Abstract
The studies concerning clinical implications of TET2 mutation in patients with primary acute myeloid leukemia (AML) are scarce. We analyzed TET2 mutation in 486 adult patients with primary AML. TET2 mutation occurred in 13.2% of our patients and was closely associated with older age, higher white blood cell and blast counts, lower platelet numbers, normal karyotype, intermediate-risk cytogenetics, isolated trisomy 8, NPM1 mutation, and ASXL1 mutation but mutually exclusive with IDH mutation. TET2 mutation is an unfavorable prognostic factor in patients with intermediate-risk cytogenetics, and its negative impact was further enhanced when the mutation was combined with FLT3-ITD, NPM1-wild, or unfavorable genotypes (other than NPM1+/FLT3-ITD− or CEBPA+). A scoring system integrating TET2 mutation with FLT3-ITD, NPM1, and CEBPA mutations could well separate AML patients with intermediate-risk cytogenetics into 4 groups with different prognoses (P < .0001). Sequential analysis revealed that TET2 mutation detected at diagnosis was frequently lost at relapse; rarely, the mutation was acquired at relapse in those without TET2 mutation at diagnosis. In conclusion, TET2 mutation is associated with poor prognosis in AML patients with intermediate-risk cytogenetics, especially when it is combined with other adverse molecular markers. TET2 mutation appeared to be unstable during disease evolution.
Introduction
Mutations in Ten-Elevan-Translocation-2 (TET2) were first discovered in myeloid malignancies by high-resolution single nucleotide polymorphism (SNP) and comparative genomic hybridization arrays.1 Subsequent studies using the same methods, direct sequencing, or next-generation sequencing confirmed that mutations in this gene were prevalent in myelodysplastic syndrome, myelodysplastic syndrome/myeloproliferative neoplasms, myeloproliferative neoplasms, and secondary acute myeloid leukemia (AML), with frequencies ∼ 10%-26%, 22%-58%, 7%-13%, and 24%-32%, respectively.2-14 The studies concerning clinical implications of TET2 mutation in patients with primary AML are scarce. There are several unresolved issues relating to TET2 mutations in primary AML. First, the association of TET2 mutations with other genetic alterations has not been fully addressed. Whereas one study showed positive association of TET2 mutation with NPM1 mutation in AML patients achieving complete remission (CR),15 other reports did not find such correlation.16,17 Second, the prognostic significance of TET2 mutation in AML is still controversial. Nibourel et al15 did not find any prognostic impact of TET2 mutation in primary AML achieving CR, whereas another study suggested an unfavorable effect of this mutation in primary cytogenetically normal AML patients bearing favorable genotypes (mutated NPM1 without FLT3-ITD or CEBPA mutation, NPM1+/FLT3-ITD− or CEBPA+).17 Third, the stability of TET2 mutations during disease evolution in AML remains unknown.
To clarify these points, we analyzed TET2 mutation in a cohort of 486 adult patients with de novo AML and correlated the result with clinical and biologic features and the status of 15 other important genetic mutations. Sequential studies were performed on 122 patients to investigate the serial changes of this mutation as the disease goes into CR and relapse. We found that the TET2 mutation was associated with several distinct clinical features not reported so far. In AML patients bearing intermediate-risk cytogenetics according to Southwest Oncology Group (SWOG) criteria,18 this mutation appears to be a poor prognostic factor for overall survival (OS). Importantly, the negative impact of TET2 mutation in this group of AML patients was significantly enhanced by combination with other unfavorable factors, including the presence of FLT3-ITD, absence of NPM1 mutation, or presence of molecular markers other than NPM1+/FLT3-ITD− or CEBPA+. A scoring system incorporating TET2 mutation with FLT3-ITD, NPM1, and CEBPA double mutations (CEBPAdouble) into survival analysis proved very useful to stratify AML patients with intermediate-risk cytogenetics into different prognostic groups (P < .0001). TET2 mutation detected at diagnosis frequently disappeared at relapse. Infrequently, this mutation was acquired at disease relapse in patients without mutation in this gene at diagnosis.
Methods
Patients
A total of 486 consecutive adult patients (≥ 15 years of age) with newly diagnosed de novo AML from 1995 to 2007 at the National Taiwan University Hospital who had adequate cryopreserved bone marrow cells for complete mutation analyses were recruited. Written informed consent in accordance with the Declaration of Helsinki was obtained from all participants, and the study was approved by the Institutional Review Board of the National Taiwan University Hospital.
Among these 486 primary AML patients, 343 (70.6%) received standard intensive chemotherapy as described previously.19 The remaining 143 patients (139 with non-M3 AML and 4 with acute promyelocytic leukemia) received palliative care or low-dose chemotherapy because of poor performance status or per patients' wish.
