Abstract
CRM1 plays an important role in the nuclear export of cargo proteins bearing nuclear exporting signal sequences. Leptomycin B (LMB), a well-known CRM1 inhibitor, possesses strong antitumor properties. However, its toxicity prevents it from being clinically useful. In this study, we demonstrate that a novel compound, CBS9106, inhibits CRM1-dependent nuclear export, causing arrest of the cell cycle and inducing apoptosis in a time- and dose-dependent manner for a broad spectrum of cancer cells, including multiple myeloma cells. CBS9106 reduces CRM1 protein levels significantly without affecting CRM1 mRNA expression. This effect could be reversed by adding bortezomib or LMB. Moreover, CBS9106-biotin allows capture of CRM1 protein by streptavidin beads in a competitive manner with LMB and vice versa. Mass spectrometric analysis shows that CBS9106 reacts with a synthetic CRM1 peptide that contains Cys528 but not with a Cys528 mutant peptide. Oral administration of CBS9106 significantly suppresses tumor growth and prolongs survival in mice bearing tumor xenograft without a significant loss in body weight. A reduced level of CRM1 protein is also observed in tumor xenografts isolated from mice treated with CBS9106. Taken together, these results indicate that CBS9106 is a novel reversible CRM1 inhibitor and a promising clinical candidate.
Introduction
In eukaryotes, the nucleus is separated from the cytoplasm by a nuclear envelope that contains nuclear pores through which proteins and mRNAs may pass, in the form of nuclear pore complexes, a process that requires the proteins importin or exportin.1,2 Importins and exportins form complexes with Ran GTPase and recognize cargo proteins that bear specific nuclear localization sequences and nuclear export sequences (NESs), respectively.1,2
CRM1/exportin1 is a receptor for the NES in both lower and higher eukaryotes and plays an important role in nucleo-cytoplasmic transport of NES-containing cargo proteins, which include transcription factors (eg, p53 and Forkhead box class Os),2,3 c-Abl/BCR-Abl,4-6 inhibitor of κB α (IκB-α).7 Because these exported molecules play key roles in proliferation and survival of cancer cells, CRM1 could be considered to be a viable therapeutic target for anticancer drug development.2,3,8-10
A well-known CRM1-specific inhibitor, leptomycin B (LMB), binds covalently to Cys528 of CRM1 by a Michael-type addition reaction and abrogates the interaction between CRM1 and its cargo protein.2,3,11 In addition, other CRM1 inhibitors, which have structures that are either similar to LMB12-14 or apparently different,15-18 also target Cys528 of CRM1. LMB shows high antitumor activity against a broad range of cancer cell lines in vitro (IC50 values in 0.1-10nM range).14 However, further clinical evaluation of LMB (Elactocin) was not recommended after a phase 1 trial because of this drug's toxicity (eg, profound anorexia and malaise) and lack of efficacy at tolerable dose levels.19 LMB analogues with much reduced toxicity have been reported,14 but, to the best of our knowledge, none of these is presently in clinical trial.
Here, we present CBS9106, a novel reversible CRM1 inhibitor with unique CRM1 degrading activity. CBS9106 shows antitumor activity, including activity against multiple myeloma (MM) cells, both in vitro and in vivo. CBS9106 is a promising clinical candidate for the treatment of cancer in general and, in particular, for the treatment of MM.
Methods
Cell lines, antibodies, and reagents
MM.1S, MM.1R, and RPMI-8226 cells were kindly provided by Prof Yasuda (Sapporo University). ARH-77 cell line was obtained from ATCC. These myeloma cell lines were maintained in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen Life Technologies). Antibodies against CRM1 (H300), PARP1/2 (H250), Cyclin B1 (H433), c-Myc (9E10), β-actin (C4), RanBP1 (C19), Ran (C20), IκB-α (C21), and LMB were obtained from Santa Cruz Biotechnology. Bortezomib was obtained from Selleck Chemicals LLC. Antibodies against p53 (no. 9282), GAPDH (no. 2118), Caspase-3 (no. 9662), Caspase-8 (no. 9746), Caspase-9 (no. 9502), as well as against rabbit IgG and mouse IgG conjugated with horseradish peroxidase (HRP) were obtained from Cell Signaling Technology. Rabbit anti–goat IgG HRP conjugated was obtained from Dako. Recombinant TNF-α was obtained from PeproTech. Block Ace was obtained from DS Pharma Biomedical Co Ltd. Triton X-100 was obtained from Nacalai Tesque Inc. CBS9106 and biotinylated compounds (LMB, CBS9106, and S08776B) were manufactured or synthesized by Cambridge Major Laboratories Europe and Mercachem, respectively. All other reagents, chemicals, and antibodies used for this study were obtained from Sigma-Aldrich unless otherwise noted.
Immunoblotting
Cells were lysed by incubating at 4°C for 30 minutes in a buffer (NaCl [100mM], Tris-HCl [50mM, pH 8.0)], DTT [1mM], NP40 [0.5% wt/vol] containing PhosSTOP Phosphatase Inhibitor Cocktail and Complete Proteinase Inhibitor Cocktail Tablets [Roche Applied Science] each diluted to the manufacturer's specified concentrations). After incubation, the lysed cell suspension was centrifuged (15 000 rpm, 4°C, 20 minutes), the supernatant (whole-cell lysate) was electrophoresed (SDS-PAGE), and the proteins were transferred from the gel onto a polyvinylidene difluoride (PVDF) membrane (Millipore) by electroblotting. After blocking with 1% Block Ace in T-TBS for 1 hour at room temperature, the PVDF membrane was incubated with primary antibody at 4°C overnight. The next day, the membrane was washed with T-TBS and incubated for 1 hour with a secondary antibody-HRP conjugate at room temperature. After washing the PVDF membranes with T-TBS, the proteins were visualized with Immobilon Western HRP detection substrate (Millipore). Images were recorded with a Luminescent Image Analyzer LAS-4000 system (Fujifilm) and quantified with Multi Gauge software (Fujifilm). The band intensity of the CRM1 at each time point was normalized to that of actin. Each normalized value was expressed as a ratio relative to that of the untreated cells.
