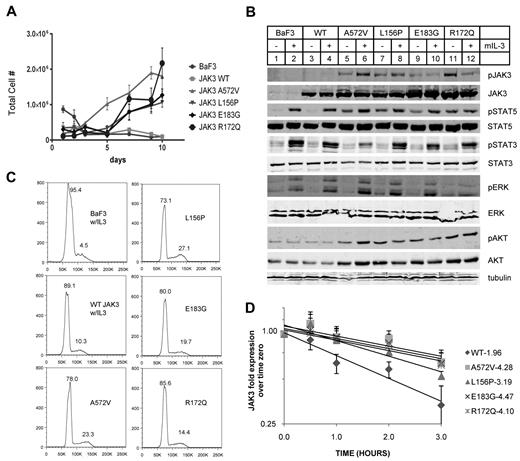
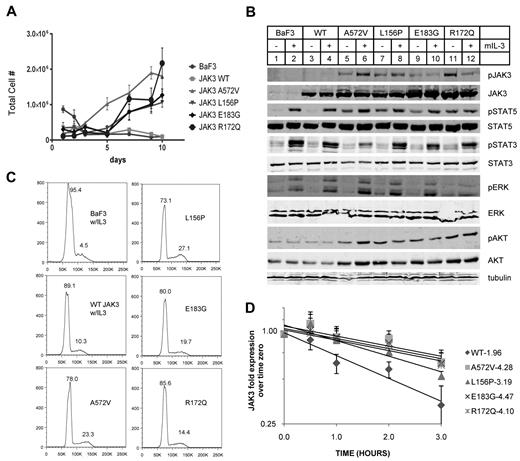
Mutant JAK3s confer cytokine independence in BaF3 cells. (A) BaF3 cells transduced with JAK3s were counted daily after IL-3 withdrawal. WT JAK3 transduced and untransduced BaF3s did not survive unless supplemented with IL-3. Those transduced with mutant JAK3s became cytokine independent by 7 days. (B) BaF3 cells expressing mutant JAK3s showed constitutive autophosphorylation and phosphorylation of STAT5 (Y694) and AKT (S473) without IL-3. All Western blot analyses were performed using whole-cell lysates except pSTAT3 and pJAK3 which were blotted from immunoprecipitates. (C) Flow cytometry histograms show Vybrant dyecycle violet staining of stable BaF3 lines. Proportion of cells in cell-cycle stages G1 or S/G2 are expressed as percentage of total cells above the peaks. Mutant JAK3-transduced BaF3 cells were maintained without IL-3. A572V, L156P, and E183G mutants showed comparable proportions of cells in S/G2 phase. R172Q is more closely comparable with WT JAK3 expressing BaF3 cells. Data shown are representative of 2 independent experiments. (D) Semi-log graph shows JAK3 protein in BaF3 cells expressing WT or mutant JAK3 after treatment with 100 μg/mL cycloheximide for 0, 0.5, 1, 2, and 3 hours. The respective half-lives were calculated from linear regression of 5 total experiments (R2 > 0.90). A572V, L156P, E183G, and R172Q are all significantly more stable than WT JAK3.
Mutant JAK3s confer cytokine independence in BaF3 cells. (A) BaF3 cells transduced with JAK3s were counted daily after IL-3 withdrawal. WT JAK3 transduced and untransduced BaF3s did not survive unless supplemented with IL-3. Those transduced with mutant JAK3s became cytokine independent by 7 days. (B) BaF3 cells expressing mutant JAK3s showed constitutive autophosphorylation and phosphorylation of STAT5 (Y694) and AKT (S473) without IL-3. All Western blot analyses were performed using whole-cell lysates except pSTAT3 and pJAK3 which were blotted from immunoprecipitates. (C) Flow cytometry histograms show Vybrant dyecycle violet staining of stable BaF3 lines. Proportion of cells in cell-cycle stages G1 or S/G2 are expressed as percentage of total cells above the peaks. Mutant JAK3-transduced BaF3 cells were maintained without IL-3. A572V, L156P, and E183G mutants showed comparable proportions of cells in S/G2 phase. R172Q is more closely comparable with WT JAK3 expressing BaF3 cells. Data shown are representative of 2 independent experiments. (D) Semi-log graph shows JAK3 protein in BaF3 cells expressing WT or mutant JAK3 after treatment with 100 μg/mL cycloheximide for 0, 0.5, 1, 2, and 3 hours. The respective half-lives were calculated from linear regression of 5 total experiments (R2 > 0.90). A572V, L156P, E183G, and R172Q are all significantly more stable than WT JAK3.