Mutation analyses
Mutation analyses for TET2 (NM_001127208) were performed by PCR largely as previously described but with mild modification.1 The primer sequences and estimated product lengths were listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Mutations on CEBPA,20 WT1,21 MLL-PTD,22 PTPN11,23,24 RUNX1,25 c-KIT,26 RAS,27 FLT3-TKD,28 IDH,29,30 IDH2,31 ASXL1,19 NPM1,32 and FLT3-ITD33,34 were performed as reported. To detect TET2 mutation at diagnosis, we used DNA amplified in vitro from patients' bone marrow cells by Illustra GenomiPhi V2 DNA amplification kit as described by the manufacturer (GE Healthcare). All the mutations detected in such samples were verified in the original nonamplified samples. All the nucleotide alterations causing premature truncation of the TET2 proteins (non-sense or frame shifting) were regarded as true mutations. Missense mutations were regarded as true only if they were documented in other papers or could be verified in somatic tissue or remission marrow samples in our patients. The patients bearing other missense mutations not known to be somatic were censored from this study.
Cytogenetic analyses and immunophenotyping
Statistics
The discrete variables of patients with and without TET2 mutation were compared using the χ2 tests or Fisher exact test. Mann-Whitney tests were used to compare continuous variables and medians of distributions. OS was measured from the date of first diagnosis to death from any cause, and relapse-free survival was calculated from the time of CR until relapse, death, or end of study. Kaplan-Meier survival curves and log-rank test were used for estimation of survival and difference between groups. Multivariate Cox proportional hazard regression analysis was used to evaluate independent prognostic factors for survival. All statistical analyses were performed using XLSTAT statistical analysis software (edition 2010 Version 5.02, Addinsoft). Whole patient population (N = 486) was included for analyses of the correlation between TET2 mutation and clinical characteristics, but only those 343 patients who received standard chemotherapy were included in analyses of survivals as previously described.19,21,25 In patients who had received allogeneic stem cell transplantation, OS was censored on the date of stem cell infusion. Bootstrapping for Kaplan-Meier estimation was conducted using SAS Version 9.0.
Results
Correlation of TET2 mutations with clinical features and laboratory data
A total of 486 patients, consisting of 212 females and 274 males with a median age of 51.5 years (range, 15-90 years) were recruited for TET2 mutation analysis. Seventy-six different TET2 mutations were detected in 64 patients (13.2%). Double mutations of TET2 were detected in 27 patients (27 of 64, 42.2%), and 6 of them had homozygous TET2 mutation. There were 27 non-sense, 33 frameshift, and 15 missense mutations, distributing across the whole coding sequence without obvious hot spots, some occurring in more than one patient (Figure 1). Some nucleotide variations were not regarded as true missense mutations: P29R, I1762V, and V218M were documented as SNPs in dbSNP (http://www.ncbi.nlm.nih.gov/snp/); P363L, L1721W, and H1778R were reported previously as SNPs15 ; and R814C, F868L, S1039L, E1513G, L1248T (resulted from in-frame deletion c.3742_3750del), and R1543P (resulted from in-frame deletion c.4627_4644del) were obviously retained in remission samples. The significance of some missense mutations, including C1374Y, H1219N, P1889H, H1868D, L1322Q, and L1326S, remained unknown, and these were thus censored from the analyses of this study. The detailed information of the 64 patients with TET2 mutations is listed in supplemental Table 2. Patients with TET2 mutation were significantly older than those without this mutation (68 years vs 48 years, P < .001); in patients older than 60 years, approximately one-fourth (24.5%, 42 of 172) had TET2 mutation, in contrast to only 7% in younger patients (60 years of age or younger; Table 1). Patients with TET2 mutations had significantly higher WBC count, higher peripheral blast count, higher serum lactate dehydrogenase level, and lower platelet count at diagnosis than those without TET2 mutations, but the levels of hemoglobin were not different (Table 1). TET2 mutations were not seen in patients with acute promyelocytic leukemia (Table 1). Leukemia cells with TET2 mutations had higher frequency of CD14 and CD56 expression (supplemental Table 3). There were no differences in various clinical and laboratory characteristics and survival between patients with a single and those with double mutations (data not shown).
The mutations of TET2. The black ticks and gray ticks represent nonsense/frameshift and missense mutations, respectively. The 2 arrows indicate conserved regions (amino acids 1134-1444 and 1842-1921). Some mutations occurred in > 1 patient.
The mutations of TET2. The black ticks and gray ticks represent nonsense/frameshift and missense mutations, respectively. The 2 arrows indicate conserved regions (amino acids 1134-1444 and 1842-1921). Some mutations occurred in > 1 patient.