Immunofluorescence microscopy
Unless otherwise indicated, all experiments were performed at room temperature. MM.1S cells were treated with LMB (1nM) or CBS9106 (0-800nM) at 37°C for 2 hours and fixed for 30 minutes with 4% paraformaldehyde phosphate buffer solution (Wako Pure Chemical Industries Ltd). Next, cell membranes were permeabilized by treatment with Triton X-100 (0.1% wt/vol) in PBS for 15 minutes. After blocking with 4% Block Ace in PBS (blocking solution) for 1 hour, cells were treated with RanBP1 antibody (2 μg/mL) in blocking solution for 1 hour. After washing with PBS, cells were treated for 1 hour with Alexa 488–conjugated donkey anti–goat antibody (2 μg/mL; Invitrogen Life Technologies) and Hoechst 43332 (1 μg/mL; Dojindo Laboratories) in blocking solution. Cells were transferred to a glass-bottomed dish containing PBS and set aside for ∼ 1 hour to allow them to sink. Photomicrographic images were recorded with the use of a confocal laser scanning microscope Fluoview FV10i (Olympus) with 60×/1.35 NA oil objective lens. Images were processed with FV10-ASW (Version 2.1) software (Olympus).
RT-PCR
Total cellular RNA was extracted with the use of an RNeasy PLUS kit (QIAGEN). It was reverse-transcribed into cDNA with the use of superscript III first-strand synthesis system for RT-PCR (Invitrogen Life Technologies). The following synthetic primers (Invitrogen Life Technologies) were used: CRM1 forward, 5′-AAT GTC CCA GCT GCT AGA GAA CCA GAA GTG-3′, and CRM1 reverse, 5′-CTC TGA GCT GCA GCT TCT TCT TGT GCA ACA-3′; GAPDH forward, 5′-TGA TTC CAC CCA TGG CAA ATT CCA TGG CAC-3′, and GAPDH reverse, 5′-AAG TCA GAG GAG ACC ACC TGG TGC TCA-3′. PCR was performed in a 20-μL reaction mixture containing cDNA (2 μL), forward and reverse primers (200nM each), nucleotide (200μM), PCR buffer (1×), and 1.25 units of KOD dash polymerase (Toyobo). For RT-PCR of CRM1 and GAPDH, the following thermocycling conditions were used: 1 cycle at 94°C for 2 minutes; 25 cycles at 94°C for 30 seconds plus 60°C for 2 seconds plus 72°C for 30 seconds; and 1 cycle at 72°C for 30 seconds. Amplified DNA fragments were electrophoresed on an agarose gel and then stained with ethidium bromide solution for 10 minutes. After washing the gel with water, images were recorded with a Luminescent Image Analyzer LAS-4000 system (Fujifilm).
Streptavidin-biotin pull-down assay
RPMI-8226 cells were treated with biotin (100nM) or biotinylated compounds (S08776B-biotin, 100nM; CBS9106-biotin, 100nM; LMB-biotin, 1nM) for 1 hour in the presence or absence of pretreatment with LMB (1nM) or CBS9106 (1μM) for 1 hour.
Whole-cell lysates were prepared from the cells and 1 mg of proteins was used for the assay. Streptavidin beads (Roche Applied Science) were added to the cell lysate and incubated at 4°C overnight. After washing the beads with lysis buffer, the captured protein was eluted from the beads by boiling them with SDS sample buffer containing 2-mercaptoethanol and was analyzed by immunoblotting.
Cell viability assay
MM cells were seeded into 96-well plates and treated for 72 hours with CBS9106. Cell viability was evaluated with the use of the WST-8 reduction assay kit mixture (Dojindo Laboratories) which was added to the wells and incubated for the last 3 hours of incubation. The absorbance of wells at 450 nm (reference wavelength, 650 nm) was measured with a microplate reader (Sunrise; Tecan). The cell viability of 60 cell lines was analyzed by Multiplexed Cytotoxicity Assay at Ricerca Biosciences LLC. Briefly, cells were seeded into 384-well plates and treated with CBS9106 24 hours after cell seeding. After a 72-hour incubation period, cells were fixed and stained with fluorescently labeled antibodies and nuclear dye. Automated fluorescence microscopy was performed with a GE Healthcare IN Cell Analyzer 1000.
NF-κB ELISA
The DNA-binding activity of NF-κB in MM.1S cells was quantified by TransAM NF-κB p65 Transcription Factor Assay Kit (Active Motif Japan), according to the manufacturer's instructions. Briefly, MM.1S cells in triplicate dishes were pretreated with CBS9106 or LMB for 5 hours and then treated with vehicle or TNF-α (50 ng/mL) for another 4 hours. Nuclear extracts were prepared with a Nuclear extraction kit (Panomics), according to the manufacturer's instructions. Nuclear extracts (20 μg/well) were incubated in 96-well plates coated with immobilized oligonucleotides containing the consensus binding site for the p65 subunit of NF-κB. The absorbance of wells at 450 nm (reference wavelength, 650 nm) was measured with the Tecan Sunrise microplate reader.
Mass spectrometric analysis
A peptide containing Cys528 of CRM1 (amino acids, 523-531) and its derivative with Cys528 replaced by Ser were synthesized at Toray Research Center Inc. These peptides were treated with or without 100μM CBS9106 in 100 μL of buffer (20mM Tris-HCl, pH 7.5, 100mM NaCl) at 37°C for 24 hours. An aliquot of this solution (0.5 μL) was mixed with an equal amount of 2,5-dihydroxybenzoic acid, and then air-dried on the sample plate before analysis. Mass spectrometric analyses were performed at APRO Life Science Institute Inc. A Voyager DE-STR matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems Japan) was used to acquire mass spectra with the use of positive ion reflector mode.
Xenograft model
SCID mice were purchased from Charles River Laboratories Japan Inc. All animal studies were conducted according to protocols approved by the institutional animal care committee of CanBas Co Ltd. Six-week-old male SCID mice were inoculated subcutaneously in the flank with a suspension of RPMI-8226 (1.0 × 107 cells). Tumor volumes, measured thrice weekly with a pair of calipers, were calculated with the following formula: volume (mm3) = [width (mm)]2 × length (mm)/2. Once the tumor growth became stably measurable (100-900 mm3 with average size of 450 mm3), mice were apportioned into 4 groups (8 mice/group) and orally treated with vehicle (5% gum arabic) or CBS9106 (31.25 mg/kg, every day × 5 weekly, 62.5 mg/kg, every day × 5 weekly, 125 mg/kg, every other day × 3 weekly) for 2 weeks. The relative tumor volume was expressed as Vt/V0 index, whereby Vt is the tumor volume on a given day and V0 is the volume of the same tumor just before initial treatment. Growth curves after treatment were generated by plotting the mean ± SE of relative tumor sizes. The changes in the body weight (thrice weekly) are expressed as a ratio relative to the weight just before initial treatment. Statistical significance of the difference between the treatment regimens was determined by Student t tests. For the survival analysis, 8-week-old male SCID mice were inoculated intravenously with a suspension of MM.1S cells (1.0 × 107 cells). Five days after the inoculation, mice were apportioned into 2 groups (10 mice/group) and orally treated with vehicle (5% gum arabic) or CBS9106 (125 mg/kg, every other day × 3 weekly) for 2 weeks. The changes in the body weight are expressed as described. Overall survival was shown by the Kaplan-Meier method.