To verify that the observed phospho-STAT5A was caused by JAK3 kinase activity, we introduced a K855A mutation that creates a catalytically inactive kinase (Figure 2B bottom panel bar 5) into a JAK3 construct with the E183G FERM domain mutation.22,35 This compound mutant expressed stable protein to the same extent as WT JAK3 (Figure 2A) but showed no phosphorylation of STAT5A when cotransfected with IL-2Rα, β, γc, and STAT5A (Figure 2A lane 10 and C bottom panel bar 5). As expected, the P132T SNP introduced into the JAK3 construct showed the same level of phospho-STAT5A as WT JAK3 (Figure 2B bottom panel bar 4). Interestingly, the Y100C SCID mutant showed less phospho-STAT5A than WT JAK3 but did not completely abolish phospho-STAT5A. Because we only observed phosphorylation of STAT5A in the presence of IL-2, the Y100C mutant may retain some binding to γc when overexpressed. Thus, reconstituted IL-2 signaling in 293T cells shows that the ATLL mutations induce increased phosphorylation of STAT5A compared with WT JAK3 protein. Increased levels of phospho-STAT5A were observed in experiments where JAK1 was cotransfected but with the same overall pattern (data not shown).
Mutant JAK3s confer cytokine independent growth in BaF3 cells
We transduced BaF3 cells using recombinant MIG retroviruses expressing mutant or WT JAK3s and sorted for GFP+ cells. BaF3 cells require phosphorylation of STAT5 through IL-3 signaling but become IL-3 independent when gain of function tyrosine kinases are expressed that signal through the same pathway.29 As expected, untransduced BaF3 cells and those transduced with WT JAK3 died after 3 days in culture without IL-3 (Figure 3A). These cells transduced with WT JAK3 were maintained in culture with the addition of IL-3. Remarkably, BaF3 cells transduced with mutant JAK3s grew without IL-3 and were continually passaged as stable cell lines (Figure 3A).
We analyzed phosphorylation of STAT5A (Y694), STAT3 (Y705), ERK1/2 (T202 and Y204), and AKT (S473) after acute withdrawal of IL-3 to identify those events that correlated with the cytokine-independent growth observed in Figure 3A. We divided the IL-3 withdrawal cultures in 2 and added IL-3 to one group before preparing whole-cell lysates or immunoprecipitates. As shown in Figure 3B, phosphorylation of Y980, a residue in the activation loop of JAK3 that is autophosphorylated, was minimal in WT JAK3 (lanes 3-4) but constitutive in all the mutant JAK3s (lanes 6-13). We did not observe phospho-Y980 in BaF3 cells expressing the E/K compound mutant JAK3 (data not shown), confirming that Y980 is autophosphorylated; and, IL-3 did not consistently affect this phosphorylation. Phospho-STAT5A was detected in all the BaF3 cells expressing mutant JAK3s (Figure 3B lanes 6-13) but not in untransduced BaF3 cells or BaF3 cells transduced with WT JAK3. Phospho-STAT5A was increased after the addition of IL-3 which induces this substrate phosphorylation through JAK2. AKT phosphorylation at S473 was increased in all the mutant JAK3-expressing BaF3 cells (Figure 3B lanes 5-12) compared with untransduced and WT JAK3-expressing BaF3s (Figure 3B lanes 1-4). ERK1/2 and STAT3 phosphorylations were present in a different pattern than JAK3, STAT5, and AKT. Phospho-ERK1/2 was not detectable in the mutant JAK3-expressing cells without the presence of IL-3 and STAT3 was minimally phosphorylated without IL-3. This basal level of phospho-STAT3 was the same in untransduced BaF3 and in cells expressing WT or mutant JAK3 and could be stimulated with IL-3 addition. In summary, the phosphorylations of JAK3, STAT5, and AKT were correlated with cytokine-independent growth of mutant JAK3-expressing cell lines whereas ERK1/2 and STAT3 phosphorylations were not correlated.
BaF3 cells that expressed mutant JAK3s grew without IL-3 and were not impaired in their proliferation as they showed cell-cycle profiles comparable with BaF3 cells maintained in IL-3 (Figure 3C). Retroviral integration site analysis showed that the stable BaF3 lines expressing mutant JAK3s were polyclonal; and, no integrations were found near genes that could account for cytokine-independent growth (data not shown).