Correlation of TET2 mutations with clinical data, FAB subtypes, and chromosomal abnormalities in AML patients (n = 486)
| Variant . | Total (N = 486) . | TET2 mutation (N = 64, 13.2%) . | TET2 wild-type (N = 422, 86.8%) . | P . |
|---|---|---|---|---|
| Median age, y (range) | 51.5 (15-90) | 68 (21-90) | 48 (15-89) | < .001 |
| Age in groups, y | < .001 | |||
| 15-20 | 23 | 0 | 23 (100) | |
| 21-30 | 57 | 3 (5.3) | 54 (94.7) | |
| 31-40 | 78 | 7 (9.0) | 71 (91.0) | |
| 41-50 | 78 | 4 (5.1) | 74 (94.9) | |
| 51-60 | 78 | 8 (10.3) | 70 (89.7) | |
| 61-70 | 78 | 15 (19.2) | 63 (80.8) | |
| 71-80 | 69 | 17 (24.6) | 52 (75.4) | |
| > 80 | 25 | 10 (40.0) | 15 (60.0) | |
| Sex | .136 | |||
| Female | 212 | 22 (10.4) | 190 (89.6) | |
| Male | 274 | 42 (15.3) | 232 (84.7) | |
| Median laboratory data (range) | ||||
| WBC, × 103/μL | 17.91 (0.12-627.8) | 43.16 (1.68-277.25) | 15.44 (0.12-627.8) | < .001 |
| Blasts, × 103/μL | 7.33 (0-456.73) | 26.53 (0-260.62) | 5.80 (0-456.73) | < .001 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 8.1 (3.5-16.2) | 8.0 (2.9-13.9) | .196 |
| Platelets, × 103/μL | 43 (3-802) | 33 (6-157) | 45 (3-802) | .017 |
| LDH, U/L | 855.5 (206-13 130) | 1248 (365-7930) | 823 (206-13 130) | .004 |
| FAB | ||||
| M0 | 10 | 0 | 10 (100) | .371 |
| M1 | 108 | 13 (12.0) | 95 (88.0) | .749 |
| M2 | 168 | 28 (16.7) | 140 (83.3) | .169 |
| M3 | 38 | 0 | 38 (100) | .005 |
| M4 | 121 | 20 (16.5) | 101 (83.5) | .290 |
| M5 | 21 | 3 (14.3) | 18 (85.7) | .748 |
| M6 | 11 | 0 | 11 (100) | .375 |
| Undetermined | 9 | 0 | 9 (100) | |
| Karyotype* SWOG | ||||
| Favorable | 96 | 5 (5.2) | 91 (94.8) | .007 |
| Intermediate | 244 | 45 (18.5) | 198 (81.5) | < .001 |
| Unfavorable | 128 | 11 (8.6) | 117 (91.4) | .091 |
| Normal karyotype | 218 | 39 (18.0) | 178 (82.0) | .006 |
| Isolated trisomy 8 | 17 | 6 (35.3) | 11 (64.7) | .017 |
| Variant . | Total (N = 486) . | TET2 mutation (N = 64, 13.2%) . | TET2 wild-type (N = 422, 86.8%) . | P . |
|---|---|---|---|---|
| Median age, y (range) | 51.5 (15-90) | 68 (21-90) | 48 (15-89) | < .001 |
| Age in groups, y | < .001 | |||
| 15-20 | 23 | 0 | 23 (100) | |
| 21-30 | 57 | 3 (5.3) | 54 (94.7) | |
| 31-40 | 78 | 7 (9.0) | 71 (91.0) | |
| 41-50 | 78 | 4 (5.1) | 74 (94.9) | |
| 51-60 | 78 | 8 (10.3) | 70 (89.7) | |
| 61-70 | 78 | 15 (19.2) | 63 (80.8) | |
| 71-80 | 69 | 17 (24.6) | 52 (75.4) | |
| > 80 | 25 | 10 (40.0) | 15 (60.0) | |
| Sex | .136 | |||
| Female | 212 | 22 (10.4) | 190 (89.6) | |
| Male | 274 | 42 (15.3) | 232 (84.7) | |
| Median laboratory data (range) | ||||
| WBC, × 103/μL | 17.91 (0.12-627.8) | 43.16 (1.68-277.25) | 15.44 (0.12-627.8) | < .001 |
| Blasts, × 103/μL | 7.33 (0-456.73) | 26.53 (0-260.62) | 5.80 (0-456.73) | < .001 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 8.1 (3.5-16.2) | 8.0 (2.9-13.9) | .196 |
| Platelets, × 103/μL | 43 (3-802) | 33 (6-157) | 45 (3-802) | .017 |
| LDH, U/L | 855.5 (206-13 130) | 1248 (365-7930) | 823 (206-13 130) | .004 |
| FAB | ||||
| M0 | 10 | 0 | 10 (100) | .371 |
| M1 | 108 | 13 (12.0) | 95 (88.0) | .749 |
| M2 | 168 | 28 (16.7) | 140 (83.3) | .169 |
| M3 | 38 | 0 | 38 (100) | .005 |
| M4 | 121 | 20 (16.5) | 101 (83.5) | .290 |
| M5 | 21 | 3 (14.3) | 18 (85.7) | .748 |
| M6 | 11 | 0 | 11 (100) | .375 |
| Undetermined | 9 | 0 | 9 (100) | |
| Karyotype* SWOG | ||||
| Favorable | 96 | 5 (5.2) | 91 (94.8) | .007 |
| Intermediate | 244 | 45 (18.5) | 198 (81.5) | < .001 |
| Unfavorable | 128 | 11 (8.6) | 117 (91.4) | .091 |
| Normal karyotype | 218 | 39 (18.0) | 178 (82.0) | .006 |
| Isolated trisomy 8 | 17 | 6 (35.3) | 11 (64.7) | .017 |
FAB indicates French-American-British; and LDH, lactate dehydrogenase.