Results
CBS9106 suppresses growth of cancer cells and nuclear export of proteins
On screening a chemical library for agents that affect the cell cycle G2 DNA damage checkpoint with the use of a cell cycle phenotype-based protocol,20,21 a novel compound, CBS9106 (Figure 1A), was identified. CBS9106 was found to sensitize cells to radiation and to suppress the growth of a variety of cancer cell lines when used alone. The estimated IC50 values ranged from 22.3 or 35.6nM in MM.1S or RPMI-8226 cells and from 3 to 278nM in 60 other human cancer cell lines (Figure 1B; Table 1).
CBS9106 inhibits cancer cell growth and CRM1-dependent nuclear export. (A) Structure of CBS9106. (B) Suppression of the cell growth of MM cells by CBS9106. The growth of 2 different MM cells were cultured with indicated doses of CBS9106 for 72 hours and analyzed by the WST-8 method. Growth rate (n = 3) was represented as relative values to those of vehicle-treated control cells. Error bars represent SD. (C) Accumulation of RanBP1 in nucleus by CBS9106 treatment. MM.1S cells were treated with vehicle, LMB (1nM), and CBS9106 (100-800nM) for 2 hours. Fixed cells were stained for RanBP1 (green) and Hoechst (blue) and analyzed by confocal microscopy.
CBS9106 inhibits cancer cell growth and CRM1-dependent nuclear export. (A) Structure of CBS9106. (B) Suppression of the cell growth of MM cells by CBS9106. The growth of 2 different MM cells were cultured with indicated doses of CBS9106 for 72 hours and analyzed by the WST-8 method. Growth rate (n = 3) was represented as relative values to those of vehicle-treated control cells. Error bars represent SD. (C) Accumulation of RanBP1 in nucleus by CBS9106 treatment. MM.1S cells were treated with vehicle, LMB (1nM), and CBS9106 (100-800nM) for 2 hours. Fixed cells were stained for RanBP1 (green) and Hoechst (blue) and analyzed by confocal microscopy.
Antiproliferative effect of CBS9106 on 60 human cancer cell lines in vitro
| . | IC50, nM . |
|---|---|
| CNS | |
| T98G | 33 |
| SK-N-AS | 28 |
| U-87 MG | 24 |
| BE(2)C | 19 |
| A172 | 16 |
| Head and neck | |
| FaDu | 64 |
| Cal 27 | 43 |
| Endocrine | |
| CAL-62 | 21 |
| CGTH-W-1 | 20 |
| Lung | |
| A427 | 46 |
| A549 | 37 |
| NCI-H460 | 29 |
| NCI-H661 | 21 |
| NCI-H292 | 10 |
| Breast | |
| Hs 578T | 112 |
| BT-549 | 98 |
| AU565 | 75 |
| MDA MB 453 | 17 |
| MDA MB 231 | 9 |
| Colon | |
| WiDr | 65 |
| HT-29 | 57 |
| SW48 | 49 |
| HCT-116 | 29 |
| DLD-1 | 26 |
| Pancreas | |
| PANC-1 | 84 |
| HuP-T4 | 57 |
| BxPC-3 | 50 |
| Mia PaCa-2 | 28 |
| Prostate | |
| BM-1604 | 126 |
| PC-3 | 28 |
| Kidney | |
| A498 | 42 |
| 786-O | 30 |
| G-401 | 11 |
| Caki-2 | 6 |
| 769-P | 3 |
| Liver | |
| HLE | 31 |
| HepG2 | 31 |
| HUH-6 Clone 5 | 28 |
| HLF | 27 |
| HuCCT1 | 16 |
| Female GU | |
| C-33A | 90 |
| SiHa | 42 |
| C-4 I | 41 |
| ES-2 | 21 |
| AN3 CA | 15 |
| Skin | |
| MeWo | 92 |
| CHL-1 | 49 |
| RPMI-7951 | 42 |
| HMCB | 31 |
| A375 | 10 |
| Soft tissue | |
| SK-UT-1 | 278 |
| SK-LMS-1 | 28 |
| HT-1080 | 19 |
| MES-SA | 15 |
| MG-63 | 14 |
| Bladder | |
| J82 | 146 |
| TCCSUP | 70 |
| BFTC-905 | 38 |
| UM-UC-3 | 33 |
| 639-V | 25 |
| . | IC50, nM . |
|---|---|
| CNS | |
| T98G | 33 |
| SK-N-AS | 28 |
| U-87 MG | 24 |
| BE(2)C | 19 |
| A172 | 16 |
| Head and neck | |
| FaDu | 64 |
| Cal 27 | 43 |
| Endocrine | |
| CAL-62 | 21 |
| CGTH-W-1 | 20 |
| Lung | |
| A427 | 46 |
| A549 | 37 |
| NCI-H460 | 29 |
| NCI-H661 | 21 |
| NCI-H292 | 10 |
| Breast | |
| Hs 578T | 112 |
| BT-549 | 98 |
| AU565 | 75 |
| MDA MB 453 | 17 |
| MDA MB 231 | 9 |
| Colon | |
| WiDr | 65 |
| HT-29 | 57 |
| SW48 | 49 |
| HCT-116 | 29 |
| DLD-1 | 26 |
| Pancreas | |
| PANC-1 | 84 |
| HuP-T4 | 57 |
| BxPC-3 | 50 |
| Mia PaCa-2 | 28 |
| Prostate | |
| BM-1604 | 126 |
| PC-3 | 28 |
| Kidney | |
| A498 | 42 |
| 786-O | 30 |
| G-401 | 11 |
| Caki-2 | 6 |
| 769-P | 3 |
| Liver | |
| HLE | 31 |
| HepG2 | 31 |
| HUH-6 Clone 5 | 28 |
| HLF | 27 |
| HuCCT1 | 16 |
| Female GU | |
| C-33A | 90 |
| SiHa | 42 |
| C-4 I | 41 |
| ES-2 | 21 |
| AN3 CA | 15 |
| Skin | |
| MeWo | 92 |
| CHL-1 | 49 |
| RPMI-7951 | 42 |
| HMCB | 31 |
| A375 | 10 |
| Soft tissue | |
| SK-UT-1 | 278 |
| SK-LMS-1 | 28 |
| HT-1080 | 19 |
| MES-SA | 15 |
| MG-63 | 14 |
| Bladder | |
| J82 | 146 |
| TCCSUP | 70 |
| BFTC-905 | 38 |
| UM-UC-3 | 33 |
| 639-V | 25 |
Results of multiplexed cytotoxicity assay on 60 cell lines. IC50 values for tumor growth inhibition are listed with mean values (n = 6).