Mutant JAK3 proteins are more stable in BaF3 cells
The JAK proteins have been shown to undergo proteasome-mediated degradation after activation.38,39 We sought to analyze the stability of WT and mutant JAK3 proteins in the BaF3 cells. We applied cycloheximide to BaF3 cells and performed Western blots for JAK3 protein. We quantified JAK3 protein using fluorescent secondary Abs and plotted the mean of 5 independent experiments (Figure 3D). Linear regression analysis of the curves showed that the mutant JAK3s clustered together with similar decay times (A572V, t1/2 = 4.3 hours; L156P, t1/2 = 3.2 hours; R172Q, t1/2 = 4.1 hour; E183G, t1/2 = 4.5 hours) whereas WT JAK3 decayed more rapidly (t1/2 = 1.96 hours). In fact, the mutant JAK3 proteins had half-lives 2-fold greater than WT JAK3 protein.
JAK3 mutations confer increased in vitro tyrosine kinase activity
We immunoprecipitated WT or mutant JAK3s from BaF3 cells and incubated the immune complexes with a peptide substrate conjugated to fluorescent beads. Phosphorylation was detected by Förster (or fluorescence) resonance energy transfer (FRET) between the fluorophore-conjugated phospho-specific Ab to the acceptor dye on the substrate. The assay parameters were established and optimized using recombinant kinase domain of JAK3. Incubation of immune complexes with the peptide substrate without ATP resulted in background counts (Figure 4A light gray bars). With the addition of 20μM ATP, counts were detected in recombinant purified JAK3, a truncated protein only expressing the kinase domain, WT JAK3, and all the mutant JAK3s. No kinase activity was detected in immunoprecipitations with isotype control IgG from WT JAK3-transduced BaF3 cells, untransduced BaF3, or the compound mutant, E183G/K855A (E/K). Most remarkably, mutant JAK3 proteins showed increased enzyme activity compared with WT JAK3 (P < .001 by Student t test). WT JAK3 activity was less than that of the truncated JAK3 protein but was increased compared with IgG control (P = .008).
JAK3 FERM domain mutations show increased kinase activity in vitro. (A) Direct determination of the kinase activity for FERM domain mutations using the Lance Ultra TR-FRET–based assay. Background counts were seen in assays without ATP addition, for BaF3 cells, and with immunoprecipitations using isotype control IgG. Recombinant human JAK3 (50 pg) was used as a positive control. Specific immune complexes of WT and mutant JAK3s showed kinase activity with ATP. The counts are shown as the average of 9 independent kinase assays (mean ± SEM). Statistical comparison between mutant JAK3s and WT JAK3 showed significantly higher kinase activity in the mutants (*P < .001). WT JAK3 showed statistically significant increased activity compared with IgG control (P = .008). (B) Shows Western blots of JAK3 from a representative immunoprecipitation and the detection of autophosphorylated JAK3 after in vitro assay. 4G10 is specific for phosphotyrosine and the third blot shows a phospho-Y980 of JAK3. Notably, a kinase-dead compound mutant E183G/ K855A showed no autophosphorylation.
JAK3 FERM domain mutations show increased kinase activity in vitro. (A) Direct determination of the kinase activity for FERM domain mutations using the Lance Ultra TR-FRET–based assay. Background counts were seen in assays without ATP addition, for BaF3 cells, and with immunoprecipitations using isotype control IgG. Recombinant human JAK3 (50 pg) was used as a positive control. Specific immune complexes of WT and mutant JAK3s showed kinase activity with ATP. The counts are shown as the average of 9 independent kinase assays (mean ± SEM). Statistical comparison between mutant JAK3s and WT JAK3 showed significantly higher kinase activity in the mutants (*P < .001). WT JAK3 showed statistically significant increased activity compared with IgG control (P = .008). (B) Shows Western blots of JAK3 from a representative immunoprecipitation and the detection of autophosphorylated JAK3 after in vitro assay. 4G10 is specific for phosphotyrosine and the third blot shows a phospho-Y980 of JAK3. Notably, a kinase-dead compound mutant E183G/ K855A showed no autophosphorylation.
A representative immunoprecipitation of JAK3 is shown in Figure 4B after the kinase assay. This was also probed using 4G10, the phosphotyrosine-specific Ab, and an Ab against phospho-Y980 of JAK3.40 Autophosphorylation of JAK3 (ie, phospho-Y980) was detected for WT and mutant JAK3s (Figure 4B lanes 1-5) but the compound mutant, E/K, did not show autophosphorylation or kinase activity (Figure 4B lane 6). Interestingly, immune complexes that were resuspended in Laemmli buffer for SDS-PAGE immediately after the immunoprecipitation procedure showed phospho-Y980 only for mutant JAK3 proteins and not WT (see Figures 3B lanes 3-12 and 6A lanes 6-21). In contrast, immune complexes that were equilibrated in ATP and kinase buffer followed by SDS-PAGE showed phospho-Y980 in both WT and mutant JAK3 proteins. Thus, the Y980 autophosphorylation assayed immediately after immunoprecipitation or in whole-cell lysates was more predictive of enzyme activity than the SDS-PAGE after the kinase assay. This also suggested that autophosphorylation was taking place during the kinase reaction for WT JAK3 protein. Previous studies have shown that Y980 autophosphorylation can be detected in JAK3 proteins with gain-of-function or loss-of-function mutations and may not be entirely predictive of substrate phosphorylation.23 In fact, immunoprecipitated JAK3 proteins from transfected 293T cells did not show consistent phospho-Y980.