Only 468 patients had available cytogenetic data at diagnosis (62 TET2-muted patients and 406 TET2-wild patients).
Correlation of TET2 mutation with karyotype and other genetic alterations
Chromosome data were available in 467 patients (Table 1). TET2 mutation occurred more frequently in patients with intermediate-risk cytogenetics according to SWOG criteria18 (18.5% vs 7.1%, P < .001), normal karyotype (18.0% vs 9.2%, P = .0064), or isolated trisomy 8 (35.3% vs 12.6%, P = .017; Table 1). TET2 mutation was mutually exclusive with IDH mutations (P < .001) but had positive association with ASXL1 mutation (P = .002) and NPM1 mutation (P = .047; Table 2).
Correlation of TET2 mutation with other gene alterations
| Mutation . | Total (n = 486) . | TET2-mutated (n = 64) . | TET2-wild (n = 422) . | P . |
|---|---|---|---|---|
| NPM1 | 102 (21.0) | 20 (31.3) | 82 (19.4) | .047 |
| FLT3-ITD | 113 (23.3) | 14 (21.9) | 99 (23.5) | .847 |
| NPM1+/FLT3-ITD− | 50 (10.3) | 10 (15.6) | 40 (9.5) | .181 |
| CEBPAdouble | 45 (9.3) | 8 (12.5) | 37 (8.8) | .353 |
| WT1 | 32 (6.6) | 2 (3.1) | 30 (7.1) | .291 |
| RUNX1 | 58 (11.9) | 8 (12.5) | 50 (11.8) | .837 |
| IDH | 80 (16.5) | 1 (1.6) | 79 (18.7) | < .001 |
| FLT3-TKD | 40 (8.2) | 4 (6.3) | 36 (8.5) | .806 |
| MLL-PTD | 25 (5.1) | 2 (3.1) | 23 (5.5) | .759 |
| KIT | 14 (2.9) | 0 | 14 (3.3) | .233 |
| NRAS | 56 (11.5) | 6 (9.4) | 50 (11.8) | .678 |
| KRAS | 17 (3.5) | 1 (1.6) | 16 (3.8) | .712 |
| ASXL1 | 53 (10.9) | 15 (23.4) | 38 (9) | .002 |
| Mutation . | Total (n = 486) . | TET2-mutated (n = 64) . | TET2-wild (n = 422) . | P . |
|---|---|---|---|---|
| NPM1 | 102 (21.0) | 20 (31.3) | 82 (19.4) | .047 |
| FLT3-ITD | 113 (23.3) | 14 (21.9) | 99 (23.5) | .847 |
| NPM1+/FLT3-ITD− | 50 (10.3) | 10 (15.6) | 40 (9.5) | .181 |
| CEBPAdouble | 45 (9.3) | 8 (12.5) | 37 (8.8) | .353 |
| WT1 | 32 (6.6) | 2 (3.1) | 30 (7.1) | .291 |
| RUNX1 | 58 (11.9) | 8 (12.5) | 50 (11.8) | .837 |
| IDH | 80 (16.5) | 1 (1.6) | 79 (18.7) | < .001 |
| FLT3-TKD | 40 (8.2) | 4 (6.3) | 36 (8.5) | .806 |
| MLL-PTD | 25 (5.1) | 2 (3.1) | 23 (5.5) | .759 |
| KIT | 14 (2.9) | 0 | 14 (3.3) | .233 |
| NRAS | 56 (11.5) | 6 (9.4) | 50 (11.8) | .678 |
| KRAS | 17 (3.5) | 1 (1.6) | 16 (3.8) | .712 |
| ASXL1 | 53 (10.9) | 15 (23.4) | 38 (9) | .002 |
Values are no. (%) of patients.
