Abstract
Inbred mice are a useful tool for studying the in vivo functions of platelets. Nonetheless, the mRNA signature of mouse platelets is not known. Here, we use paired-end next-generation RNA sequencing (RNA-seq) to characterize the polyadenylated transcriptomes of human and mouse platelets. We report that RNA-seq provides unprecedented resolution of mRNAs that are expressed across the entire human and mouse genomes. Transcript expression and abundance are often conserved between the 2 species. Several mRNAs, however, are differentially expressed in human and mouse platelets. Moreover, previously described functional disparities between mouse and human platelets are reflected in differences at the transcript level, including protease activated receptor-1, protease activated receptor-3, platelet activating factor receptor, and factor V. This suggests that RNA-seq is a useful tool for predicting differences in platelet function between mice and humans. Our next-generation sequencing analysis provides new insights into the human and murine platelet transcriptomes. The sequencing dataset will be useful in the design of mouse models of hemostasis and a catalyst for discovery of new functions of platelets. Access to the dataset is found in the “Introduction.”
Introduction
Platelets stop bleeding and promote wound healing, essential activities that are conserved across a variety of species, including the human and the mouse. Because mice are amenable to genetic manipulation and environmental control, mouse models are commonly used as surrogates to understand the function of human platelets. Indeed, several mouse models recapitulate human phenotypes of platelet dysfunction.1-3 Differences between mouse and human platelets also exist, however. Although some differences between mouse and human platelets, such as platelet counts and size, are obvious, variances at the molecular level are not as apparent. One striking example is the expression of protease activated receptor 1 (PAR1), the quintessential receptor for thrombin signaling in human platelets. Knockout of Par1 did not prevent thrombin from activating mouse platelets.4,5 Only afterward was it discovered that mouse platelets lack protein for Par1.6
Gene expression profiling is frequently used to identify the mRNA pool in cells and potential molecular differences between cell populations or lineages. In humans, it is well appreciated that thousands of transcripts are present in platelets.7,8 Gnatenko et al recently used an “mRNA chip” to distinguish between platelets from patients with essential thrombocythemia, reactive thrombocytosis, or healthy persons.9-11 mRNA expression profiling has been used to identify functional differences in platelets isolated from patients with sickle cell anemia or systemic lupus erythematosus.12,13 Analysis of platelet transcripts identified mRNA patterns associated with platelet reactivity, body mass index, and cardiovascular disease.14-16 mRNA profiling also identified MRP-8/14 as a risk factor for future vascular events in women.17 Together, these studies demonstrate that mRNA expression analyses are a sensitive tool for identifying functional changes in human platelets. Furthermore, they suggest that similarities and differences in function between human and mouse platelets may also be identified by examining them at the mRNA level. The transcriptome of mouse platelets has not yet been characterized by any available approaches.
Here, we use next-generation RNA sequencing (RNA-seq, for definitions of RNA-seq related terms, see supplemental data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article)18-23 to provide the first detailed analysis of the mouse platelet transcriptome. RNA-seq is also applied, for the first time, to human platelets, adding unprecedented depth and breadth to the known human platelet transcriptome. In direct comparisons, we identify both conserved and differential expression patterns between human and mouse platelet transcriptomes. Some of the expression differences confirm and extend previous observations, whereas others are previously unrecognized. The sequencing datasets from 2 independent isolations of mouse platelets (C57bl/6, 1 male pool and 1 female pool, 4-8 mice per pool), and platelets from 1 male and 1 female healthy human donor can be accessed at www.bioserver.hci.utah.edu:8080/DAS2DB/genopub (Login: guest. Password: guest. For human data: Homo sapiens > H_sapiens_ Feb_2009 > Weyrichlab > RNA-Seq. For mouse data: Mus musculus > M_musculus_Jul_2007 > WeyrichLab > Mouse_platelets). Additional details regarding access to and visualization of this dataset are found in the supplemental data.
Methods
Cell isolation
Human platelets and polymorphonuclear leukocytes (PMNs): Whole blood was collected from healthy subjects using protocols approved by the University of Utah Institutional Review Board, and all human participants gave written informed consent in accordance with the Declaration of Helsinki. Platelets were isolated using established methods that deplete CD45+ leukocytes from the preparations before analyses.24 Human PMNs were prepared by positive CD15 selection as recently described.25
Mouse platelets
Platelets were isolated at room temperature from C57bl/6 mice using approved Institutional Animal Care and Use Committee protocols and guidelines. In brief, whole blood was collected from the carotid artery, anticoagulated with acid-citrate-dextrose, and diluted with warm PIPES saline glucose containing 300μM of prostaglandin E1 (PSG/PGE). The blood was centrifuged (115g, 10 minutes) to collect platelet-rich plasma, which was diluted further in PSG/PGE. Then, the platelet-rich plasma was centrifuged (500g, 10 minutes) and platelet pellets were resuspended in PSG/PGE in the presence of anti–Ter-119 and anti-CD45 beads (Miltenyi Biotec) to remove residual red blood cells and leukocytes, respectively. After this depletion step, purified platelets were washed in PSG/PGE before processing. Of note, platelets from 4-8 C57bl/6 mice were pooled to obtain sufficient quantities (1 μg) of high-quality RNA.
RNA isolation
Platelets (∼ 1-3 × 109) were lysed in Trizol (Invitrogen) as previously described24 or in miRVANA (Ambion) lysis buffer, which is henceforth referred to as column isolation. RNA was isolated as described by each manufacturer, but additional wash steps were included in the procedure. RNA was resuspended in RNAse free dH2O and treated with TurboDNAse (Ambion). The DNAse-treated RNA was precipitated with ethanol (3× volume) and sodium acetate (one-tenth volume) followed by rigorous ethanol (70%) washes. The integrity of the RNA was evaluated on an Agilent Bioanalyzer, and samples with RNA integrity numbers > 7.0 were prepared for sequencing. Poly(A)–tailed RNA was subsequently prepared by the University of Utah Core facility using the mRNA Seq Sample Prep Kit (Illumina) and used to create libraries for the deep sequencing studies. Additional details regarding sample preparation and sequencing are found in the supplemental data.