Molecular modeling suggests autoregulatory function for the JAK3 FERM domain
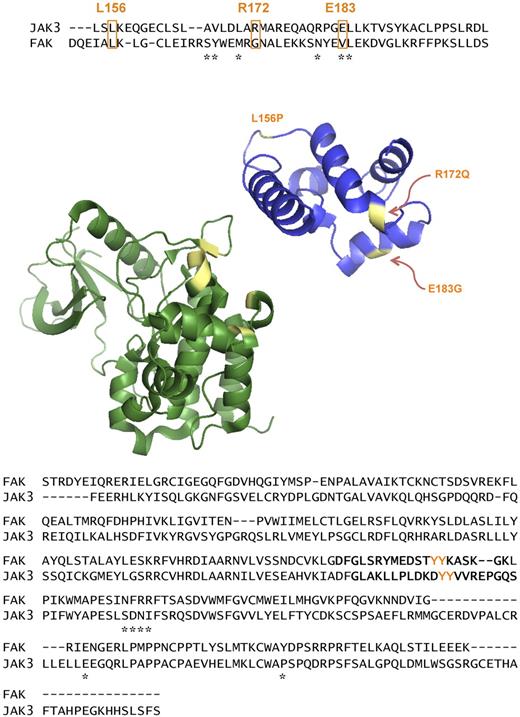
The cell-based assays and the in vitro kinase results support a gain of function activity for the mutant JAK3s. Interestingly, the JAK3 FERM domain's potentiation of kinase activity was suspected by O'Shea and colleagues.35 In their studies, FERM domain mutations from SCID patients abolished γc binding and kinase activity in vitro and they found that an intact FERM domain was required for JAK3 binding of an ATP analog. The crystal structure of the FERM domain of focal adhesion kinase (FAK) revealed the structural basis for this kinase autoregulation. Amino acid side chains from the FAK FERM were shown to directly contact the intramolecular kinase domain causing occlusion of the substrate pocket.41 The JAK3 crystal structure was solved for a protein without the FERM domain.42 Therefore, we modeled the JAK3 FERM by homology to the FAK FERM domain crystal structure. As shown in Figure 5, the residues of the FAK FERM domain that contact the FAK kinase domain (denoted by asterisks) align in the same region as the ATLL mutations (boxed in orange). Interestingly, this region is mainly α helical and the ATLL mutations are predicted to be helix disrupting.
ATLL FERM domain mutations align to the autoregulatory region of the FAK FERM based on homology modeling. ATLL mutations (orange) align with the F2 subdomain of the FAK FERM (blue) that autoregulates the kinase domain of FAK. Asterisks (*) denote residues of FAK FERM that contact the intramolecular FAK kinase domain. Molecular structure is based on the crystal structure of the FAK FERM domain. Those residues that are mutated in ATLL patients are highlighted (yellow). The JAK3 kinase domain (green) is shown as it would be oriented to the FERM domain. Below it, the respective kinase domains of FAK and JAK3 are aligned. The activation loop of the 2 proteins is in bold and the autoregulatory tyrosines, Y980 and Y981, are highlighted in orange. Asterisks (*) show the residues of FAK that contact the intramolecular FERM. The residues of JAK3 that align with these are shown in yellow.
ATLL FERM domain mutations align to the autoregulatory region of the FAK FERM based on homology modeling. ATLL mutations (orange) align with the F2 subdomain of the FAK FERM (blue) that autoregulates the kinase domain of FAK. Asterisks (*) denote residues of FAK FERM that contact the intramolecular FAK kinase domain. Molecular structure is based on the crystal structure of the FAK FERM domain. Those residues that are mutated in ATLL patients are highlighted (yellow). The JAK3 kinase domain (green) is shown as it would be oriented to the FERM domain. Below it, the respective kinase domains of FAK and JAK3 are aligned. The activation loop of the 2 proteins is in bold and the autoregulatory tyrosines, Y980 and Y981, are highlighted in orange. Asterisks (*) show the residues of FAK that contact the intramolecular FERM. The residues of JAK3 that align with these are shown in yellow.