Survival analysis
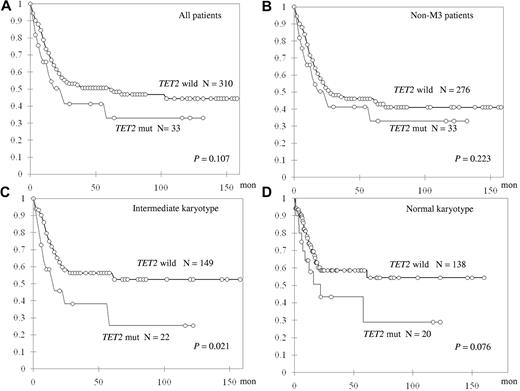
TET2-mutated patients showed only a trend of shorter OS in all 343 patients who received standard intensive chemotherapy (median, 61.1 vs 22.2 months, P = .107; Figure 2A). Subgroup analysis showed that, among 310 non-M3 AML patients, TET2 mutation did not have a significant impact on OS or relapse-free survival (Figure 2B; and data not shown). However, TET2 mutations were associated with shorter OS in the patients with intermediate-risk cytogenetics (N = 171) and a trend of shorter OS in those with a normal karyotype (N = 158, median not reached [NR] vs 14.7 months, P = .021 and NR vs 22.1 months, P = .076, respectively; Figure 2C-D). There was no difference in induction-related mortality, CR rate, or relapse-free survival between TET2-mutated patients and TET2-wild patients in any subgroup (data not shown).
Kaplan-Meier curves for OS stratified by the status of TET2 mutations in different subgroups of patients.
Kaplan-Meier curves for OS stratified by the status of TET2 mutations in different subgroups of patients.
Univariate analyses in patients with intermediate-risk cytogenetics (N = 171) showed that TET2 mutation, as well as older age, higher WBC count, and mutations of RUNX1 and ASXL1, was an unfavorable prognostic factor for OS, whereas CEBPAdouble was a good prognostic factor and mutation of NPM1 without FLT3-ITD (NPM1+/FLT3-ITD−) had a trend to be a favorable factor for OS (Table 3). In multivariate analysis, only 3 factors remained to be independent prognostic factors for OS: WBC count, NPM1+/FLT3-ITD−, and CEBPAdouble; old age and TET2 mutation had a trend toward poor OS (Table 4). TET mutation was not an independent prognostic factor for OS in patients with a normal karyotype, either, probably because of smaller patient number. We also analyzed the effects of 3 common SNPs in TET2, including P29R (N = 168), I1762V (N = 164), and V218M (N = 66) on OS but could not find any significant prognostic impact of these SNP variants (data not shown).
Univariate analysis of the impact of clinical parameters and molecular alterations on OS in AML patients with intermediate-risk karyotype (N = 171)
| Variable . | OS, mo, median ± SD . | P . |
|---|---|---|
| Age, y | < .001 | |
| ≤ 60 | NR | |
| > 60 | 12.3 ± 4.6 | |
| WBC | .001 | |
| ≤ 50 000/μL | NR | |
| > 50 000/μL | 14.5 ± 3.6 | |
| NPM1+/FLT3-ITD− | .080 | |
| Yes | NR | |
| Others | 61.0 | |
| CEBPA | .010 | |
| Double mutation | NR | |
| Others | 22.0 ± 16.7 | |
| RUNX1 | .015 | |
| Mutated | 14.5 ± 3.8 | |
| Wild | NR | |
| TET2 | .021 | |
| Mutated | 14.7 ± 7.8 | |
| Wild | NR | |
| ASXL1 | .014 | |
| Mutated | 10.0 ± 3.2 | |
| Wild | NR |
| Variable . | OS, mo, median ± SD . | P . |
|---|---|---|
| Age, y | < .001 | |
| ≤ 60 | NR | |
| > 60 | 12.3 ± 4.6 | |
| WBC | .001 | |
| ≤ 50 000/μL | NR | |
| > 50 000/μL | 14.5 ± 3.6 | |
| NPM1+/FLT3-ITD− | .080 | |
| Yes | NR | |
| Others | 61.0 | |
| CEBPA | .010 | |
| Double mutation | NR | |
| Others | 22.0 ± 16.7 | |
| RUNX1 | .015 | |
| Mutated | 14.5 ± 3.8 | |
| Wild | NR | |
| TET2 | .021 | |
| Mutated | 14.7 ± 7.8 | |
| Wild | NR | |
| ASXL1 | .014 | |
| Mutated | 10.0 ± 3.2 | |
| Wild | NR |
Only variables with P < 0.1 are shown. KRAS, NRAS, WT1, MLL-PTD, FLT3-TKD, cKIT, and IDH1 mutations, and many other clinical parameters, such as hemoglobin, platelet counts, and sex, were not significantly associated with survival (P > 0.1) in this group of patients with intermediate-risk karyotype, and so were not listed in the table.
Multivariate analysis (Cox regression) for the OS in AML patients with intermediate-risk karyotype (N = 171)
| Variable . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| Age* | 1.692 | 0.967-2.961 | .065 |
| WBC† | 2.216 | 1.296-3.787 | .004 |
| NPM1+/FLT-ITD− | 0.335 | 0.140-0.798 | .014 |
| CEBPAdouble | 0.285 | 0.119-0.685 | .005 |
| TET2 | 1.804 | 0.934-3.484 | .079 |
| Variable . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| Age* | 1.692 | 0.967-2.961 | .065 |
| WBC† | 2.216 | 1.296-3.787 | .004 |
| NPM1+/FLT-ITD− | 0.335 | 0.140-0.798 | .014 |
| CEBPAdouble | 0.285 | 0.119-0.685 | .005 |
| TET2 | 1.804 | 0.934-3.484 | .079 |
Age older than 60 years versus age 60 years or younger.