Sequencing and analysis
Sequencing and analysis were performed on pools of platelets from 2 independent groups of mice (1 female pool, 1 male pool) and on platelets from 2 independent human donors (1 female, 1 male). Samples were sequenced for 36 cycles, paired-end, on the Illumina GAIIx sequencer. Sequence reads were processed with the help of the University of Utah Bioinformatics core. FASTQ sequence reads were aligned with Novocraft's Novoalignment program (Novocraft Technologies; www.novocraft.com). Human alignments were to the Hg19 February 2009 GRCh37 build. Mouse alignments were to the mm9 July 2007 NCBI build 37. The following Novoalignment options were set: -t60 -r0.2 -q5 -i 250 10 000 -a AGATCGGAAGAGCGGTTCAG. All other options were left at default (www.novocraft.com/wiki/tiki-index.php?page = Novoalign%20command%20line %20Options). This method allows for reporting only high-quality, nearly unique alignments (alignments are further filtered using NovoAlignParser to report only unique alignments). More alignment details are found in supplemental data.
Downstream analysis was performed using a combination of programs found in the University of Utah's Useq analysis package26 and ad hoc perl programs. Although all analyses were performed on samples from both isolations (and were generally similar between isolations), when analyses were performed on individual samples, the majority of the analyses and figures represented are from the Trizol isolations and done without regard to gender, unless specified otherwise. Alignments were parsed using Useq's NovoalignParser program. Distribution of reads across different regions were determined using Useq's FilterPointData application. Reads across introns and exons were based on a combined University of California Santa Cruz (UCSC) transcripts, RefSeq transcripts, and ensemble transcript table which were downloaded from UCSC table browser (www.genome.ucsc.edu).27 Where reads mapped to an intron overlapping an exon of another transcript, they were counted only as an exon read based on the assumption that the majority of reads map to exons. Novel transcripts were assigned by running USeq package ScanSeqs followed by EnrichedRegionMaker. All other reads were assigned as intergenic reads.
Read coverage files and paired end-read files were generated using USeq analysis programs ReadCoverage and NovoalignPairParser. Paired end-reads and read coverage files were uploaded onto Genopub (www.bioserver.hci.utah.edu:8080/DAS2DB/genopub) for access and visualization in the Integrated Genome Browser.28 Details regarding access and use of the data are found in the Supplemental data.
RPKM assignment, ortholog and isoform choice, real-time comparison
RefSeq refgenes, which contain both protein-coding and non–protein-coding gene predictions, were downloaded from UCSC table browser27,29 (www.genome.ucsc.edu). RPKMs (reads per kilobase of exon model per million mapped reads) were calculated according to the formula published by Mortazavi et al: RPKM = 1 × 109 × (total exon reads)/(mapped reads (millions) × exon length (BP).19 RefSeq genes contain both protein-coding and non–protein-coding gene predictions. In this manuscript, the RefSeq annotated exons of a single “best expressed” transcript isoform for each gene prediction are used as the “exon model” in the RPKM calculation. Because a single mature transcript is chosen to represent each gene, “transcript,” “gene,” and “mRNA” are used interchangeably throughout to refer to the representative transcript derived from the gene prediction. RPKM assignments were further refined using published criteria that excludes the 3′-untranslated region from RPKM calculations.30 It has been demonstrated that removal of the 3′-untranslated region for RPKM calculations improves abundance estimates. We also found a slight improvement in abundance estimates after excluding the 3′-untranslated region from the calculation. RPKMs for each truncated (no 3′-untranslated region) transcript isoform were initially assigned using USeq26 package's DefinedRegionScanSeqs (DRSS) program, without removal of overlapping regions. Because the functional ortholog match of individual isoforms is poorly characterized between species, a “gene-wise” comparison between human and mouse platelets was performed. To this end, a single isoform with the highest RPKM (best expressed isoform) from each set transcripts arising from a single gene region was selected. In cases where more than one isoform of a gene had identical RPKMs, the representative isoform for analysis was chosen according to maximum total exon size. Each nonredundant isoform was again assigned RPKMs, using USeq application DRSS, this time removing any portions of exons overlapping other annotated exons from the analysis. Human-mouse orthologs were downloaded from HGNC's HCOP website (www.genenames.org/cgi-bin/hcop.pl).31 Orthologs were selected based on matches to the RefSeq name2 (gene_id) field. Preference was given to orthologs predicted by MGI and UCSC. In the case where there were multiple ortholog matches, priority was given first to those having exact RefSeq name matches. Gene descriptions were obtained using DAVID gene ID conversion.32,33 Additional details regarding analyses are found in the Supplemental data.
Ubiquitous genes were obtained from a list of 7897 RefSeq genes that were previously characterized as being ubiquitously expressed in RNA-seq results across a diverse set of tissues.30
Real-time PCR was performed using an SABiosciences RT2 Profiler Hematopoietic Stem Cells and Hematopoiesis PCR Array (QIAGEN) available for both human and mouse.
CD68 staining and flow cytometry
Platelets (human and mouse) or RAW 264.7 mouse macrophages (ATCC) were fixed with BD FACS Lysing (for sample fixation and red blood cell lysis) Solution (BD Biosciences) and then permeabilized (BD Perm buffer 2; BD Biosciences). Anti–human or anti–mouse Fc block (eBioscience) was added to the cells and before the addition of anti–human CD68 PE (clone Y1/82A, eBioscience) or FITC anti–mouse CD68 (clone FA-11; BioLegend). PE mouse IgG2bk (isotype), or FITC rat IgG2a (isotype) were used as controls for the mouse and human platelets, respectively. Stained cells were diluted with BD FACS Lysing Solution before flow analysis on a BD FACScan analyzer.