The crystal structure of the JAK3 kinase domain is also shown with an alignment to FAK. The asterisks denote those residues in FAK that directly contact the intramolecular FERM domain. The homologous residues in JAK3 are colored in yellow in the ribbon diagram and are at the surface available for binding and not buried within the molecule but only one of the 6 contact residues are conserved. The modeled JAK3 FERM structure suggests that the ATLL mutations could perturb the secondary structure required for autoregulation of kinase activity. Interestingly, a SCID mutation has been described in this region as well, D169E, which causes decreased γc association and loss of kinase activity.43 This SCID mutation is only 3 residues away from a mutation (R172Q) that shows gain of function in our assays. These data suggest that the secondary structure of the JAK3 FERM domain is very sensitive to amino acid substitutions perhaps because of intramolecular contact with the kinase domain, analogous to FAK.
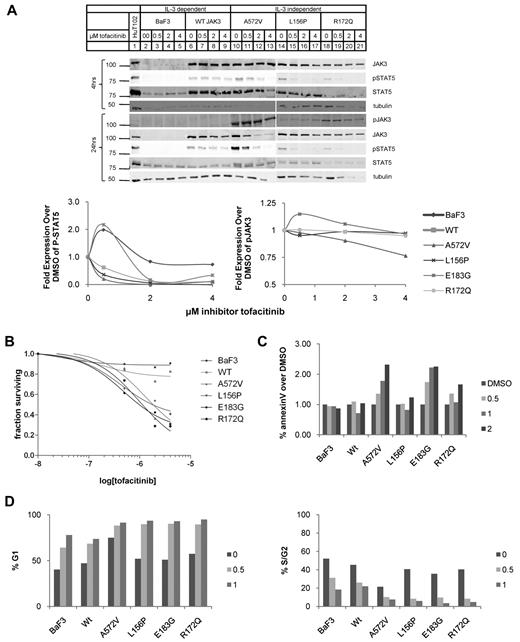
Mutant JAK3s are sensitive to specific inhibitors
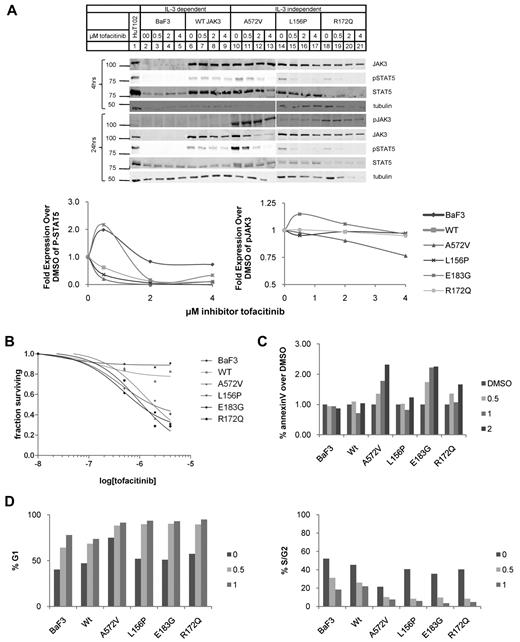
Next, we tested a JAK3-specific inhibitor, tofacitinib, on the BaF3 cells stably expressing mutant JAK3s.24,25,44 We cultured BaF3 cells expressing WT JAK3 or mutant JAK3s with 0.5, 2, or 4μM of the inhibitor or with solvent (DMSO) alone and probed the levels of JAK3 protein, phospho-Y980 of JAK3, and phospho-STAT5A at 4 and 24 hours by quantitative Western blot analysis (Figure 6A). The JAK3 and phospho-STAT5A protein levels were expressed as fold difference with BaF3 cells treated with solvent alone. The BaF3 lines stably expressing mutant JAK3s showed marked dose-dependent decrease in phospho-STAT5A (quantified in Figure 6A bottom panel). At 2μM, phospho-STAT5A levels were decreased by 80% for all BaF3 cells stably expressing mutant JAK3s. Untransduced BaF3 cells showed much lower phospho-STAT5A at steady state (Figure 6A lane 2) and this quantity only decreased 30% at much higher concentrations (4μM) of tofacitinib; phospho-STAT5A in BaF3 cells transduced with WT JAK3 was similarly resistant to the effects of inhibitor. JAK3 and tubulin were quantified as well. BaF3 cells have STAT5 phosphorylation secondary to the IL-3/JAK2 signaling which may have variable sensitivity to tofacitinib in cell-based assays even though JAK2 kinase activity is inhibited at concentrations comparable with JAK3 inhibition in cell-free enzyme assays.24
Mutant JAK3s are sensitive to JAK3-specific tyrosine kinase inhibitor, tofacitinib. (A) BaF3 cells transduced with mutant JAK3s were treated with tofacitinib at concentrations of 0, 0.5, 2.0, and 4.0μM and analyzed by Western blot analysis at 4 and 24 hours. The Western blots and graphs that display their quantification are shown. Protein levels are expressed as fold over levels seen in cells treated with DMSO (solvent) alone. JAK3 (106 kDa) protein expression was comparable with DMSO alone but decreased in the mutant JAK3-transduced BaF3 cells. P-STAT5A (92 kDa) levels decreased markedly with increasing concentrations of inhibitor. P-JAK3 (106 kDa) levels were relatively stable. JAK3 is not phosphorylated in WT-transduced cells so it could not be quantified. Tubulin (55 kDa), a loading control, remained relatively stable. (B) Live cells were quantified by the MTT assay. Cell survival decreased with increasing concentrations of tofacitinib. BaF3 cells expressing mutant JAK3s were susceptible