WBC > 50 × 103/μL versus ≤ 50 × 103/μL.
The prognostic values of TET2 mutation in context with other mutations for patients with intermediate-risk cytogenetics
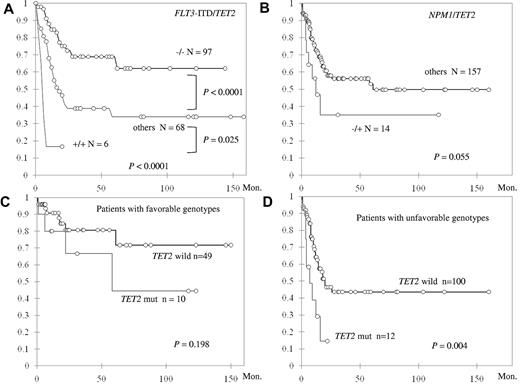
When the patients with intermediate-risk cytogenetics were stratified by the status of TET2 mutation and FLT3-ITD, those with TET2 wild-type and FLT3-ITD wild-type (TET2−/FLT3-ITD−) had the longest OS, those with TET2 mutation and FLT3-ITD (TET2+/FLT3-ITD+) had the shortest OS, and others (TET2−/FLT3-ITD+ and TET2+/FLT3-ITD−) had OS in between (NR vs 5.0 vs 16.9 months, P < .0001; Figure 3A). In addition, the negative impact of TET2+/FLT3-ITD+ genotype on OS appeared to be independent of age, WBC count, and CEBPAdouble and NPM1 mutation (hazard ratio = 3.84; 95% CI, 1.46-10.69; P = .007). TET2 mutation combined with absence of NPM1 mutation (NPM1−/TET2+) also showed a strong trend of shorter OS compared with other genotypes (median 12.3 vs 61 months, P = .055; Figure 3B), and the negative effect was also independent of age, WBC count, and CEBPAdouble (hazard ratio = 2.25; 95% CI, 1.05-4.83; P = .037). Although among patients with favorable molecular markers (NPM1+/FLT3-ITD− or CEBPAdouble), TET2 mutation did not have significant impact on OS (Figure 3C), those without these favorable genotypes fared much worse if they also harbored TET2 mutation (median 7 vs 20 months, P = .004; Figure 3D).
Kaplan-Meier curves for OS in 172 patients with intermediate-risk cytogenetics stratified by status. (A) TET2 mutation and FLT3-ITD. (B) TET2 and NPM1 mutations. The OS was also stratified by the status of TET2 mutation in patients with favorable molecular genotypes (NPM1+/FLT3-ITD− or CEBPAdouble; C) and in patients without these favorable genotypes (D). Both the genotypes TET2+/FLT3-ITD+ and TET2+/NPM1− are independent unfavorable factors for OS.
Kaplan-Meier curves for OS in 172 patients with intermediate-risk cytogenetics stratified by status. (A) TET2 mutation and FLT3-ITD. (B) TET2 and NPM1 mutations. The OS was also stratified by the status of TET2 mutation in patients with favorable molecular genotypes (NPM1+/FLT3-ITD− or CEBPAdouble; C) and in patients without these favorable genotypes (D). Both the genotypes TET2+/FLT3-ITD+ and TET2+/NPM1− are independent unfavorable factors for OS.
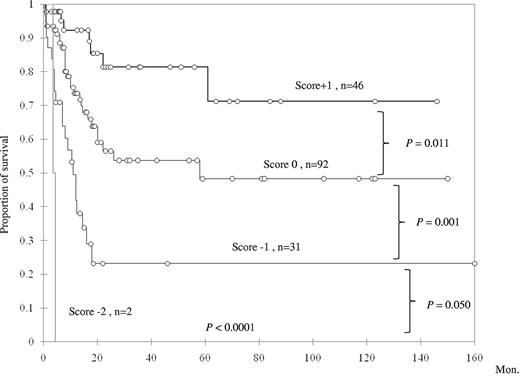
To further explore the prognostic impact of TET2 mutation in primary AML patients with intermediate-risk cytogenetics, a scoring system was generated based on the status of TET2 mutation and 3 other genetic alterations (NPM1 mutation, CEBPAdouble mutation, and FLT3-ITD), which were all well-documented prognostic factors in AML. The good prognostic mutations (NPM1 mutation and CEBPAdouble) were scored +1 point each, and poor prognostic mutations (FLT3-ITD and TET2 mutation) −1 point each. All the scores were summated as the final score for each patient. This scoring system could well separate AML patients with intermediate cytogenetics into 4 groups of distinct prognosis; the median survival was NR (N = 46), 58 months (N = 92), 11 months (N = 31), and 4.3 months (N = 2) in patients with a score of +1, 0, −1, and −2 points, respectively (P < .0001, Figure 4). We validated this algorithm by carrying out 100 times bootstrapping for this Kaplan-Meier estimation with SAS Version 9.0. The results confirmed that this scoring system was reliable to predict the prognosis of primary AML patients with intermediate-risk cytogenetics (supplemental Table 4). These results suggested a close interaction among TET2 mutation and other mutations in affecting patients' prognosis.