GU indicates genitourinary; and CNS, central nervous system.
To identify the mechanism of action of CBS9106, we focused on 3 notable effects observed on treatment with this compound: (1) the induction of cell cycle arrest at G1 and G2/M phase, (2) the stabilization of p53, and (3) the reduction of G2/M accumulation (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Stabilization of p53 was not considered to be the predominant mode of action because CBS9106-induced cell cycle arrest was seen regardless of the p53 status of the cell. We noted that a similar phenotype was reported for LMB, a specific CRM1 inhibitor.22,23 Therefore, we considered the possibility that CBS9106 could be a novel CRM1 inhibitor. To investigate this, we analyzed the subcellular localization of RanBP1, which is known to be exported from the nucleus to the cytosol in a CRM1-dependent manner.14 As shown in Figure 1C, RanBP1 is found exclusively in the cytosol in vehicle-treated cells. In contrast, treatment with LMB led to strong accumulation of RanBP1 in the nucleus. With CBS9106, it was found that the treatment also led to nuclear accumulation of RanBP1 in dose- and time-dependent manners (supplemental Figure 2). CRM1-dependent effects on nuclear transport by LMB and CBS9106 were also detected with a luciferase reporter based cellular assay for HIV Rev function. The complex between HIV Rev and the RNA-containing Rev-response element is also known to be recognized by CRM1 and RanGTP.15,24 Both LMB and CBS9106 inhibit HIV Rev function with IC50 values of 0.5nM and 5.5nM, respectively (supplemental Figure 3). These results suggest that CBS9106 functions as a potent CRM1 inhibitor.
CBS9106 decreases CRM1 protein expression levels
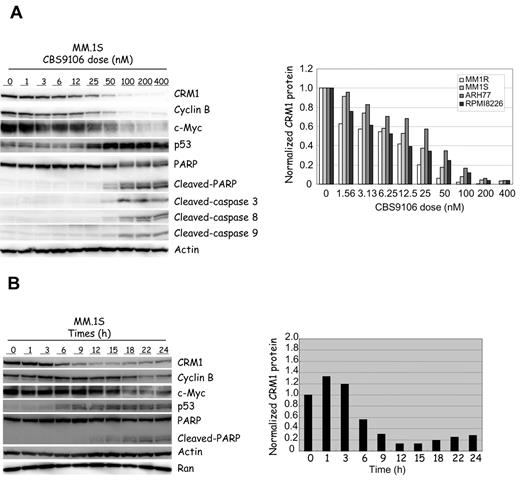
The effect of CBS9106 on the expression level of CRM1 protein in MM.1S cells was analyzed. The level of CRM1 protein expression became markedly reduced in a dose-dependent manner on treatment with CBS9106 (Figure 2A). This effect was reproduced in virtually all of the cancer cell lines analyzed, including 4 MM cells (Figure 2A; supplemental Figure 4A,C). Accompanying the decrease in CRM1 were decreases in the amounts of Cyclin B1 and c-Myc proteins, stabilization of p53, and the induction of apoptosis (cleavage of PARP and caspases). In MM cells, ∼ 80% reduction in CRM1 levels on treatment with CBS9106 coincided with the induction of apoptosis in vitro (Figure 2A; supplemental Figure 4A-B). A time course analysis showed revealed that the reduction of CRM1 protein levels and the up-regulation of p53 are first observed at 3-6 hours after CBS9106 treatment at 100nM. The reduction in cyclin B1 and c-Myc followed this, first becoming apparent ∼ 18 hours after treatment (Figure 2B). This suggests that the reduction of cyclin B1 and c-Myc by CBS9106 is a secondary result of the cell cycle arrest at G1 phase. Indeed, the reduction in cyclin B1 levels by CBS9106 could be reversed by DNA-damaging agents that cause arrest at G2/M phase rather than at G1 phase (data not shown). These results suggest that the inhibition of CRM1 activity by CBS9106 results from the reduction in CRM1 protein expression.
CBS9106 reduces CRM1. Immunoblot (left) of MM.1S cells and the relative expression level of CRM1 (right). (A) MM cells were treated with vehicle or CBS9106 (1-400nM) for 24 hours, and the whole-cell lysates were analyzed by immunoblotting for the presence of indicated proteins with corresponding antibodies. Expression levels of CRM1 protein in 4 MM cell lines were normalized relative to a loading control band of actin and are expressed as ratios relative to the corresponding control. (B) MM.1S cells were treated with CBS9106 (100nM) for 0-24 hours, and the whole-cell lysates were analyzed by immunoblotting (left). Expression levels of CRM1 protein were normalized with a loading control band of actin and are expressed as ratios relative to the corresponding control (right).
CBS9106 reduces CRM1. Immunoblot (left) of MM.1S cells and the relative expression level of CRM1 (right). (A) MM cells were treated with vehicle or CBS9106 (1-400nM) for 24 hours, and the whole-cell lysates were analyzed by immunoblotting for the presence of indicated proteins with corresponding antibodies. Expression levels of CRM1 protein in 4 MM cell lines were normalized relative to a loading control band of actin and are expressed as ratios relative to the corresponding control. (B) MM.1S cells were treated with CBS9106 (100nM) for 0-24 hours, and the whole-cell lysates were analyzed by immunoblotting (left). Expression levels of CRM1 protein were normalized with a loading control band of actin and are expressed as ratios relative to the corresponding control (right).
CBS9106 inhibits NF-κB activity by protection against IκB-α degradation
NF-κB plays important roles in MM cells, and it is one of the important targets for antimyeloma therapy.25 LMB is known to inhibit NF-κB activity by preventing IκB-α degradation through CRM1 inhibition.7,10 Therefore, we considered whether CBS9106 might also inhibit NF-κB activity. Figure 3A shows the inhibition of TNF-α–induced IκB-α degradation in MM.1S cells by CBS9106. TNF-α treatment rapidly induces IκB-α degradation, but this effect is avoided by adding either CBS9106 or LMB. Similar results are observed in RPMI-8226 cells (data not shown). Next, we evaluated the effect of CBS9106 on NF-κB activity by analyzing the DNA-binding activity of the NF-κB p65 subunit with the use of an ELISA-based kit (Figure 3B). Basal levels of NF-κB activity were not changed by either CBS9106 or LMB treatment. In contrast, treatment with CBS9106 or with LMB both lead to inhibition of TNF-α–induced NF-κB activation. These results indicate that CBS9106 may inhibit NF-κB activity.