to tofacitinib (IC50: A572V, 0.22nM; L156P, 0.27nM; E183G, 0.087nM; R172Q, 0.082nM). (C) BaF3 cells transduced with mutant JAK3 constructs treated with 0, 0.5, 1, and 2μM tofacitinib for 24 hours and analyzed for apoptosis by staining for annexin V and propidium iodide; percentages of cells staining positive for annexin V relative to DMSO only are shown. The percentage of cells stained for annexin V only as determined by flow cytometry when treated with tofacitinib. Graph shows increase in annexin V staining with inhibitor treatment in all JAK3-transduced BaF3 cells except L156P mutant JAK3. Data shown are representative of 4 independent experiments. (D) Cell-cycle profiles cells treated with DMSO; 0.5 or 1μM tofacitinib were stained with Vybrant dye cycle-violet and analyzed by flow cytometry. Graph shows the percentage of cells in G1 phase. BaF3 and WT-transduced cells have < 75% of cells in G1 after 24 hours on 1μM tofacitinib. All mutant-transduced BaF3 cells have > 90% G1 arrest with 1μM tofacitinib. Graph shows proportion of cells in G1 increases in a dose-dependent manner for all mutant JAK3s. BaF3 cells and those transduced with WT JAK3 were grown in IL-3. Data shown are representative of 2 independent experiments.
Mutant JAK3s are sensitive to JAK3-specific tyrosine kinase inhibitor, tofacitinib. (A) BaF3 cells transduced with mutant JAK3s were treated with tofacitinib at concentrations of 0, 0.5, 2.0, and 4.0μM and analyzed by Western blot analysis at 4 and 24 hours. The Western blots and graphs that display their quantification are shown. Protein levels are expressed as fold over levels seen in cells treated with DMSO (solvent) alone. JAK3 (106 kDa) protein expression was comparable with DMSO alone but decreased in the mutant JAK3-transduced BaF3 cells. P-STAT5A (92 kDa) levels decreased markedly with increasing concentrations of inhibitor. P-JAK3 (106 kDa) levels were relatively stable. JAK3 is not phosphorylated in WT-transduced cells so it could not be quantified. Tubulin (55 kDa), a loading control, remained relatively stable. (B) Live cells were quantified by the MTT assay. Cell survival decreased with increasing concentrations of tofacitinib. BaF3 cells expressing mutant JAK3s were susceptible to tofacitinib (IC50: A572V, 0.22nM; L156P, 0.27nM; E183G, 0.087nM; R172Q, 0.082nM). (C) BaF3 cells transduced with mutant JAK3 constructs treated with 0, 0.5, 1, and 2μM tofacitinib for 24 hours and analyzed for apoptosis by staining for annexin V and propidium iodide; percentages of cells staining positive for annexin V relative to DMSO only are shown. The percentage of cells stained for annexin V only as determined by flow cytometry when treated with tofacitinib. Graph shows increase in annexin V staining with inhibitor treatment in all JAK3-transduced BaF3 cells except L156P mutant JAK3. Data shown are representative of 4 independent experiments. (D) Cell-cycle profiles cells treated with DMSO; 0.5 or 1μM tofacitinib were stained with Vybrant dye cycle-violet and analyzed by flow cytometry. Graph shows the percentage of cells in G1 phase. BaF3 and WT-transduced cells have < 75% of cells in G1 after 24 hours on 1μM tofacitinib. All mutant-transduced BaF3 cells have > 90% G1 arrest with 1μM tofacitinib. Graph shows proportion of cells in G1 increases in a dose-dependent manner for all mutant JAK3s. BaF3 cells and those transduced with WT JAK3 were grown in IL-3. Data shown are representative of 2 independent experiments.
Tofacitinib did not inhibit Y980 phosphorylation to the same extent as STAT5A phosphorylation. Phospho-Y980 in the A572V mutant JAK3 was most sensitive but only decreased to 75% of basal levels at 4μM.
The inhibitor-induced decrease in phospho-STAT5A correlated with decreased growth of these cells (Figure 6B). The IC50 curves demonstrate marked difference between BaF3 cells expressing mutant JAK3s versus those expressing WT JAK3 and untransduced BaF3. Untransduced BaF3 cells and those expressing WT JAK3 were not as sensitive to tofacitinib and curve-fitting did not generate models with high correlation (ie, R2 > 0.90, see Table 1). Mutant JAK3-expressing BaF3s showed consistent sensitivity to tofacitinib with well correlated curves (Table 1). The decreased cell numbers were because of increased apoptosis and G1 cell-cycle arrest (Figure 6C-D). Apoptosis was modestly increased only in the A572V and E183G mutant-expressing cells. All the mutant JAK3 cell lines showed marked G1 arrest compared with untransduced and WT JAK3-expressing BaF3 cells (Figure 6D). Interestingly, the BaF3 cells expressing mutant JAK3s were sensitive to PI3K and mTOR inhibitors but not to ERK1/2 inhibitor (Table 1, supplemental Figures 5-7). This finding is consistent with the STAT5A and PI3K/AKT pathways being active in the BaF3 cells expressing mutant JAK3 protein (Figure 3B). Phospho-STAT5A in both HuT-102 and MT-2 cells, both derived from ATLL patients, was sensitive to the tofacitinib (supplemental Figure 4). We did not find mutations in the JAK3 gene in HuT-102 or MT-2 cells although both lines use JAK3 for signal transduction.
Discussion
Data accumulated over many years suggest a central role for JAK3/γc-associated cytokines in ATLL pathogenesis. In this study, we analyzed the JAK3 gene and found 3 missense mutations in the N-terminal FERM domain in 11% of ATLL patients. These mutations cause gain of function in JAK3 kinase activity in cell-based assays and in in vitro kinase assays and may contribute to the development of ATLL. The mutant JAK3s caused increased phosphorylation of STAT5A in reconstituted IL-2 signaling in 293T cells and induced cytokine-independent growth in BaF3 cells. One important question is whether the mutant JAK3s require receptor and ligand for signal transduction. The phosphorylation of STAT5A in the 293T cell assay required IL-2 and hematopoietic stem and progenitor cells transduced with mutant JAK3s did not differentiate or proliferate in the absence of IL-7. In contrast, BaF3 cells transduced with mutant JAK3s grew and cycled without cytokines suggesting that the JAK3s did not require ligand binding to receptors. These data imply some cell type specificity in the requirement for ligand in mutant JAK3 signaling; in this respect, both the FERM mutants and the pseudokinase mutation, A572V, required IL-2 in the 293T cell assay. Other proteins recruited to the receptors and JAK3/γc could contribute to this. Recently, marked overexpression of the SYK and LYN tyrosine kinases was observed in ATLL patients.45 This may have pathogenic significance because these kinases are recruited by IL-2Rβ and γc. We cotransfected JAK1 in the 293T assay but saw no change in IL-2 dependence consistent with prior reports.46 The observation of ligand-dependent or -independent growth is clinically important because if FERM domain mutations are found to require JAK3/γc using ligand in T cells, then cytokines must signal either by autocrine or paracrine mechanisms. HTLV-1–infected T cells do show increased expression of IL-2 and IL-9.13 There is also evidence for IL-9 secretion from monocytes isolated from ATLL patients; and, in T-cell neoplasms that resemble ATLL such as cutaneous T-cell lymphoma (CTCL), IL-7 secretion by keratinocytes may support tumor cell growth.47,48 We are investigating whether this could be the case for ATLL as well, whether paracrine JAK3/γc cytokines from supporting tissues could be trophic or promote growth of ATLL cells, and, accordingly whether mutant JAK3s allow growth of primary T cells without cytokines.
The ATLL mutations are also highly informative for understanding the biochemistry of JAK3 kinase function. Our homology model based on the crystal structure of FAK showed that the ATLL mutations align to the FAK FERM F2 subdomain that occludes and inhibits the activity of the FAK kinase domain. Therefore, the JAK3 FERM may have a similar autoregulatory role. JAK2 activation induced by the erythropoietin receptor induces autophosphorylation of the activation loop and the FERM domain at Y119; this phosphorylation causes dissociation of the FERM from the erythropoietin receptor thus exerting negative feedback on the signal.49 A Y119F mutation introduced into JAK2 creates an enzyme that is more active and more stable. Alanine substitution at the comparable residue in JAK3, Y105, did not abolish JAK3 binding to IL2RG and did not affect enzymatic activity.35 Hence, JAK3's FERM has different regulatory properties from the FERM domain of JAK2. Interestingly, the FERM domain mutations described here also increase the overall protein stability of JAK3. It is not clear whether the increased stability is a cause of the mutations or an effect of JAK3 activation. The increased stability may be related to the increased activity of JAK3 because the FERM domain mutant proteins had comparable half-lives to the A572V mutant protein. It is possible that activation induces an allosteric change that makes JAK3 more stable. A detailed crystal structure of the JAK3 FERM with its kinase domain will provide significant insight into this mechanism. Our results also raise the possibility of FERM domain mutations in other JAK kinases implicated in cancer.
The incidence and timing of JAK3 mutations will be important to investigate in future studies; it would be important to examine whether JAK3 mutations occur early in the chronic phase of the disease or late in the acute setting. Recently, a cohort of 20 Japanese ATLL patients were screened for JAK1 and JAK3 mutations and none were found.50 This could signify a difference between Caribbean and Asian ATLL disease pathogenesis. A more thorough mutational analysis of the whole pathway would be highly informative in both sets of patients. As noted, the only other recurrent rearrangement found in ATLL was a deletion of an E-box binding homeobox transcription factor (ZEB1) that represses expression of the IL2 gene. ZEB1 loss of function presumably deregulates IL-2 which could promote inappropriate activation of the pathway.51 Also, NOTCH1 mutations have been described in ATLL but are not as prevalent as they are in T-lymphoblastic leukemias.52
Our results implicate the JAK3/γc pathway as an important therapeutic target in ATLL. Thus far, we have not been able to establish a human ATLL cell line with a gain-of-function JAK3 mutation to formally test oncogene dependence on the pathway. Nevertheless, tofacitinib, a specific JAK3 inhibitor inhibited growth of 2 HTLV-1–expressing cell lines that had no mutations in JAK3 and primary ATLL cells whose JAK3 status was unknown.53 This result suggests that there could be oncogene dependence on JAK3 even without a gain of function mutation. For example, the HuT-102 line requires exogenous IL-2 for optimal growth. The MT-2 cells also have deregulation of the JAK3/γc axis in the form of HTLV-1 integration 5′ of the IL9R gene. This retroviral insertion induces overexpression of the receptor such that high levels of IL-9R may signal through JAK3 without ligand.54,55 Other components of the JAK3/γc pathway may be mutated in ATLL or other mechanisms may deregulate the pathway because not all patients in our cohort had JAK3 mutations.11 Finally, JAK3-specific inhibitors have been clinically applied in phase 1 and phase 2 studies of autoimmune disease and may be worth testing in ATLL.25
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Brian Druker for sharing JAK3 constructs, and Dr Juan Carlos Zúñiga-Pflücker for OP9-DL1 and OP9-GFP cells. They also thank Drs Carlos Arteaga, Mark Boothby, Steve Brandt, Mike Engel, Scott Hiebert, and Vivian Siegel for critically reviewing the manuscript and for helpful discussions. In addition, they thank the Flow Cytometry Core of Vanderbilt University.
The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). This work was supported by award K08HL089403 from the National Heart, Lung, and Blood Institute. This work was also supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award (U.P.D.); the Leukemia & Lymphoma Society; the Vanderbilt Ingram Cancer Center (P30 CA68485); a Monforton family grant; the T. J. Martell Foundation (U.P.D.); and U54 National Institutes of Health grants CA101598, CA101388, and NIH P30CA0113696 (V.G.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: N.E.E. performed research, analyzed data, and helped write the manuscript; S.M.C. designed and performed research, analyzed data, and helped write the manuscript; V.G. contributed vital new reagents; J.J. and T.A.W. performed research and contributed vital new reagents; and U.P.D. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Utpal P. Davé, MD, Division of Hematology/Oncology, Departments of Medicine and Cancer Biology, Vanderbilt University Medical Center, 777 Preston Research Bldg, Nashville, TN 37232-6307; e-mail: utpal.dave@vanderbilt.edu.