Kaplan-Meier survival curves according to a scoring system based on mutation status of NPM1, CEBPAdouble, TET2, and FLT3-ITD.CEBPAdouble and NPM1 mutations, 2 favorable prognostic factors, were scored +1 each, whereas the other 2 unfavorable mutations were each scored −1.
Kaplan-Meier survival curves according to a scoring system based on mutation status of NPM1, CEBPAdouble, TET2, and FLT3-ITD.CEBPAdouble and NPM1 mutations, 2 favorable prognostic factors, were scored +1 each, whereas the other 2 unfavorable mutations were each scored −1.
Sequential mutation analyses of TET2
Sequential analysis of TET2 mutation was performed in 122 patients: 23 with TET2 mutation at diagnosis and 99 without. Among the 23 TET2-mutated patients who had ever obtained a CR and had available samples for study, the initial TET2 mutation disappeared at CR in 18 patients, but 5 (patients 2, 4, 28, 40, and 43) retained it (supplemental Table 5); all these 5 patients relapsed finally, suggesting the presence of residual leukemia cells. The mutations in 4 of these 5 patients were frameshift or non-sense mutations and the remaining one (patient 28) was missense mutation in the conserved region and the skin tissue from this patient was devoid of this mutation, so they were all significant mutations but not somatic changes. Ten of 23 patients remained in continuous CR at the time of this study, and none had detectable TET2 mutation after achieving CR. Eight of them had concurrent other genetic changes at diagnosis, which also disappeared in CR. Interestingly, among the 13 patients who got relapse, 6 patients (patients 2, 4, 19, 20, 47, and 58) lost the original TET2 mutations at relapse by sequencing. Patient 4 retained the original TET2 mutation at the first relapse, but the mutation was no more detectable at the second relapse; patient 2 gained a novel mutation in TET2 (c.2366_2367insG, p.N789KfsX13) but lost the original one (c.1799delT, p.M600RfsX39). Because direct sequencing might not be sensitive enough to detect the low level of TET2 mutation signal, we therefore sequenced 20 TA clones of the PCR product from these 6 patients and searched for any mutant clone. All the relapsed samples from these patients were devoid of mutant clone, except for the one from patient 47, in which 1 mutant clone of 20 was detected. To further explore whether low amounts of TET2 mutants, undetectable by direct sequencing or TA cloning, existed in these 5 patients, we set out allele-specific PCR, with sensitivities ranging from 1 of 625 to 1 of 125 (data not shown), to test the relapsed DNA from these 5 patients (supplemental Table 6). We found that 2 (patients 2 and 20) of these 5 patients still retained the TET2 mutant by this sensitive method; however, the other 3 were still devoid of the original TET2 mutations, even by this PCR assay. We also analyzed TET2 mutation in the relapsed samples from 99 patients without this mutation at diagnosis; only 1 of them gained TET2 mutation at relapse (data not shown). These findings suggested that TET2 mutation was not stable as the disease evolved.
Discussion
This study recruits a large cohort of adults with de novo AML for TET2 mutation for analysis. It reveals several distinct clinical and biologic features not reported before in patients with this gene mutation.
TET2 protein functions as an enzyme catalyzing conversion of methylcytosine to hydroxymethylcytosine, with ferrous iron and α-ketoglutarate as cofactors.37,38 The mutation pattern in our patients indicated that TET2 mutation was a loss-of-function mutation. Other reports that showed uniparental disomy or loss of corresponding chromosomal region of TET2 gene also provide strong evidence for this hypothesis.1-3,39 However, the findings that most TET2-mutated patients had single allele mutation and there was no difference in clinical characteristics between patients with single and those with double mutations of TET2 in this study suggested a role of haploinsufficiency of TET2 in the development of leukemia.
The mutual exclusion between TET2 mutation and IDH mutation, as shown in other reports40-42 and in this study, suggests that mutations of the 2 genes may involve a common pathway in leukemogenesis. This is supported by a recent report showing that TET2 loss-of-function mutations and IDH mutations are associated with similar abnormal global hypermethylation.42 Another report also demonstrated that 2-hydroxyglutarate converted from α-ketoglutarate in IDH-mutated cells inhibited TET2-mediated hydroxymethylation of cytosine, indicating overlapping effects of these 2 mutations in cells.40 However, some mysteries remain to be answered: although IDH mutations and TET2 mutations show some common biologic effects in AML, many different clinical features exist between patients with TET2 and IDH mutations. For example, IDH1/2 mutations are closely associated with higher hemoglobin levels, higher platelet counts, and lower lactate dehydrogenase levels, but not higher WBC counts and older age, which are very different from the associations between these factors and TET2 mutations. The impacts of IDH and TET2 mutations on survival are also different.31 TET2 mutation and IDH mutation may have other different biologic effects, which await further studies. Other molecular alterations accompanied with TET2 or IDH mutation may also influence the clinical characteristics in patients with either mutation.
We noted a close correlation of TET2 mutation with ASXL1 mutation. In human, the exact functions of ASXL1 remain to be defined, but it can bind steroid receptor coactivator 1 to activate retinoic acid pathway,43 and is involved in regulation of histone methylation by cooperation with heterochromatin protein-1 to modulate the activity of LSD1,44 a histone demethylase for H3K4 and H3K9.45 It is interesting to see simultaneous mutations of TET2 and ASXL1 genes in the same patient, both related to epigenetic regulation but through different mechanisms. How these 2 mutations act together to influence epigenetic regulation of the whole genome remains to be defined.
In a report of the Cancer and Leukemia Group B, among cytogenetically normal AML patients with favorable genetic changes (mutated CEBPA or NPM1+/FLT3-ITD−), TET2 mutations adversely affected CR rate and OS.17 However, these findings could not be demonstrated in our current study as well as in an AML Study Group.46 The reason that our results were different from those from Cancer and Leukemia Group B study was not clear but might be because of differences in patient characteristics between the 2 studies. The patients in Cancer and Leukemia Group B study were older than ours, especially in the TET2-wild group (median, 60 vs 48 years). This was also reflected in the higher prevalence of TET2 mutations in their study than in ours (23% vs 18.2% in normal cytogenetic AML). The ethnic difference might be another factor. However, because our study was based on a retrospective analysis, further studies are necessary to confirm the results.
By combined analyses of TET2 and other mutations, we found both FLT3-ITD+/TET2+ and NPM1−/TET2+ genotypes were independent poor prognostic factors, suggesting synergism of negative impact between TET2 mutation and other adverse prognostic molecular markers. The impact of TET2 mutation on prognosis in AML patients with intermediate cytogenetics was further highlighted by our scoring system.
We noted a frequent loss of TET2 mutation in relapsed samples. Given the high blast percentages in the relapsed samples, it is highly probable that TET2 mutation was really lost in the bulk of the tumor cells, although some residual cells may retain the mutation. The tendency of loss of TET2 mutation in relapsed samples is analogous to FLT3-ITD and RUNX1 mutation, but in contrast to NPM1, IDH1, and IDH2 mutations, which usually persist in relapsed samples.25,29-31,34,47,48 These findings suggest that TET2 mutation may not be a good marker for monitoring minimal residual disease. TET2 mutation may be a secondary event during initial leukemogenesis, and leukemic cells with this mutation can be selected away by chemotherapy, leaving TET2-wild residual leukemia cells to grow subsequently into major population at relapse. Alternatively, TET2 mutation may be important for initiation of leukemogenesis, but not necessary for maintenance of the phenotype.
In conclusion, we showed distinct clinical and biologic characteristics of de novo AML with TET2 mutation in a large cohort of adult patients. TET2 mutation was a poor prognostic factor in patients with intermediate-risk cytogenetics, and a scoring system integrating TET2 mutation with FLT3-ITD, NPM1, and CEBPAdouble mutations could stratify these patients into 4 distinct prognostic groups.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Science Council, Taiwan (grants 96-2628-B002-013-MY2, 97-2314-B002-015-MY3, 97-2628-B-002-002-MY3, 98-2314-B-002-033-MY3, 100-2325-B-002-032, and 100-2325-B-002-033), the National Health Research Institute (NHRI-EX97-9731BI), the Department of Health, Taiwan (DOH100-TD-C-111-001), National Taiwan University Hospital (NTUH 98-S1052 and NTUH 98-S1383), and YongLin Healthcare Foundation.
Authorship
Contribution: W.-C.C. and H.-F.T. designed the experiment; W.-C.C., S.-C.C., H.-F.T., and C.-Y.C. analyzed the data and wrote the paper; H.-A.H., Y.-C. Chang, F.-Y.L., M.-C. Liu, C.-W.L., Yuan-Yeh Kuo, M.-C. Lee, Yi-Yi Kuo, M.-H.T., and C.-F.H. performed the experiment; J.-L.T., M.Y., W.T., B.-S.K., S.-J.W., S.-Y.H., S.-C.H., K.-T.K., and Y.-C. Chen provided important materials; and C.-Y.L., S.-C.C., and W.-C.C. performed statistical analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan, 100; e-mail: hftien@ntu.edu.tw; and Wen-Chien Chou, Department of Laboratory Medicine and Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan, 100; e-mail: wchou@ntu.edu.tw.
References
Author notes
W.-C.C. and S.-C.C. contributed equally to this study.