CBS9106 inhibits TNF-α–induced IκB-α degradation in MM cells. (A) MM.1S cells were treated with vehicle, LMB (10nM), and CBS9106 (50, 200, or 800nM) for 8 hours, followed by vehicle or TNF-α (20 ng/mL) treatment for 25 minutes. The whole-cell lysates were analyzed by immunoblotting. (B) MM.1S cells were treated with vehicle, LMB (10nM), and CBS9106 (200 or 800nM) for 5 hours, followed by vehicle or TNF-α (50 ng/mL) treatment for another 4 hours. The NF-κB activity of the cells was determined by TransAM NF-κB p65 Transcription Factor Assay Kit, according to the manufacturer's instructions. The obtained values (n = 3) are plotted as ratios relative to the corresponding control (vehicle-treated sample). Error bars represent SD.
CBS9106 inhibits TNF-α–induced IκB-α degradation in MM cells. (A) MM.1S cells were treated with vehicle, LMB (10nM), and CBS9106 (50, 200, or 800nM) for 8 hours, followed by vehicle or TNF-α (20 ng/mL) treatment for 25 minutes. The whole-cell lysates were analyzed by immunoblotting. (B) MM.1S cells were treated with vehicle, LMB (10nM), and CBS9106 (200 or 800nM) for 5 hours, followed by vehicle or TNF-α (50 ng/mL) treatment for another 4 hours. The NF-κB activity of the cells was determined by TransAM NF-κB p65 Transcription Factor Assay Kit, according to the manufacturer's instructions. The obtained values (n = 3) are plotted as ratios relative to the corresponding control (vehicle-treated sample). Error bars represent SD.
CBS9106-mediated CRM1 depletion requires ubiquitin/proteasome pathway
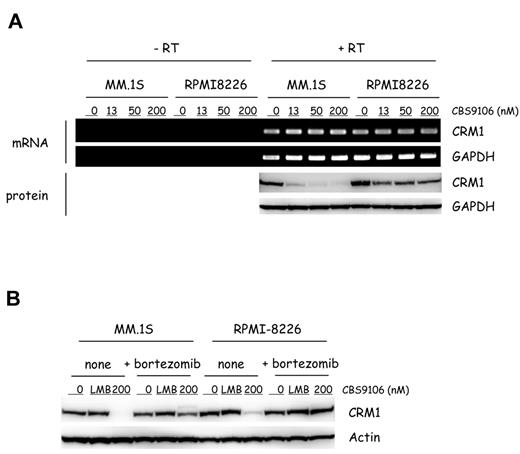
Although the half-life of CRM1 is known to be long,26,27 the levels of CRM1 protein can be reduced by CRM1 small interfering RNA.28 Therefore, we considered whether the effect of CBS9106 might have stemmed from the reduced levels of CRM1 mRNA. To investigate this, CRM1 mRNA and protein levels were analyzed by RT-PCR and immunoblotting. CBS9106 treatment led to the reduction in CRM1 protein levels in a dose-dependent manner. However, the levels of CRM1 mRNA were not changed in both MM.1S and RPMI-8226 cells (Figure 4A). These results indicate that the effects of CBS9106 do not derive from reduced levels of CRM1 mRNA.
CBS9106-induced CRM1 protein reduction depends on ubiquitin/proteasome. (A) MM.1S and RPMI-8226 cells were treated with vehicle or CBS9106 (13, 50, or 200nM) for 5 hours, and the changes of CRM1 mRNA and protein were determined by RT-PCR and immunoblotting, respectively. (B) MM.1S and RPMI-8226 cells were treated with vehicle, LMB (2nM), and CBS9106 (200nM) in the presence or absence of bortezomib (10nM) for 8 hours, and the whole-cell lysates were analyzed by immunoblotting.
CBS9106-induced CRM1 protein reduction depends on ubiquitin/proteasome. (A) MM.1S and RPMI-8226 cells were treated with vehicle or CBS9106 (13, 50, or 200nM) for 5 hours, and the changes of CRM1 mRNA and protein were determined by RT-PCR and immunoblotting, respectively. (B) MM.1S and RPMI-8226 cells were treated with vehicle, LMB (2nM), and CBS9106 (200nM) in the presence or absence of bortezomib (10nM) for 8 hours, and the whole-cell lysates were analyzed by immunoblotting.
The degradation of damaged or misfolded intracellular proteins commonly proceeds through the ubiquitin/proteasome pathway29 ; therefore, it was possible that the reduction in CRM1 protein levels by CBS9106 requires this pathway. To investigate this, we analyzed the effect of bortezomib, a proteasome inhibitor, on CBS9106-induced CRM1 protein reduction. MM.1S or RPMI-8226 cells were treated with LMB (2nM) or CBS9106 (200nM) for 8 hours in the presence or absence of bortezomib (10nM). Reduced CRM1 protein levels were observed in CBS9106-treated cells but not in LMB-treated cells. In contrast, in the presence of bortezomib, depletion of CRM1 protein by CBS9106 was almost completely abolished (Figure 4B). Similar results were obtained with the use of MG132, another proteasome inhibitor (data not shown). These results suggest that CBS9106-induced CRM1 depletion requires the ubiquitin/proteasome pathway.
CBS9106 interacts directly with CRM1 in a competitive manner with LMB
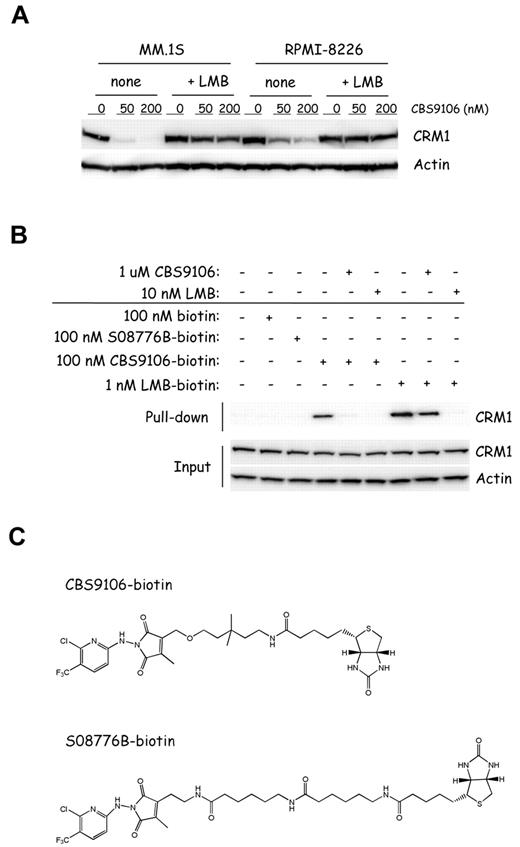
CRM1 inhibitors, including LMB, are known to bind covalently to Cys528 of CRM1 and to inhibit the binding between CRM1 and cargo proteins that have the NES sequence.12-14,16-18 Therefore, we examined whether CBS9106 acted on CRM1 similarly to LMB. Because CBS9106, but not LMB, reduces CRM1 protein levels in the treated cells, we investigated the effect of LMB treatment on the CRM1 reduction by CBS9106. As shown in Figure 5A, the reduction in CRM1 protein levels by CBS9106 was prevented in the presence of LMB, which suggests that CBS9106 and LMB bind competitively with each other for CRM1.