Correlations
To allow for log adjustment, genes with 0 RPKM are assigned a value of 0.002. Correlations were determined using the cor.test function in R34 with options set alternative = “greater” and method = “Spearman.” For correlations over a range of RPKMs as in Figure 3, an ad hoc function in R was used that bins genes represented in the correlation analysis based on increasing RPKM thresholds.
Additional online supplementary methods
Additional methods details can be found in the supplemental data.
Results
Deep sequence reads distribute throughout the genome and reflect transcript abundance
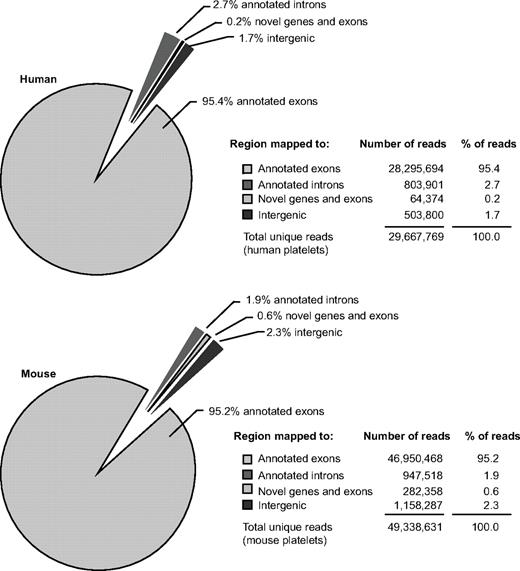
Next-generation RNA-seq was used to comprehensively identify and characterize transcripts in freshly isolated human and mouse platelets. Sequencing and analysis were performed on pools of platelets from 2 independent groups of mice and on platelets from 2 independent human donors. From individual samples, we obtained 29 667 769 and 49 338 631 reads (eg, 36 bp) that mapped to the human and mouse genome, respectively (Figure 1). Consistent with RNA-seq data obtained from other eukaryotic cells,19 the majority (95%) of sequencing reads mapped to annotated exons in both human and mouse platelets. The remaining reads mapped to introns (2.7%/1.9%, human/mouse), predicted novel genes and exons (0.2%/0.6%, human/mouse, a conservatively low estimate), or other uncharacterized intergenic regions (1.7%/2.3%, human/mouse; Figure 1). The distribution of reads was similar between 2 independent RNA isolation procedures (see “RNA isolation”), although the number of reads in column isolated samples were considerably less than Trizol-isolated samples (supplemental Figure 1A; data not shown). The datasets can be manually and programmatically accessed through a DAS/2 server (www.bioserver.hci.utah.edu:8080/DAS2DB/genopub; for access instructions, see supplemental data).
Distribution of sequencing reads in human and mouse platelets. Pie charts represent the number and percentage of sequencing reads from human platelets (top) or mouse platelets (bottom) mapping to indicated genomic regions. Only high-quality alignments following Novoalignment are represented. Although the majority of reads map to known intronic and exonic regions (combined RefSeq, UCSC, and Ensembl annotations), the remaining 1.9% (human) and 2.9% (mouse) of reads map to previously unannotated regions. Novel gene and exon regions are defined as unannotated regions that are enriched, at a conservative threshold, in sequencing reads. All other reads are termed intergenic. Intergenic reads may therefore contain some reads that map to novel genes and exons expressed below the arbitrary threshold.
Distribution of sequencing reads in human and mouse platelets. Pie charts represent the number and percentage of sequencing reads from human platelets (top) or mouse platelets (bottom) mapping to indicated genomic regions. Only high-quality alignments following Novoalignment are represented. Although the majority of reads map to known intronic and exonic regions (combined RefSeq, UCSC, and Ensembl annotations), the remaining 1.9% (human) and 2.9% (mouse) of reads map to previously unannotated regions. Novel gene and exon regions are defined as unannotated regions that are enriched, at a conservative threshold, in sequencing reads. All other reads are termed intergenic. Intergenic reads may therefore contain some reads that map to novel genes and exons expressed below the arbitrary threshold.
Next, we assigned reads to genes according to RefSeq annotations. Reads mapped to 14 189 human RefSeq genes (of 21 845) and 11 199 mouse RefSeq genes (of 21 529) in human and mouse platelets, respectively (supplemental Figure 1B-C). RPKMs are measures of individual transcript abundance in RNA-seq datasets and have been shown to be highly accurate across multiple cell types.18,19 We assigned RPKMs to a representative transcript (see “RPKM assignment, ortholog and isoform choice, real-time comparison”) for each RefSeq gene using previously published criteria.30,35 A wide range of RPKM values were found in human and mouse platelet transcriptomes with a median RPKM of 0.62 (combining isolations) in both human and mouse platelets. Applying a previously defined optimal threshold (0.3 RPKM) for gene expression,30 we found 9538 RefSeq genes and 6493 RefSeq genes expressed in human and mouse platelets, respectively (combining isolations; supplemental Figure 1D-E). The most highly expressed gene in human platelets (Trizol isolation), β2-microglobulin, accounted for 7% of the transcript pool, whereas thymosin β 4x (Tmsb4x) accounted for 11% of the total transcripts in the mouse platelet pool (Trizol isolation). Broken down further, the 20 most highly expressed mRNAs were composed of 36% and 49% of total transcripts in these human and mouse platelets. We observed similar results from 2 additionally sequenced human and mouse platelet isolations where the top 20 most highly expressed transcripts accounted for 38% and 39% (human), and 49% and 50% (mouse) of the total transcripts (data not shown). These data indicate that the platelet transcriptome is a diverse mixture of transcripts, but, as shown in supplemental Figure 2, is less complex than the transcriptome of human PMNs in which the 100 most highly expressed genes account for 40% of the total transcript represented in this sample. The RPKM values, read counts, and chromosomal coordinates for all RefSeq genes identified in mouse and human platelet transcriptomes are found in the “DRSS tables” (supplemental data). These tables are hyperlinked to Integrated Genome Browser for rapid visualization of gene expression, structure, and sequence.