CBS9106 directly binds to CRM1 in a competitive manner with LMB. (A) MM.1S and RPMI-8226 cells were treated with vehicle or CBS9106 (50 or 200nM) in the presence or absence of LMB (2nM) for 8 hours. The whole-cell lysates were analyzed by immunoblotting for CRM1 and actin. (B) RPMI-8226 cells were treated with biotinylated compounds, including biotin (100nM), S08776B-biotin (100nM), CBS9106-biotin (100nM), or LMB-biotin (1nM) for 1 hour in the presence or absence of pretreatment with LMB (10nM) or CBS9106 (1μM) for 1 hour. The whole-cell lysates were subjected to pull-down analysis with the use of streptavidin beads. Captured proteins were analyzed by immunoblotting. (C) The structures of biotinylated compounds CBS9106 (top) and S08776B, an inactive analog of CBS9106 (bottom).
CBS9106 directly binds to CRM1 in a competitive manner with LMB. (A) MM.1S and RPMI-8226 cells were treated with vehicle or CBS9106 (50 or 200nM) in the presence or absence of LMB (2nM) for 8 hours. The whole-cell lysates were analyzed by immunoblotting for CRM1 and actin. (B) RPMI-8226 cells were treated with biotinylated compounds, including biotin (100nM), S08776B-biotin (100nM), CBS9106-biotin (100nM), or LMB-biotin (1nM) for 1 hour in the presence or absence of pretreatment with LMB (10nM) or CBS9106 (1μM) for 1 hour. The whole-cell lysates were subjected to pull-down analysis with the use of streptavidin beads. Captured proteins were analyzed by immunoblotting. (C) The structures of biotinylated compounds CBS9106 (top) and S08776B, an inactive analog of CBS9106 (bottom).
Next, we performed CRM1 pull-down analysis with the use of biotinylated compounds LMB-biotin,11 CBS9106-biotin (Figure 5C top), and S08776B-biotin, an inactive form of CBS9106 (Figure 5C bottom). As predicted, CRM1 was pulled down from the lysates of the cells treated with LMB-biotin and CBS9106-biotin but not S08776B-biotin (Figure 5B lanes 3,4,7). The binding between CRM1 and CBS9106-biotin or LMB-biotin was prevented by the addition of free CBS9106 or LMB, respectively. However, the competitive effect of free LMB was higher than that of free CBS9106 (Figure 5B lanes 5-6 and lane 8-9). These results indicate that binding of CBS9106 and LMB to CRM1 is mutually exclusive and that CBS9106 binds to CRM1 at or near the LMB covalent binding site at Cys528.
CBS9106 binds Cys528 of CRM1 and the binding is reversible
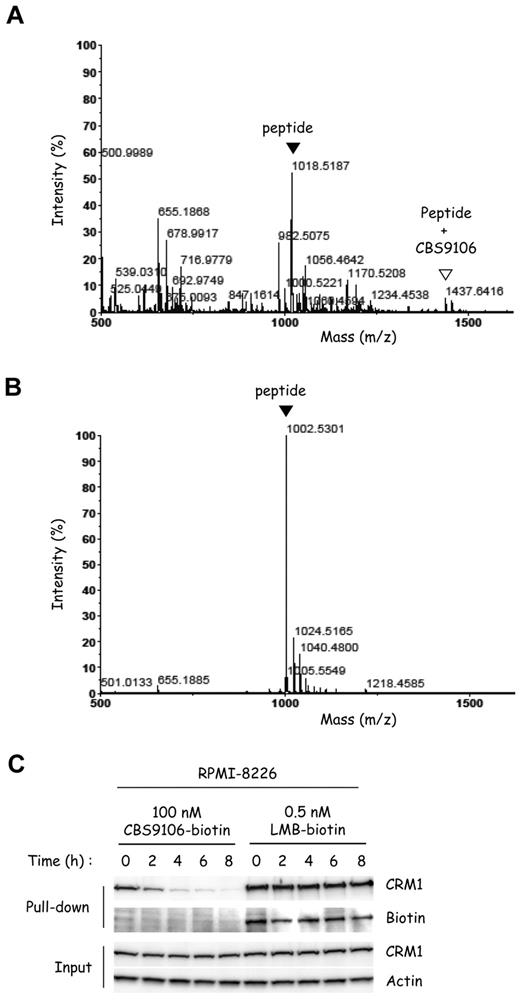
LMB binds CRM1 irreversibly by a Michael-type addition11 ; however, this has been thought to be an undesirable mode of action because of its toxic side effect.22 Therefore, it was important to examine whether CBS9106 also binds covalently or tightly to CRM1. First, we examined whether CBS9106 also binds covalently to Cys528 of CRM1, as LMB does,11 by mass spectrometry (MS). Synthetic peptide fragments of CRM1 containing either Cys (CRM1 wild-type peptide, WT) or Ser (CRM1 mutant peptide, MT) at the position corresponding to Cys528 in CRM1 were treated with CBS9106 and analyzed by MALDI-TOF MS. Peaks at both m/z 1018.5187 and 1437.6416, which correspond to the CRM1 WT peptide and its CBS9106 adduct, respectively, could be detected in the sample derived from CRM1 WT peptide (Figure 6A). However, no peak corresponding to a CBS9106 adduct could be observed near m/z 1421.6695, for the sample derived from the CRM1 MT peptide (Figure 6B). The result indicates that CBS9106 can bind covalently to CRM1 at Cys528.
The covalent binding of CBS9106 to Cys528 of CRM1 is reversible. Synthetic peptides (CRM1 WT peptide) containing Cys528 of CRM1 (A) or the mutant peptides (CRM1 MT peptide) in which Cys528 was substituted with Ser (B) were treated with CBS9106 at 37°C for 24 hours and analyzed by MALDI-TOF MS. The black arrowheads correspond to the free peptide, and the white arrowhead corresponds to the adduct of the peptide with CBS9106. (C) RPMI-8226 cells were treated with biotinylated compounds CBS9106-biotin (100nM) or LMB-biotin (0.5nM) for 1 hour, changed to fresh medium, and incubated for the indicated period (0-8 hours). The whole-cell lysates were subjected to pull-down analysis with the use of streptavidin beads. Captured proteins were analyzed by immunoblotting.