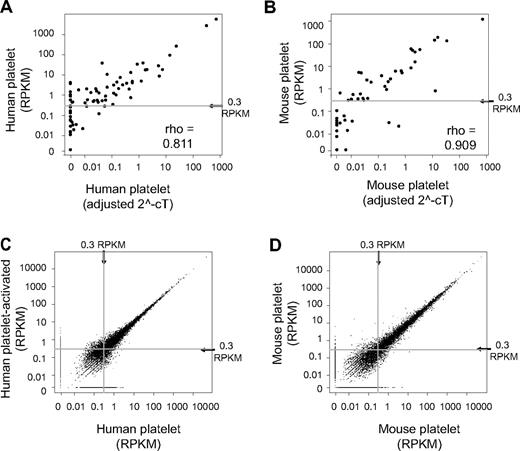
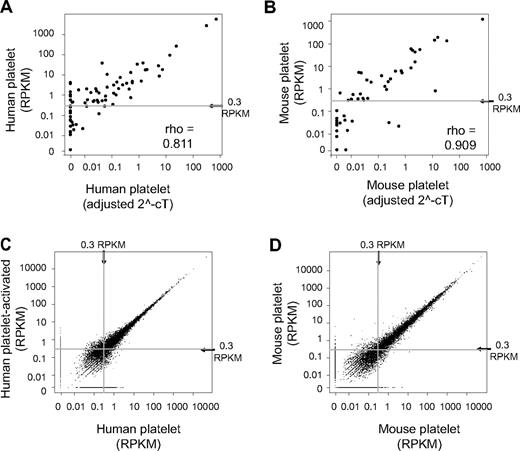
RPKM measurements confirmed the abundant expression, in both human and mouse, of transcripts previously identified in platelets, including actin B (ACTB), β2-microglobulin, integrin αIIb (ITGA2B), neurogranin (NGN), platelet factor 4 (PF4), and proplatelet basic protein (PPB) (supplemental Tables 1-2). We further validated the RPKM assignments of 84 genes involved in the development of blood cell lineages by real-time PCR, where the platelet mRNA was isolated from independent human donors or independent groups of mice. In every case, the real-time PCR results confirmed the presence of mRNAs that were originally identified by the RNA-seq analyses. Strong correlations were also observed between RPKM and real-time Ct expression estimates for human (ρ = 0.81) and mouse (ρ = 0.91) platelets (Figure 2A-B).
Abundance (RPKM) measured by RNA-seq in platelets correlates to real-time PCR results and is reproducible between independent platelet isolations. (A-B) Scatter plots comparing RNA-seq–derived RPKM measurements with real-time PCR results from independently isolated platelet preparations from (A) human and (B) mouse. The adjusted 2−Ct value for each gene as measured by real-time PCR are plotted along the x-axis versus the same gene's RPKM on the y-axis. (C-D) Scatter plots demonstrate the within-species correlation of RPKM measurements between independent samples. (C) Human platelet sample split and stimulated with thrombin (y-axis) or left unstimulated (x-axis) before RNA isolation and sequencing. (D) Comparison of RPKM measurements from 2 independent mouse platelet isolations. ρ indicates the Spearman rank correlation coefficient.
Abundance (RPKM) measured by RNA-seq in platelets correlates to real-time PCR results and is reproducible between independent platelet isolations. (A-B) Scatter plots comparing RNA-seq–derived RPKM measurements with real-time PCR results from independently isolated platelet preparations from (A) human and (B) mouse. The adjusted 2−Ct value for each gene as measured by real-time PCR are plotted along the x-axis versus the same gene's RPKM on the y-axis. (C-D) Scatter plots demonstrate the within-species correlation of RPKM measurements between independent samples. (C) Human platelet sample split and stimulated with thrombin (y-axis) or left unstimulated (x-axis) before RNA isolation and sequencing. (D) Comparison of RPKM measurements from 2 independent mouse platelet isolations. ρ indicates the Spearman rank correlation coefficient.
We also found that RNA-seq reliably estimated mRNA expression patterns in platelets regardless of their activation status, whether they were isolated from 2 independent donors or whether different RNA isolation procedures were used. As shown in Figure 2C (also see supplemental Table 3), RPKMs between human platelets that were isolated from the same donor, split in half, and then stimulated with or without thrombin, were highly correlated with one another (ρ = 0.94 at a threshold of 0.3 RPKM). The correlation was similarly high (ρ = 0.97 at a threshold of 0.3 RPKM) between 2 independent mouse platelet preparations (Figure 2D; supplemental Table 3) or 2 different RNA isolation procedures that were performed on independent human and mouse platelet isolations (supplemental Figure 3A-B; supplemental Table 3).
There are similarities and differences in mRNA expression patterns between human and mouse platelets
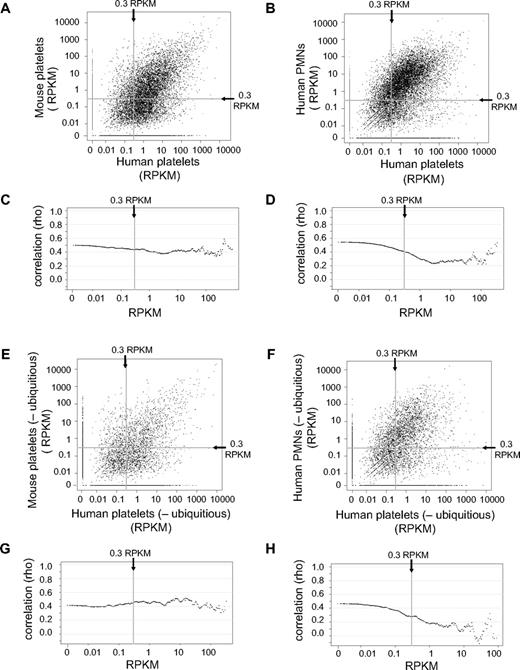
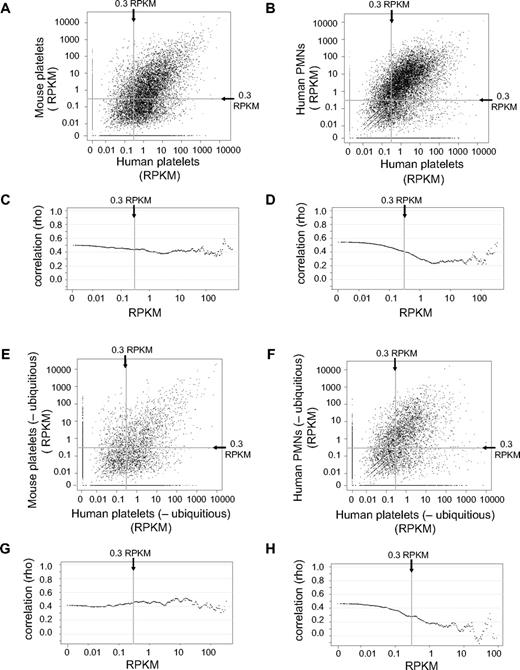
To begin comparing mRNA expression patterns between human and mouse platelets, human-mouse orthologs were identified as described in “RPKM assignment, ortholog and isoform choice, real-time comparison.” In this regard, at a threshold of 0.3 RPKM, 8532 and 6012 predicted orthologs (of 16 950) were identified in human and mouse platelets, respectively. When we compared human and mouse orthologs that were expressed above a 0.3 RPKM threshold, mRNA expression levels correlated at a ρ of 0.44 (Figure 3A; supplemental Table 3). In addition, ρ values did not significantly waver through a range of RPKM thresholds (Figure 3C). In contrast, the correlation of gene expression levels between human platelets and human PMNs was similar at the 0.3 RPKM threshold (ρ = 0.41) but decreased as the RPKM cutoff increased (Figure 3B,D; supplemental Table 3). We also found that removal of ubiquitous genes, which is composed of ∼ 75% of all mRNAs in nucleated cells,30 did not significantly alter the correlation between human and mouse platelet mRNA expression levels at all RPKM thresholds tested (Figure 3E,G; supplemental Table 3). Conversely, removal of ubiquitous genes decreased the correlation (to below ρ = 0 at some RPKM thresholds) between gene expression levels of human platelets and human PMNs (Figure 3F,H; supplemental Table 3).
Conservation of gene expression levels between human and mouse platelets or between human platelets and PMNs. (A-B) Scatter plots comparing the RPKMs of human platelet genes (x-axis) plotted against the RPKM (y-axis) of the corresponding gene in mouse platelets (A) or in human PMNs (B). Only genes with a human-mouse ortholog are represented. (C-D) The RPKM cutoff used in a Spearman rank correlation analysis (x-axis), plotted against the corresponding value of the ρ correlation coefficient calculated when comparing (C) human and mouse platelets or (D) human platelets and human PMNs. For example, the y-value of the far left point (0 RPKM) represents the ρ correlation coefficient between the RPKMs of all genes expressed in either sample. The y-value of the point at 0.3 RPKM on the x-axis (the vertical line) represents the ρ correlation coefficient calculated for all genes with an RPKM > 0.3 in both samples. The far right point includes only the 20 most highly expressed genes in the calculation of the corresponding correlation coefficient. (E-F) Scatter plots comparing, after removal of all ubiquitously expressed genes, the RPKMs of human platelet genes (x-axis) versus the RPKM of the corresponding gene (y-axis) in mouse platelets (E) or in human PMNs (F). (G-H) After removal of all ubiquitously expressed genes, the RPKM cutoff used in a Spearman rank correlation analysis (x-axis) is plotted against the corresponding value of the ρ correlation coefficient (y-axis) calculated comparing (G) human and mouse platelets or (H) human platelets and human PMNs. All plots shown represent samples isolated via Trizol.
Conservation of gene expression levels between human and mouse platelets or between human platelets and PMNs. (A-B) Scatter plots comparing the RPKMs of human platelet genes (x-axis) plotted against the RPKM (y-axis) of the corresponding gene in mouse platelets (A) or in human PMNs (B). Only genes with a human-mouse ortholog are represented. (C-D) The RPKM cutoff used in a Spearman rank correlation analysis (x-axis), plotted against the corresponding value of the ρ correlation coefficient calculated when comparing (C) human and mouse platelets or (D) human platelets and human PMNs. For example, the y-value of the far left point (0 RPKM) represents the ρ correlation coefficient between the RPKMs of all genes expressed in either sample. The y-value of the point at 0.3 RPKM on the x-axis (the vertical line) represents the ρ correlation coefficient calculated for all genes with an RPKM > 0.3 in both samples. The far right point includes only the 20 most highly expressed genes in the calculation of the corresponding correlation coefficient. (E-F) Scatter plots comparing, after removal of all ubiquitously expressed genes, the RPKMs of human platelet genes (x-axis) versus the RPKM of the corresponding gene (y-axis) in mouse platelets (E) or in human PMNs (F). (G-H) After removal of all ubiquitously expressed genes, the RPKM cutoff used in a Spearman rank correlation analysis (x-axis) is plotted against the corresponding value of the ρ correlation coefficient (y-axis) calculated comparing (G) human and mouse platelets or (H) human platelets and human PMNs. All plots shown represent samples isolated via Trizol.
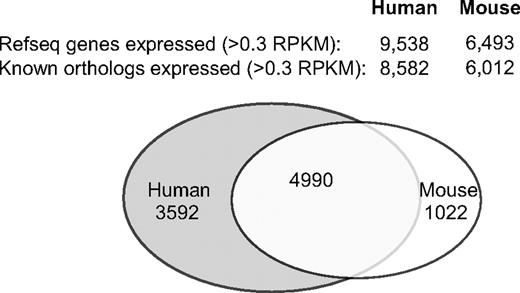
The results displayed in Figure 3 demonstrate that the expression levels of many transcripts correlate well between human and mouse platelets, but some do not. When examined in more detail, cross-species comparisons of orthologs revealed that 4990 transcripts are expressed by both human and mouse platelets at an RPKM threshold of 0.3 (Figure 4). Specifically, within the sequenced samples, 58% of the mRNAs expressed by human platelets are also found in the mouse platelets (eg, 4990 of 8582 orthologs), whereas 83% of transcripts expressed by mouse platelets are found in human platelets (eg, 4990 of 6012 orthologs), although the overlaps vary considerably according to the RPKM threshold used (data not shown).
Venn diagram of genes expressed in human versus mouse platelets. Venn diagram represents the overlap in gene expression (> 0.3 RPKM) of all genes with an ortholog match in human versus mouse platelets. The diagram was generated from a combined list of genes expressed in either the Trizol or column isolations. The total numbers of genes expressed with an ortholog match (known orthologs expressed) or, regardless of an ortholog match (RefSeq genes expressed), are given above the diagram.
Venn diagram of genes expressed in human versus mouse platelets. Venn diagram represents the overlap in gene expression (> 0.3 RPKM) of all genes with an ortholog match in human versus mouse platelets. The diagram was generated from a combined list of genes expressed in either the Trizol or column isolations. The total numbers of genes expressed with an ortholog match (known orthologs expressed) or, regardless of an ortholog match (RefSeq genes expressed), are given above the diagram.
Of the 4990 orthologs expressed in mouse and human platelets, the most abundant nonubiquitous mRNAs detected in the 2 individual human and 2 pooled mouse platelets are neurogranin and proplatelet basic protein (Tables 1–2). At a 0.3 RPKM cut-off, 36 of the 40 most highly expressed nonubiquitous transcripts expressed in the 2 individual human platelets samples are present in the 2 pooled mouse platelet samples (Table 1). Similarly, 39 of the 40 most highly expressed nonubiquitous transcripts in the 2 mouse platelet pools are found in the individual human platelet samples (Table 2). Nevertheless, transcripts that are common to human and mouse platelets often exhibit diverse expression levels. As an example, the RPKM value for CCL5 (RANTES) is 3371.6 and 4.8 for human and mouse platelets, respectively (Table 1). Full lists comparing RPKM measurements of orthologs and nonubiquitous orthologs expressed in human and mouse platelets are found in the Supplemental data (tables titled “All_Orthologs” and “Non_ubiquitous_orthologs”).
To identify differentially expressed transcripts, we examined the ratio of platelet mRNA expression levels between human and mouse orthologs. Tables 3 and 4 provide a short list of mRNAs that are expressed in either human or mouse platelets but are absent, or present at low levels, in platelets from the other species. Visual representations of differentially expressed transcripts, which were chosen based on previously published data inferring functional differences between human and mouse platelets,2,5,6,36-38 are shown in Figure 5. These transcripts include PAR1 (F2R), PAR3 (F2RL2), PTAFR (platelet activating factor receptor), and factor V (F5). For comparative purposes, the expression profile of ITGA2B, a transcript that is common to both human and mouse platelets, is shown in parallel (Figure 5A). mRNA for PAR1, which serves as the primary thrombin receptor in human platelets but is not active in mouse platelets,4,5 is 13.5-fold higher in human platelets than in mouse platelets (Figure 5B). In contrast, the mRNA for the alternative mouse receptor6 PAR3 is highly expressed in both mouse platelet isolations but is not detected in human platelets (Figure 5C). Consistent with previous studies showing that human, but not mouse, platelets respond to platelet activating factor (PAF),2,39 mRNA for PTAFR is detected in human platelets and absent in mouse platelets (Figure 5D). Lastly, only mouse platelets expressed mRNA for FV (Figure 5E), a result that is compatible with the notion that mouse megakaryocytes principally produce FV and transfer it to platelets,36 whereas human platelets internalize FV produced by the liver.37
Representative images of sequencing reads across genes expressed in human or mouse platelets. (A-E) Pictures taken from Integrated Genome Browser of (A) ITGA2B, (B) PAR1, (C) PAR3, (D) PTAFR, and (E) F5 genes expressed in human (left panels) or mouse (right panels) platelets. The height of bars represents the relative accumulated number of 36-bp reads spanning a particular sequence. The number at the top of each plot is the maximum of the y-axis scale for each plot. Gene symbols and RefSeq gene annotations are shown on the bottom of each panel. Thick horizontal lines represent exons; and thin horizontal lines, introns.
Representative images of sequencing reads across genes expressed in human or mouse platelets. (A-E) Pictures taken from Integrated Genome Browser of (A) ITGA2B, (B) PAR1, (C) PAR3, (D) PTAFR, and (E) F5 genes expressed in human (left panels) or mouse (right panels) platelets. The height of bars represents the relative accumulated number of 36-bp reads spanning a particular sequence. The number at the top of each plot is the maximum of the y-axis scale for each plot. Gene symbols and RefSeq gene annotations are shown on the bottom of each panel. Thick horizontal lines represent exons; and thin horizontal lines, introns.
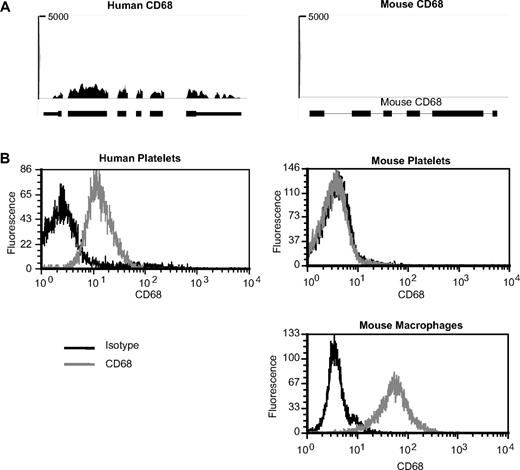
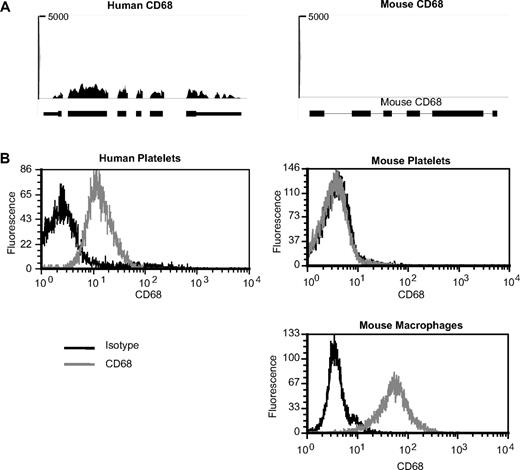
In an additional set of studies, we determined whether the expression of a candidate differentially expressed mRNA, CD68, predicted the presence of its corresponding protein in platelets. As shown in Figure 6A, CD68 mRNA (RPKM = 248) is highly expressed in human platelets but undetectable in mouse platelets. Likewise, human, but not mouse, platelets express CD68 protein (Figure 6B). As expected, CD68 protein is present in murine macrophages (Figure 6B).
CD68 expression in human and mouse platelets. (A) Pictures taken from Integrated Genome Browser of CD68 expression in humans (left panel) or mouse (right panel) platelets. The height of bars represents the relative accumulated number of 36-bp reads spanning a particular sequence. The number at the top of each plot is the maximum of the y-axis scale for each plot. Gene symbols and RefSeq gene annotations are shown on the bottom of each panel. Thick horizontal lines represent exons; and thin horizontal lines, introns. (B) Histograms representing detection of CD68 in human (left panel) platelets or mouse (right panels) platelets or RAW 264.7 macrophages. CD68 or respective isotype controls were detected by intracellular antibody staining followed by analysis on a flow cytometer.
CD68 expression in human and mouse platelets. (A) Pictures taken from Integrated Genome Browser of CD68 expression in humans (left panel) or mouse (right panel) platelets. The height of bars represents the relative accumulated number of 36-bp reads spanning a particular sequence. The number at the top of each plot is the maximum of the y-axis scale for each plot. Gene symbols and RefSeq gene annotations are shown on the bottom of each panel. Thick horizontal lines represent exons; and thin horizontal lines, introns. (B) Histograms representing detection of CD68 in human (left panel) platelets or mouse (right panels) platelets or RAW 264.7 macrophages. CD68 or respective isotype controls were detected by intracellular antibody staining followed by analysis on a flow cytometer.
Discussion
In the present study, we use next-generation RNA-seq to provide the first unbiased comprehensive comparison of mouse and human platelet transcriptomes. We achieved thorough transcriptome coverage30 with > 10 million unique mapped reads in every mouse or human sample. Sequence read distributions in anucleate platelets resemble patterns in nucleated cells where reads predominantly map to exons but also map to predicted novel genes, intergenic regions, or introns.19 Reads mapped to an assortment of annotated transcripts varying in chromosomal location, length, and expression level. This indicates that, similar to other cells and tissues, mouse and human megakaryocytes invest platelets with a diverse repertoire of transcripts.
mRNA expression patterns reflect functional differences between mouse and human platelets
Mouse platelets are commonly used as surrogates to study in vivo human platelet function. Nevertheless, there is often uncertainty regarding the functional differences and similarities between mouse and human platelets and the applicability of inbred mice as models of disease. Our RNA-seq analysis adds considerable insight into these issues. Not surprisingly, we found that expression of many mRNAs is conserved between mouse and human platelets. Among this group is PF4, which is the primary gene used in Cre-based systems to control gene expression in the mouse megakaryocyte lineage. mRNA transcripts for integrin αIIbβ3 and P-selectin are also conserved between species. In this regard, platelets from mice lacking αIIbβ3 or P-selectin have abnormalities that are consistent with the known function of each adhesion molecule in human platelets.40,41
In addition to identifying conserved expression, however, transcript analysis also confirmed discrepancies between mouse and human platelets. As discussed in the “Introduction,” PAR1 functions in human but not in mouse platelets. For reasons that are not completely understood, PAR3 serves as an alternative receptor in mouse platelets6,42,43 and has no known function in human platelets. Our transcript profiles in platelets from the 2 species are consistent with and explain these observations. We also found that mouse platelets do not express mRNA for PTAFR, whereas human platelets do. These results are consistent with previous observations demonstrating that PAF does not activate mouse platelets.2,39 In contrast, PAF is a well-recognized agonist of human platelet activation.44
RNA-seq analysis also revealed that CD68 mRNA is differentially expressed between mouse and human platelets. Specifically, CD68 mRNA is robustly expressed by human platelets but undetectable in mouse platelets. Previous studies demonstrated that CD68 protein is expressed by human platelets and is a marker of lysosomal translocation in response to activation.45,46 Similar to these reports, we detected mRNA and protein for CD68 in human platelets. However, neither the mRNA nor protein was detectable in murine platelets. The disparity in expression patterns between species and the functional significance of CD68 expression in human platelets is not clear. Nonetheless, these data suggest that knockout of CD68 in the murine megakaryocyte lineage would lack an appreciable phenotype. Conversely, transgenic expression of CD68 in murine platelets may yield insights into CD68 function similar to gain of function studies observed with FcγRIIA.47
Our in-depth RNA analysis may also have utility in determining whether platelet proteins are derived from megakaryocyte transcripts or are endocytosed. A prime example is factor V. In humans, factor V is produced by the liver and sequestered by circulating platelets.37 This suggests that factor V is not transferred from megakaryocytes to platelets; and, consistent with this conclusion, we did not detect factor V transcripts in human platelets. In contrast, we detected factor V transcripts in mouse platelets, a finding that coincides with a previous report demonstrating that factor V is produced by megakaryocytes in mice.36
Points to consider while interpreting the data
Cross-species ortholog comparisons have limitations that need to be recognized. These include: (1) many genes do not have a predicted ortholog; (2) predicted orthologs are occasionally incorrect; (3) orthologs are chosen based on comparisons between the primary transcripts, which does not account for multiple transcripts or isoforms that are derived from the same primary transcript; and (4) RPKM measurements that are based on transcript length may be spuriously inflated or reduced depending on which isoform is used for the expression analysis. These potential discrepancies are minimized by our analysis strategy: we conservatively chose orthologous transcripts and restricted our analysis to one isoform for each primary gene (“RPKM assignment, ortholog and isoform choice, real-time comparison”). In addition, because we did not necessarily expect a linear relationship between RPKM values of mouse and human, correlations are based on the ranks of gene expression (Spearman). Inspecting both the relative rank order of gene expression and absolute RPKM given in the provided tables can be informative when inferring functional differences between species.
We based our abundance analyses solely on RefSeq genes. Genes exclusively archived in other annotation sets or previously unannotated (novel) genes, which are numerous in platelets (J.W.R., A.J.O., and A.S.W., unpublished data, June 2010), are accessible within our public datasets. In our comparative analysis, we found 6493 and 9538 RefSeq genes expressed in mouse and human platelets, respectively. These estimates are based on a 0.3 RPKM cut-off, which previously has been used as an expression threshold that balances the number of false positives with the number of false negatives.30 Consistent with this, we have noted that we can reproducibly detect, by real-time PCR, most transcripts expressed above the 0.3 RPKM threshold in platelets. On the other hand, whereas many genes expressed below 0.3 RPKM are detectable by real-time PCR, these results are often more difficult to consistently reproduce (J.W.R., unpublished observations, October 2010).
With regard to RNA-seq data, mapped reads can yield false-positives when transcripts overlap adjacent transcripts or overlap other genes expressed from the opposite strand. In our analysis, sense-antisense overlapping exons are excluded from abundance calculations. False positives may still arise from overlap of genes with expressed regions that are not annotated as part of a mature transcript. Such is the case with Rasl10a, a gene that appeared to be abundantly expressed in mouse platelets and absent in human platelets. On further inspection, reads were falsely assigned to RasI10a because it overlaps with an expressed intronic region of Gas211, which is abundantly expressed in mouse platelets. In other cases, false negatives may arise when the exons of a gene completely overlap the exons of another transcript. Nevertheless, these exceptions are rare and readily identified by visual inspection of the sequencing reads made accessible in Genopub.
Catalyst for future studies
Our approach using RNA-seq in platelets should catalyze future studies in mouse and human platelets, providing a framework for future RNA-seq-based studies and a starting point for the dissection of molecular pathways and the development of appropriate platelet model systems. Our data can immediately be used as a tool for novel gene and gene feature discovery in platelets. As part of our analysis, we demonstrate that platelets from healthy human persons display similar expression patterns. This indicates that RNA-seq applied to clinical platelet isolates may identify targets that are differentially expressed between healthy and diseased populations. It should be noted that variations in RNA expression levels in platelets from persons differing in factors, such as gender, age, ethnicity, and health status, are not captured within our limited sample set. Similar to what has been done using other platforms, genome-wide association studies will require sequencing the transcriptome of thousands to tens of thousands of persons to establish a representative baseline transcriptome sequence. As the cost (currently rivaling or less than microarray costs), technical difficulty, and turnaround time of next-generation sequencing continues to decline, the possibility of using next-generation sequencing for platelet genome-wide association studies is becoming more feasible. Because of its reproducibility and ability to run multiple bar-coded samples simultaneously, and its unbiased ability to detect single nucleotide polymorphisms, novel genes, and sequence features, next-generation sequencing will probably replace microarray technology as the platform of choice for genome-wide gene expression studies. Many fewer samples are needed for clinical studies where serial draws, both before and after disease or treatment, can be made.
Platelets have critical roles in multiple processes and diseases, including inflammation, immunity, cancer metastasis, and angiogenesis.48 There is evolving evidence that the molecular signature of platelets may be changed in disease conditions where these processes are altered. RNA-seq can be used to shed light on how diseases alter platelet function at the molecular level and how molecularly “reprogrammed” platelets might reciprocally influence the development and progression of disease. The ability to perform parallel types of studies in murine models, where the environment can be controlled, will be a valuable tool to elucidate the molecular mechanisms that affect platelet function. In this regard, transcriptome profiling may also help clarify secondary changes in transcripts that may contribute indirectly to phenotypic differences in platelets from knockout animals, and guide the design of future studies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Diana Lim for preparing the figures and Jenny Pierce for assistance in submission of the manuscript.
This work was supported by the National Institutes of Health (R01: HL066277, HL044525, and HL091754; K08: HD049699; and T32: HL105321).
National Institutes of Health
Authorship
Contribution: J.W.R. performed the majority of experiments and computer analyses, and drafted and prepared the manuscript; A.O. performed bioinformatics and computer programming; N.D.T., B.H., E.N.L., and C.C.Y. performed experiments; D.A.N. provided bioinformatics support; G.A.Z. designed the experiment and reviewed the manuscript; and A.S.W. directed all aspects of the study and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew S. Weyrich, Department of Internal Medicine, University of Utah, Program in Molecular Medicine, Eccles Institute of Human Genetics, Bldg 533, Rm 4220, 15 N 2030 E, Salt Lake City, UT 84112; e-mail: andy.weyrich@u2m2.utah.edu.