The covalent binding of CBS9106 to Cys528 of CRM1 is reversible. Synthetic peptides (CRM1 WT peptide) containing Cys528 of CRM1 (A) or the mutant peptides (CRM1 MT peptide) in which Cys528 was substituted with Ser (B) were treated with CBS9106 at 37°C for 24 hours and analyzed by MALDI-TOF MS. The black arrowheads correspond to the free peptide, and the white arrowhead corresponds to the adduct of the peptide with CBS9106. (C) RPMI-8226 cells were treated with biotinylated compounds CBS9106-biotin (100nM) or LMB-biotin (0.5nM) for 1 hour, changed to fresh medium, and incubated for the indicated period (0-8 hours). The whole-cell lysates were subjected to pull-down analysis with the use of streptavidin beads. Captured proteins were analyzed by immunoblotting.
Next, knowing that LMB binds irreversibly to CRM1,11 we examined whether CBS9106 also binds irreversibly. We measured the time course of changes in the amount of biotinylated compound-associated CRM1 after switching cells from compound-containing to compound-free media. Immediately after the exchange of the cells to compound-free medium, CRM1 could be pulled down from lysates of the cells treated with either LMB-biotin or CBS9106-biotin. However, the amount of CRM1 protein pulled down with CBS9106-biotin became reduced in a time-dependent manner (Figure 6C). In contrast, the amount of CRM1 protein pulled down with LMB-biotin did not change. Moreover, biotin could be detected at a position corresponding to the molecular size of CRM1 only for protein pulled down with LMB-biotin, but not with CBS9106-biotin. These results are consistent with reversible binding by CBS9106 to CRM1 and show a feature of binding by CBS9106 to CRM1 that differs markedly from the irreversible binding exhibited by LMB.
Effects of CBS9106 in vivo
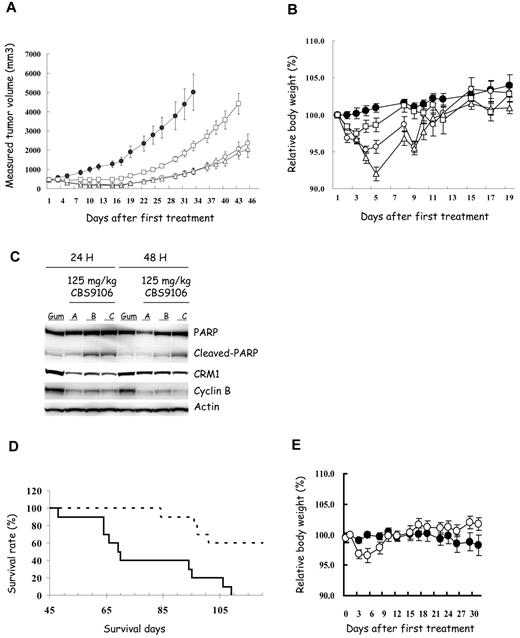
SCID mice were inoculated subcutaneously with RPMI-8226 cells for a tumor growth study (Figure 7A-C) and were inoculated intravenously with MM.1S cells for a survival study (Figure 7D-E). Orally administered CBS9106 at a dose level of 31.25 mg/kg 5 times weekly showed slight inhibition of tumor growth that was not statistically significant relative to vehicle administration. However, at dose levels of 62.5 mg/kg (5 doses/week) or 125 mg/kg (3 doses/week), CBS9106 showed statistically significant tumor growth inhibition (P < .01; Figure 7A). To examine the in vivo relevance of the in vitro results, the effects of CBS9106 on protein expression were analyzed in tumor cells isolated from mice. The xenograft tumors were harvested from mice at 24 or 48 hours after treatment with an orally administered, single dose of CBS9106 (125 mg/kg). CBS9106-induced reduction in CRM1 and cyclin B and cleavage of PARP were observed at 24 hours after drug administration, and these effects began to be reversed at 48 hours after drug administration (Figure 7C). Thus, the effects of CBS9106 observed in in vitro experiments could also be observed in vivo. In addition, as shown in a Kaplan-Meier graph, a statistically significant prolongation of survival time could be observed in mice treated with CBS9106 (125 mg/kg/d, 3 doses/week; P < .001; Figure 7D). Most importantly, administration of CBS9106 was well tolerated in mice and resulted in no significant body weight loss in both studies (Figure 7B,E). These results suggest that CBS9106 is a promising clinical candidate for the treatment of MM cells.
Efficacy of CBS9106 in human MM xenograft models. Subcutaneous tumor growth curves (A) and the body weight change (B) of subcutaneous xenograft model of RPMI-8226 (n = 8). Mice were treated orally with vehicle (Gum Arabic, ●) or CBS9106 (31.25 mg/kg, 5 times weekly, □; 62.5 mg/kg, 5 times weekly, ▵; or 125 mg/kg, 3 times weekly, ○) for 2 weeks. (C) Tumors from RPMI-8226 cell xenografts were harvested at 24 or 48 hours from the mice treated orally with vehicle (Gum Arabic, Gum) or CBS9106 (125 mg/kg, A-C). Protein extracts prepared from the tumors were analyzed by immunoblotting to detect indicated proteins. (D) Survival analysis of the MM.1S xenograft model mice (n = 10) with oral CBS9106 treatment (125 mg/kg, 3 times weekly, dotted line) for 2 weeks. (E) Body weight change for panel D. Error bars represent SE.
Efficacy of CBS9106 in human MM xenograft models. Subcutaneous tumor growth curves (A) and the body weight change (B) of subcutaneous xenograft model of RPMI-8226 (n = 8). Mice were treated orally with vehicle (Gum Arabic, ●) or CBS9106 (31.25 mg/kg, 5 times weekly, □; 62.5 mg/kg, 5 times weekly, ▵; or 125 mg/kg, 3 times weekly, ○) for 2 weeks. (C) Tumors from RPMI-8226 cell xenografts were harvested at 24 or 48 hours from the mice treated orally with vehicle (Gum Arabic, Gum) or CBS9106 (125 mg/kg, A-C). Protein extracts prepared from the tumors were analyzed by immunoblotting to detect indicated proteins. (D) Survival analysis of the MM.1S xenograft model mice (n = 10) with oral CBS9106 treatment (125 mg/kg, 3 times weekly, dotted line) for 2 weeks. (E) Body weight change for panel D. Error bars represent SE.
Discussion
This is the first detailed report on a novel compound, CBS9106, and on its proposed mechanism of action, as a unique reversible CRM1 inhibitor. We demonstrate that CBS9106 suppresses growth of a variety of tumor cells, including MM cells, and reduces CRM1 protein levels both in vitro and in vivo (Figures 2A, 7C, and supplemental Figure 4A,C). Pull-down analysis shows that CBS9106 binds directly to CRM1 and that the interaction of CRM1 with CBS9106 is blocked by LMB and vice versa (Figure 5B). Similar to observations with LMB,30 the activity of CBS9106 can be suppressed by N-acetylcysteine (data not shown), which indicates that CBS9106 interacts selectively with thiols. Mass spectrometric analysis showed that CBS9106 could bind directly to a synthetic CRM1-derived peptide containing Cys528, but not to a mutated peptide (Cys528Ser) lacking Cys (Figure 6A-B). However, binding of CBS9106 to CRM1 appears to be reversible in contrast to LMB-CRM1 binding. A biotinylated complex with the molecular size of CRM1 could be detected in pull-down experiments with LMB-biotin but could not be detected in similar experiments with CBS9106-biotin (Figure 6C). These combined results suggest that, although CBS9106 binding to CRM1 is covalent on Cys528 of CRM1, it is reversible. The reversibility of CBS9106 binding offers an explanation for the reduced efficiency of CBS9106 versus LMB in competitive pull-down assays (Figure 5B). Whereas CBS9106 and most other CRM1 inhibitors target Cys528 of CRM1,12-18 to the best of our knowledge, only CBS9106 induces CRM1 depletion via the ubiquitin/proteasome pathway (Figures 4B and 5A). This CBS9106-induced CRM1 depletion was suppressed in the presence of LMB (Figure 5A), suggesting that binding to Cys528 of CRM1 is indispensable for CBS9106-induced CRM1 depletion.
The differences between CBS9106 and LMB in their interaction with CRM1 may lead to key differences in pharmacologic effectiveness. Although LMB possesses strong antitumor activity in vitro, phase 1 trials of LMB were discontinued because of its toxicity and lack of apparent efficacy in the tolerable dose ranges.19 Mutka et al14 reported that some LMB analogues were less toxic, exhibiting improved tolerance in vivo by up to a factor of 16 over LMB, and the loss of body weight observed on treatment with these LMB-analogues was < 10%. Those researchers also noted that LMB has off-target effects against proteins other than CRM1 and that these off-target effects might contribute to LMB's side effects (∼ 20% body weight loss). In contrast, multiple doses of CBS9106 are well tolerated (< 5% body weight loss), and CBS9106 exhibits high antitumor activity in mouse xenograft models (Figure 7). Similar activity is also observed in other solid tumor xenograft mouse models (NCI-H226 and 22RV1 cells; data not shown). Orally administered CBS9106 at a dose level of 125 mg/kg, 3 times weekly, was found to be the most effective protocol for such in vivo experiments in our laboratory, and under these conditions CBS9106 is well tolerated (∼ 5%-10% body weight loss; data not shown).
The reduction in CRM1 protein levels, which is observed with CBS9106 but not with LMB, shows a strong correlation with the efficacy of CBS9106 in vitro. When the CRM1 protein levels in vitro are depleted to < 20% of that found for untreated cells, apoptosis was induced in all analyzed MM cell lines (Figure 2A; supplemental Figure 4A-B). CBS9106-induced CRM1 depletion and the induction of apoptosis were reproduced in studies with an in vivo xenograft model (Figure 7C). Therefore, the expression level of CRM1 protein may serve as a surrogate marker to determine a suitable dosing schedule of CBS9106 in the future clinical studies.
Finally, we showed that CBS9106 and LMB both inhibit intracellular processes that require on the nuclear export function of CRM1. Examination of the effects on HIV Rev activity provided a quantitative estimate of the strength of this interaction and further show CBS9106's effectiveness and reduced toxicity relative to LMB. Such activity, which requires the presence of active CRM1, is inhibited by both LMB and CBS9106 with IC50 values of 0.5nM and 5.5nM, respectively. However, the 50% toxicity concentration values of LMB and CBS9106 were 1.4nM and 126nM, respectively (supplemental Figure 3). Thus, CBS9106's suppression of HIV Rev exhibits a wider therapeutic window than that of LMB in vitro whereby CRM1 suppression is believed to be the mechanism for suppression of HIV Rev activity. These results suggest that CBS9106 should also be expected to have a better toxicity profile than LMB in vivo for this activity.
Another CRM1-dependent process affected by both LMB and CBS9106 was found to be the activation of NF-κB via protection against TNF-α–induced IκB-α degradation in MM.1S cells (Figure 3). NF-κB plays important roles in MM cells and anti-MM agents used in the clinic such as dexamethasone and thalidomide/immunomodulatory drugs inhibit NF-κB activity.25,31-33 Therefore, inhibition of NF-κB activity is a reasonable explanation for antimyeloma activity of CBS9106. Because of the variety of NES-containing proteins transported by CRM1, including IκB-α,1-10 CBS9106 is expected to affect numerous intracellular pathways. Matsuyama et al34 showed that CRM1 was responsible for the intracellular localization of 285 proteins in the fission yeast. Current anti-MM agents, including dexamethasone, bortezomib, and thalidomide/immunomodulatory drugs, also affect numerous pathways.31,33,35 Thus, NF-κB seems to be a key factor that may be responsible for the antimyeloma effect of CBS9106. Further investigation is needed to pinpoint unambiguously the specific factors that contribute to the anti-MM effect of CBS9106.
In summary, results of this study show that CBS9106 is a novel CRM1 inhibitor that acts by depleting cells of CRM1 protein reversibly, as shown both in vitro and in vivo. Taken together, CBS9106 is a novel anticancer drug candidate for the treatment of diverse malignancies, including MM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr W. G. Dunphy for scientific discussion.
Authorship
Contribution: K.S. designed and performed the experiments and drafted the manuscript; N.S., T.S., A.S., and Y.H. performed experiments; T.I. arranged the large-scale synthesis of CBS9106; D.W.K., D.D.V., and T.K. discussed the plan and results of the experiments; D.W.K., D.D.V., J.M.F., and T.K. critically reviewed and wrote the manuscript; and T.K. supervised the research.
Conflict-of-interest disclosure: T.S., Y.H., and T.K. are among the inventors of a product related to the work described in this study. K.S., N.S., T.S., A.S., Y.H., J.M.F., T.I., and T.K. are employed by and D.W.K and D.D.V. are consultants to CanBas Co Ltd, whose product was studied in the present work.
Correspondence: Takumi Kawabe, CanBas Co Ltd, 2-2-1, Otemachi, Numazu 410-0801, Japan; e-mail: takumi@canbas.co.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal