Abstract
Chronic myelomonocytic leukemia (CMML), a myelodysplastic/myeloproliferative neoplasm, is characterized by monocytic proliferation, dysplasia, and progression to acute myeloid leukemia. CMML has been associated with somatic mutations in diverse recently identified genes. We analyzed 72 well-characterized patients with CMML (N = 52) and CMML-derived acute myeloid leukemia (N = 20) for recurrent chromosomal abnormalities with the use of routine cytogenetics and single nucleotide polymorphism arrays along with comprehensive mutational screening. Cytogenetic aberrations were present in 46% of cases, whereas single nucleotide polymorphism array increased the diagnostic yield to 60%. At least 1 mutation was found in 86% of all cases; novel UTX, DNMT3A, and EZH2 mutations were found in 8%, 10%, and 5.5% of patients, respectively. TET2 mutations were present in 49%, ASXL1 in 43%, CBL in 14%, IDH1/2 in 4%, KRAS in 7%, NRAS in 4%, and JAK2 V617F in 1% of patients. Various mutant genotype combinations were observed, indicating molecular heterogeneity in CMML. Our results suggest that molecular defects affecting distinct pathways can lead to similar clinical phenotypes.
Introduction
Chronic myelomonocytic leukemia (CMML) is a distinct entity, a myelodysplastic/myeloproliferative neoplasm (MDS/MPN) characterized by morphologic dysplasia and monocytosis. Pathomorphologic similarities exist between more advanced forms of CMML, including CMML-2 and CMML-derived secondary acute myeloid leukemia (sAML), and some primary forms of AML with monocytoid differentiation.
Unlike chronic myelogenous leukemia, characterized by a BCR-ABL1 fusion, the molecular pathogenesis of closely related CMML remains unclear.1,2 Recurrent reciprocal translocations, involving PDGFRA and PDGFRB, are rare in CMML; if present, they indicate that myeloblast/monocyte proliferation can be mediated by activation of phospho-tyrosine kinase pathways.3 Nevertheless, large, unbiased tyrosine kinase sequencing projects performed in CMML to identify kinase or regulatory domain mutations have not been revealing.4 Recurrent chromosomal abnormalities are mostly unbalanced and are similar to those in typical MDS.5-7 Despite reports of new chromosomal defects and mutations, a significant proportion of patients are found to have normal karyotype by metaphase cytogenetics (MC).
RAS family and RUNX1 mutations have been found in a proportion of CMML cases.8,9 Recently, several genes have been found mutated in myeloid malignancies, including CMML.10-13 These discoveries were facilitated by single nucleotide polymorphism array (SNP-A) karyotyping, which enables detection of somatic regions of copy number neutral loss of heterozygosity (CN-LOH), also called uniparental disomy (UPD). In CMML homozygous mutations in CBL and TET2 are associated with regions of UPD.6,14-19 In addition, mutations in the EZH2 gene have been detected in a substantial fraction of patients with myeloid malignancies characterized by the presence of CN-LOH 7q.20-22 In contrast, mutations in ASXL1 and IDH1/2 genes are mostly heterozygous.11,23-25
TET2 mediates the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in DNA.26-28 DNMT3A mutations, recently identified by next-generation sequencing in de novo AML,29,30 exemplify another type of mutation affecting epigenetic DNA modification. Similar to DNA modifying genes, mutations of genes regulating histone methylation have been found in myeloid malignancies. For instance, ASXL1, a polycomb and mixed lineage leukemia/trithorax chromatin modifier, was found to be mutated in CMML.11,23 ASXL1, in association with LSD1, is involved in H3K4 demethylation and thereby chromatin remodeling.31 Although ASXL1 mutations are ubiquitous among myeloid malignancies, a knockout mouse model displayed mild defects in myelopoiesis and did not develop MDS or AML.32 Trimethylation of H3K27 can be affected by EZH2, a H3K27 methyltransferase,33 also mutated in myeloid malignancies.20-22 Alteration of this histone mark may contribute to the pathogenesis of malignant evolution. Indeed, UTX, which encodes for a demethylase also specific for H3K27, has been found mutated in some hematopoietic cell lines of myeloid origin.34,35
It is probable that multiple molecular defects can be present in an individual patient, explaining clinical heterogeneity, or conversely, distinct mutational events and chromosomal aberrations may produce similar clinical/pathomorphologic phenotypes, leading to unifying features. To address these issues, we have performed comprehensive cytogenetic analysis and targeted sequencing of various genes known to be mutated in CMML, including some newer candidate genes, particularly UTX (KDM6A) and DNMT3A, in a cohort of 72 CMML and CMML-derived AML cases. Rational molecular screening may successfully show mutational patterns and may identify surrogate phenotypic markers associated with mutations or their combinations.
Methods
Patients
Informed consent for sample collection was obtained according to protocols approved by the institutional review boards of Cleveland Clinic and Johns Hopkins University, following the Declaration of Helsinki. Diagnosis was assigned according to the World Health Organization (WHO) classification criteria, with all diagnoses confirmed at the 2 participating institutions.1,2 SNP-A karyotyping was performed on 70 patients and mutational screening on 72 patients with CMML and AML derived from CMML (Table 1). Clinical parameters examined included survival, blood counts and pathomorphologic criteria. The mutational status of CBL, TET2, RAS, and EZH2 were previously reported in 40, 28, 1, and 24 patients of 72 present in this study, respectively.14,15,19,22
SNP-A analysis
Mutational analysis
Screening of selected genes at known mutational hotspot regions and consensus splicing sites, for example, CBL (exons 8-9), KRAS and NRAS (exons 1-2), IDH1 and IDH2 (exons 4), DNMT3A (exons 18-23), TET2, UTX, and EZH2 (all coding exons) was performed with direct genomic sequencing by standard techniques on the ABI 3730×l DNA analyzer (Applied Biosystems) as described.14,15,19,22 UTX mutations were detected with either exon-specific primers or cDNA primers as previously described and were scored as pathogenic on the basis of their absence in 400 male controls.35 All mutations were detected by bidirectional sequencing and were scored as pathogenic if not detected in normal or available nonclonal CD3 samples and absent in published SNP databases36 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Furthermore, canonical mutations (ie, described as somatic in the literature and those associated with somatic CN-LOH) were not further confirmed. Frameshift mutations were validated by cloning and sequencing individual colonies (TOPO TA cloning; Invitrogen). Novel missense mutations were confirmed when possible; for germ line confirmation (when constitutional DNA available), only exons containing mutations were tested. Screening for JAK2 V617F mutation was performed as previously described.37
Measurement of 5hmC levels
The 5hmC levels in genomic DNA from patients (N = 36) and healthy controls (N = 17) were measured by bisulfite conversion and dot blot with anti-CMS antiserum as described.28 Results were normalized, and patients were divided into groups that were based on high or low 5hmC level as previously described.28
Immunohistochemical detection of pSTAT5
Staining was performed on a Benchmark XT platform (Ventana Medical Systems) according to the manufacturer's instructions, using mouse monoclonal anti-phospho-STAT5a/b (Y694/99; Advantex BioReagents LLP) at 1:500 dilution. All stains were scored without knowledge of the clinical diagnosis or mutational status. Phospho-STAT5–positive staining (nMEG pSTAT5) was defined as previously reported.37
Statistical analysis
When appropriate, Kaplan-Meier statistics were applied to assess overall survival (OS) and compared with the log-rank test. For comparison of the frequency of mutation or other clinical features between diseases groups, categorical variables were analyzed with the Fisher exact test. A P value < .05 was set as the threshold of clinical significance.
Results
Characteristics and genetics of patients with CMML
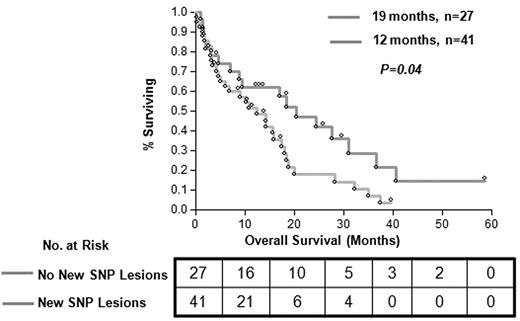
A total of 72 patients were studied, 36 with CMML-1, 16 with CMML-2, and 20 with AML derived from the above conditions (Table 1). Among the patients with CMML, 46% cases had myelodysplastic CMML and 44% had proliferative CMML, as classified by the French-American-British group.38 With the use of conventional MC, 30 patients (43%) showed an abnormal karyotype; the most common shared defects included monosomy 7 (9%), trisomy 8 (6%), and losses of the Y or X chromosomes (7%). Results were noninformative in 3 patients. SNP-A karyotyping confirmed defects identified by MC (except for some balanced translocations that are undetectable by SNP-A). New lesions were found in 60% of patients (42 of 70); previously cryptic somatic areas of CN-LOH were found in 30 of 42 patients (71%) and microdeletions in 19 of 42 patients (45%; supplemental Figure 1). The presence of multiple chromosomal defects as detected by SNP-A had a significant effect on patients' OS compared with patients without novel SNP lesions (16 vs 21 months; P = .04) by Kaplan-Meier analysis and univariate analysis (odds ratio [OR], 1.81; 95% confidence interval [CI], 1.02-3.35; Figure 1).
Baseline characteristics of patients with CMML and with CMML-derived AML
| Characteristic . | No. . |
|---|---|
| No. of patients | 72 |
| Sex | |
| Male | 48 |
| Female | 24 |
| Age, y | |
| Median | 70 |
| Range | 38-89 |
| Metaphase cytogenetics (MC) | |
| Normal | 39 |
| Abnormal | 30 |
| No growth by MC | 3 |
| SNP-A karotyping | 70 |
| Clinical data not available | 2 |
| WHO classification | |
| CMML-1 | 36 |
| CMML-2 | 16 |
| sAML to CMML-1/2 | 20 |
| Characteristic . | No. . |
|---|---|
| No. of patients | 72 |
| Sex | |
| Male | 48 |
| Female | 24 |
| Age, y | |
| Median | 70 |
| Range | 38-89 |
| Metaphase cytogenetics (MC) | |
| Normal | 39 |
| Abnormal | 30 |
| No growth by MC | 3 |
| SNP-A karotyping | 70 |
| Clinical data not available | 2 |
| WHO classification | |
| CMML-1 | 36 |
| CMML-2 | 16 |
| sAML to CMML-1/2 | 20 |
WHO indicates World Health Organization.
Clinical effect of new lesions detected by SNP-A karyotyping analysis. Patients with novel lesions detected by SNP-A and normal karyotype (MC) showed worse OS (median, 12 months; OR, 1.81; 95% CI, 1.02-3.35) versus patients without novel SNP lesions (median, 19 months; OR, 0.55; 95% CI, 0.29-0.98; P = .04). Patients with new SNP lesions and those without are represented by gray line (N = 41) and orange line (N = 27), respectively.
Clinical effect of new lesions detected by SNP-A karyotyping analysis. Patients with novel lesions detected by SNP-A and normal karyotype (MC) showed worse OS (median, 12 months; OR, 1.81; 95% CI, 1.02-3.35) versus patients without novel SNP lesions (median, 19 months; OR, 0.55; 95% CI, 0.29-0.98; P = .04). Patients with new SNP lesions and those without are represented by gray line (N = 41) and orange line (N = 27), respectively.
Identification of novel mutations in UTX, EZH2, and DNMT3A
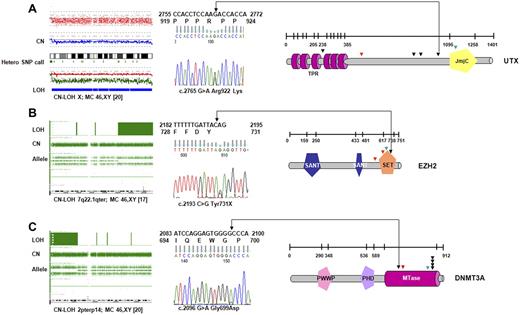
When all 72 patients were screened, novel UTX variants were found in 4 patients (of 52; 8%) with CMML-1/2 and 2 patients with sAML (of 20; 10%); these mutations were spread over several exons (Figure 2A; Table 2). One case showed a homozygous missense UTX mutation associated with CN-LOH of chromosome X. Another female patient with CMML-1 was found to harbor a somatic frameshift mutation, and a male patient had a nonsense mutation. Moreover, we detected 3 novel missense variants for which no constitutional DNA was available. However, these alterations have not been seen in controls (with a frequency of < 1/400 of male controls35 ) or described as polymorphisms in available databases. In addition to UTX variants, we have identified a female patient with a hemizygous deletion involving the entire UTX gene on one X chromosome homologue. Collectively, deletions and mutations of UTX were present in 10% of the affected persons.
UTX, EZH2, and DNMT3A mutations identified in CMML. (A) In a cohort of 72 patients with CMML and AML-derived CMML, 2 somatic, inactivating UTX mutations were detected by sequencing. Four additional missense variants that could correspond to rare polymorphisms were identified. Profile of CN-LOH of X chromosome by SNP-A in one male patient with a UTX missense variant. Mutations/variants are denoted and relative to the UTX sequence NM_021140.1. The location of the TRP (tetratrico peptide repeat region) and JmjC (Jumonji C) domains is shown. (B) Identification of variations in EZH2-domain structure and positions of mutations in patients. All mutations were found in the SET domain or in close proximity. In 3 subjects mutations were identified along with CN-LOH7q encompassing EZH2 as shown for representative patient (SET indicates suppressor of variegation 3-9, enhancer of zeste and trithorax domain; SANT-DNA–binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB). (C) Mutations identified in methyltransferase domain of DNMT3A in 7 patients with CMML and AML-derived CMML. Homozygous DNMT3A mutations were associated with CN-LOH2p involving DNMT3A. The location of the PWWP (a highly conserved proline-tryptophan-tryptophan-proline motif), PHD (plant homeodomain finger, a zinc-fingerlike motif), and MTase (methyltransferase) domains is shown. Missense mutations are indicated by black; frameshift by red, and nonsense mutations by green arrows.
UTX, EZH2, and DNMT3A mutations identified in CMML. (A) In a cohort of 72 patients with CMML and AML-derived CMML, 2 somatic, inactivating UTX mutations were detected by sequencing. Four additional missense variants that could correspond to rare polymorphisms were identified. Profile of CN-LOH of X chromosome by SNP-A in one male patient with a UTX missense variant. Mutations/variants are denoted and relative to the UTX sequence NM_021140.1. The location of the TRP (tetratrico peptide repeat region) and JmjC (Jumonji C) domains is shown. (B) Identification of variations in EZH2-domain structure and positions of mutations in patients. All mutations were found in the SET domain or in close proximity. In 3 subjects mutations were identified along with CN-LOH7q encompassing EZH2 as shown for representative patient (SET indicates suppressor of variegation 3-9, enhancer of zeste and trithorax domain; SANT-DNA–binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB). (C) Mutations identified in methyltransferase domain of DNMT3A in 7 patients with CMML and AML-derived CMML. Homozygous DNMT3A mutations were associated with CN-LOH2p involving DNMT3A. The location of the PWWP (a highly conserved proline-tryptophan-tryptophan-proline motif), PHD (plant homeodomain finger, a zinc-fingerlike motif), and MTase (methyltransferase) domains is shown. Missense mutations are indicated by black; frameshift by red, and nonsense mutations by green arrows.
Summary of identified mutations in UTX, EZH2, and DNMT3A genes in patients with CMML and with CMML-derived AML
| Sex . | Age, y . | WHO Dx . | Metaphase cytogenetics . | SNP-A lesions . | UTX mutation . | EZH2 mutation . | DNMT3A mutation . | ||
|---|---|---|---|---|---|---|---|---|---|
| Gain . | Loss . | CN-LOH . | |||||||
| M | 69 | sAML | 46,XX[20] | 22q11.1 | 7q22.1 | 4q21.21qter 11q13.5qter | c.1324 C > T p.Gln442X* | Negative | Negative |
| M | 78 | CMML-1 | 46,XY,inv(3)(q21q26)[20] | N | N | N | c.2373 A > G p.Asn791Ser (homozygous)* | Negative | Negative |
| F | 53 | CMML-2 | 47,XX,+21[19]/46,XX[1] | N | N | 11q22 3q25 | c.2502 G > T p.Glu834Asp* | Negative | Negative |
| M | 78 | sAML | 46,XY[20] | N | N | X | c.2765 G > A p.Arg922Lys* | Negative | c.2645 G > A p.Arg882His† |
| F | 65 | CMML-2 | 46,XX[20] | N | 12q24.31 | 7q11.23qter 14q23.2qter | c.3551 delA p.Asn1184fsX2* | Negative | Negative |
| F | 60 | CMML-1 | 46,XX,der(16)t(1;16)(q12;q11.2) [17]/46,XX[3] | 1p11.2qter 8q12.1 | 16p11.1qter | N | c.808 A > G p.Ile270Val* | Negative | Negative |
| F | 76 | CMML-1 | 46,XX[20] | 8p23.2 | N | N | Negative | c.1747 C > T p.Arg583X† | Negative |
| M | 60 | sAML | 45,X,-Y[20] | N | N | 4q23qter 7q11.23qter | Negative | c.2006-2063 (dup57nt) p.Ser669fs (homozygous) | Negative |
| F | 78 | CMML-1 | 47, XX, +8 | 8p23.2-q24.3 | 7q34 | 7q11.21-q36.3 | Negative | c.2069 G > A p.Arg690His (homozygous)† | Negative |
| M | 69 | CMML-1 | 46,XY[17] | N | N | 7q22.1qter | Negative | c.2193 C > G p.Tyr731X (homozygous)† | Negative |
| M | 72 | sAML | 46,XY[20] | Yq11.23 | N | 2pterp14 | Negative | Negative | c.2096 G > A p. Gly699Asp (homozygous) |
| F | 69 | CMML-2 | 46,XX[20] | N | N | 2p11.2p25.3 | Negative | Negative | c.2446 C > T p. Gln816X (homozygous) |
| M | 68 | sAML | 46,XY[20] | N | N | 13 | Negative | Negative | c.2644 C > T p.Arg882Cys† |
| M | 72 | sAML | 46,XY[20] | N | N | 3p21.31p21.1 | Negative | Negative | c.2645 G > A p.Arg882His† |
| F | 74 | sAML | 46,XX,−7[3]/46,XX,add(12)(p13)[4]/46,XX,−7, add(12)(p13)[2]/46,XX[11] | Yp11.2 | 7 | N | Negative | Negative | c.2191 del (TTC) p.Phe731 |
| F | 58 | CMML-2 | 46,XX,del(17)(q24)[6]/46,XX[24] | 21q22 13q22.2 | 2q24 3q32.1 | 1p36.13pter | Negative | Negative | c.2645 G > C p.Arg882Pro† |
| Sex . | Age, y . | WHO Dx . | Metaphase cytogenetics . | SNP-A lesions . | UTX mutation . | EZH2 mutation . | DNMT3A mutation . | ||
|---|---|---|---|---|---|---|---|---|---|
| Gain . | Loss . | CN-LOH . | |||||||
| M | 69 | sAML | 46,XX[20] | 22q11.1 | 7q22.1 | 4q21.21qter 11q13.5qter | c.1324 C > T p.Gln442X* | Negative | Negative |
| M | 78 | CMML-1 | 46,XY,inv(3)(q21q26)[20] | N | N | N | c.2373 A > G p.Asn791Ser (homozygous)* | Negative | Negative |
| F | 53 | CMML-2 | 47,XX,+21[19]/46,XX[1] | N | N | 11q22 3q25 | c.2502 G > T p.Glu834Asp* | Negative | Negative |
| M | 78 | sAML | 46,XY[20] | N | N | X | c.2765 G > A p.Arg922Lys* | Negative | c.2645 G > A p.Arg882His† |
| F | 65 | CMML-2 | 46,XX[20] | N | 12q24.31 | 7q11.23qter 14q23.2qter | c.3551 delA p.Asn1184fsX2* | Negative | Negative |
| F | 60 | CMML-1 | 46,XX,der(16)t(1;16)(q12;q11.2) [17]/46,XX[3] | 1p11.2qter 8q12.1 | 16p11.1qter | N | c.808 A > G p.Ile270Val* | Negative | Negative |
| F | 76 | CMML-1 | 46,XX[20] | 8p23.2 | N | N | Negative | c.1747 C > T p.Arg583X† | Negative |
| M | 60 | sAML | 45,X,-Y[20] | N | N | 4q23qter 7q11.23qter | Negative | c.2006-2063 (dup57nt) p.Ser669fs (homozygous) | Negative |
| F | 78 | CMML-1 | 47, XX, +8 | 8p23.2-q24.3 | 7q34 | 7q11.21-q36.3 | Negative | c.2069 G > A p.Arg690His (homozygous)† | Negative |
| M | 69 | CMML-1 | 46,XY[17] | N | N | 7q22.1qter | Negative | c.2193 C > G p.Tyr731X (homozygous)† | Negative |
| M | 72 | sAML | 46,XY[20] | Yq11.23 | N | 2pterp14 | Negative | Negative | c.2096 G > A p. Gly699Asp (homozygous) |
| F | 69 | CMML-2 | 46,XX[20] | N | N | 2p11.2p25.3 | Negative | Negative | c.2446 C > T p. Gln816X (homozygous) |
| M | 68 | sAML | 46,XY[20] | N | N | 13 | Negative | Negative | c.2644 C > T p.Arg882Cys† |
| M | 72 | sAML | 46,XY[20] | N | N | 3p21.31p21.1 | Negative | Negative | c.2645 G > A p.Arg882His† |
| F | 74 | sAML | 46,XX,−7[3]/46,XX,add(12)(p13)[4]/46,XX,−7, add(12)(p13)[2]/46,XX[11] | Yp11.2 | 7 | N | Negative | Negative | c.2191 del (TTC) p.Phe731 |
| F | 58 | CMML-2 | 46,XX,del(17)(q24)[6]/46,XX[24] | 21q22 13q22.2 | 2q24 3q32.1 | 1p36.13pter | Negative | Negative | c.2645 G > C p.Arg882Pro† |
EZH2 mutations were identified in 3 of 52 patients (6%) with CMML (all with CMML-1; N = 36; 8%) and 1 patient with AML antecedent to CMML (N = 20; 5%). In 3 patients mutations were associated with LOH7q encompassing EZH2, whereas in 4 patients bearing CN-LOH7q EZH2 mutations were not found. No mutations have been detected in patients with loss of chromosome 7 and/or 7q (−7/7q; N = 6). In 1 patient, a heterozygous nonsense mutation was encountered. All detected mutations disrupted highly conserved amino acids of EZH2. Notably, the SET domain, essential for the methyltransferase activity of EZH2, was either altered or truncated in ≥ 1 allele in all mutant cases (Figure 2B; Table 2).
DNMT3A mutations were detected in 2 of 52 patients with CMML (4%), all with CMML-2 (2 of 16; 12%). In addition, 5 mutant cases were identified in patients with CMML-derived AML (6 of 20; 25%). The DNMT3A mutations were mostly heterozygous and affected the R882 position (Figure 2C; Table 2), a mutational hotspot previously reported to be of somatic origin.29,30 This mutation is predicted to act in a dominant-negative fashion and to reduce methyltransferase activity by > 50% in vitro.29 Most mutant cases displayed a normal karyotype by MC (5 of 7; 71%). CN-LOH encompassing DNMT3A was found in 3 patients; 2 of these patients harbored novel homozygous DNMT3A mutations.
Frequency and distribution of other mutations
To gain potential insights into pathway abnormalities and relationships, we performed a targeted mutational screen of 11 candidate genes in our cohort. In patients with CMML-1/2 (N = 52), TET2 mutations were identified in 44% (N = 23), ASXL1 in 46% (N = 24), CBL in 13% (N = 7), IDH1/2 in 4% (N = 2), KRAS in 8% (N = 4), and NRAS in 2% (N = 1). In patients with CMML-derived AML (N = 20), TET2 mutations were detected in 60% (N = 12), ASXL1 in 35% (N = 7), CBL in 15% (N = 3), IDH1/2 in 5% (N = 1), KRAS in 5% (N = 1), NRAS in 10% (N = 2). JAK2 V617F was found in 1% of patients (Figure 3; supplemental Table 1).
Schematic representation of the localization of molecular mutations at the protein level. Localization of all mutations detected in 72 persons with CMML and CMML-derived AML within the protein. Genomic sequencing of the protein-coding region and splice donor/acceptor sites showed missense (black triangles), nonsense (green triangles), and frameshift mutations (red triangles). Mutations found in CBL and TET2 genes resulted in new splice variants (blue triangles). Most mutations were found in conserved domains, and specific known conserved motifs and domains are shown for each protein. DSBH indicates 2OG-dependent dioxygenase domain; IMDH, isocitrate/isopropylmalate dehydrogenase, conserved sites; PEST sequence, P indicates proline (P), glutamic acid (E), serine (S), and threonine (T); PHD, the plant homeodomain; TKB, tyrosine kinase binding domain; RF, ring finger; L, linker sequence; PPP, proline-rich region; LZ/UBA, leucine zipper/ubiquitin-associated domain; and RAS, subfamily of RAS small GTPases binding domain.
Schematic representation of the localization of molecular mutations at the protein level. Localization of all mutations detected in 72 persons with CMML and CMML-derived AML within the protein. Genomic sequencing of the protein-coding region and splice donor/acceptor sites showed missense (black triangles), nonsense (green triangles), and frameshift mutations (red triangles). Mutations found in CBL and TET2 genes resulted in new splice variants (blue triangles). Most mutations were found in conserved domains, and specific known conserved motifs and domains are shown for each protein. DSBH indicates 2OG-dependent dioxygenase domain; IMDH, isocitrate/isopropylmalate dehydrogenase, conserved sites; PEST sequence, P indicates proline (P), glutamic acid (E), serine (S), and threonine (T); PHD, the plant homeodomain; TKB, tyrosine kinase binding domain; RF, ring finger; L, linker sequence; PPP, proline-rich region; LZ/UBA, leucine zipper/ubiquitin-associated domain; and RAS, subfamily of RAS small GTPases binding domain.
TET2 was the most frequently mutated gene in our study. TET2 mutations were found throughout the entire coding region, including the N-terminus and the conserved Cys-rich and DSBH 2OG-Fe(II)–dependent dioxygenase domains26 (Figure 3). In 13 patients, compound heterozygous (biallelic) TET2 mutations were found; in 7 patients mutations were associated with LOH spanning the entire coding region of TET2. Among all mutations, 11 (26%) were single-base substitutions, 22 (52%) were frameshift, 11 (26%) were nonsense mutations, and 4 mutations (9%) targeted acceptor/donor splice sites (N = 48). Except for 4 patients, missense mutations were associated with homozygosity or compound heterozygosity. In contrast to a diversity of TET2 lesions, 31 ASXL1 mutations were all heterozygous, including 19 frameshift and 7 nonsense mutations presumed to truncate the PHD. In addition, the controversial ASXL1 variant, c.1934dupG p.Gly646TrpfsX12, was found in 15% of all patients.39 Missense mutations in ASXL1 were found only in 5 patients, and no LOH encompassing ASXL1 gene was detected. Both homozygous and hemizygous mutations in CBL were identified (60% of mutant cases showed CN-LOH11q or loss of 11q23.3), as well as heterozygous mutations. All RAS mutations were heterozygous, except for one NRAS homozygous variant found in a patient with UPD1p. No LOH involving 2q (IDH1 gene) and 15q (IDH2 gene) was found, confirming the heterozygous nature of the corresponding mutations (Figure 3; supplemental Table 1).
Pathogenetic relationships
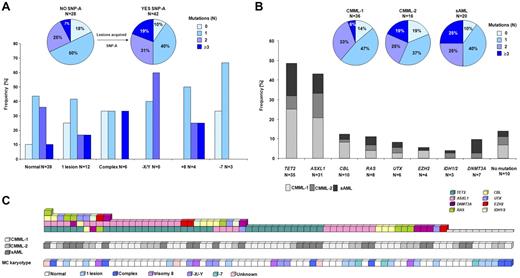
Comprehensive mutational analysis of the CMML-1/2 and CMML-derived AML cohorts resulted in the identification of patients with multiple mutational events and defined mutational patterns (Figure 4). Most of the patients with normal cytogenetics (by MC) showed the presence of ≥ 1 of the mutations studied (35 of 39; 90%). Among those patients with chromosomal defects, more mutations were detected (≥ 3 mutations in 19% vs 7% in those without chromosomal defects by SNP array; P = .29; Figure 4A). Furthermore, as expected patients with more advanced disease accumulated more mutations (≥ 3 mutations in 25% vs 19% and 6% in CMML-derived AML vs CMML-2 and CMML-1; P = .70 and P = .08, respectively; Figure 4B).
Frequencies and distributions of molecular mutations. Identified mutations and their frequencies in patients annotated according to karyotype by MC and SNP-A (A) and diagnostic criteria of CMML-1, CMML-2, and sAML (B). Most identified mutations were detected in patients with a normal karyotype by MC. SNP-A improved the detection rate, and only 10% of patients with uncovered SNP-A lesions did not carry any mutation. Among these patients an accumulation of mutations was observed (≥ 3 mutations in 19% vs 7% in those without new SNP-A lesions). Additional accumulation of mutations was found in AML derived from CMML compared with CMML-1/2. (C) Systematic mutational sequencing shows almost all possible combinations of genetic states for studied genes in patients with CMML and AML derived from CMML. Of 72 patients, only 14% of patients did not display mutations in any of the studied genes.
Frequencies and distributions of molecular mutations. Identified mutations and their frequencies in patients annotated according to karyotype by MC and SNP-A (A) and diagnostic criteria of CMML-1, CMML-2, and sAML (B). Most identified mutations were detected in patients with a normal karyotype by MC. SNP-A improved the detection rate, and only 10% of patients with uncovered SNP-A lesions did not carry any mutation. Among these patients an accumulation of mutations was observed (≥ 3 mutations in 19% vs 7% in those without new SNP-A lesions). Additional accumulation of mutations was found in AML derived from CMML compared with CMML-1/2. (C) Systematic mutational sequencing shows almost all possible combinations of genetic states for studied genes in patients with CMML and AML derived from CMML. Of 72 patients, only 14% of patients did not display mutations in any of the studied genes.
Novel UTX mutations were present as a sole event (of those tested) in one patient, whereas 5 of 6 cases harbored UTX mutations associated with other mutations (ASXL1, TET2, or CBL). Although the sample size is a limitation, UTX mutations were more frequent in advanced CMML (CMML-2, CMML-AML). EZH2 variants were also accompanied by other molecular mutations affecting TET2 and ASXL1 or RAS genes. Intriguingly, UTX and EZH2 mutations were not identified simultaneously in any of the patients studied. DNMT3A mutations were detected as the sole abnormality in 3 patients and in 4 cases were associated with other mutational events. Among IDH1/2 mutant cases, one patient harbored a nonsense TET2 mutation (Figure 4C).
In 86% of all patients, > 1 mutation was found (62 of 72). One mutation was found in 31 patients (43%); concomitant second or ≥ 3 mutations were found in 21 (29%) and 11 (14%) patients, respectively (Figure 4). In the CMML cohort, > 1 mutation was found in 85% (44 of 52 patients), whereas in the CMML-derived AML cohort, multiple lesions were detected in 90% of patients (18 of 20). Concomitant second or ≥ 3 mutations in CMML-1/2 were found in 16 (31%) and 11 (21%) patients, respectively, and in AML antecedent CMML was found in 5 (25%) patients each. The most frequently observed mutation combinations included TET2 and ASXL1, as well as TET2 and CBL, in 22% and 7% of all patients, respectively. TET2/ASXL1 combinations were present in 46% of all TET2 mutants (16 of 35), whereas TET2/CBL were present in 14% of all TET2 mutant cases (5 of 35).
Combined analysis of clinical effect of mutations
We analyzed the association of specific mutations within the whole cohort and within individual risk groups of CMML (CMML-1/2 and AML-derived CMML; supplemental Table 2A-C). Although few mutant cases within each group were found, patients with CMML-derived AML harbored DNMT3A mutations more frequently than patients with other subtypes (25% [5 of 20] vs 4% [2 of 52]; P = .015). We have also compared patients carrying a specific mutation (eg, in ASXL1 or TET2 gene, etc) to the remaining patients without mutation of the corresponding gene with respect to a number of patient factors (supplemental Table 3A-C). Because of a significant overlap in the distribution of the 11 gene mutations tested, specific mutations cannot be compared with each other, and analysis would only be able to detect very pervasive features. Nevertheless, we were able to note certain biologic trends. For instance, there was a significant association of TET2 and CBL mutations with more advanced age (P = .007 and P = .04, respectively). These associations remain significant even after patients with CMML-1/2 were analyzed separately (P = .013 and P = .043, respectively). In the whole cohort, CBL mutant cases more frequently displayed splenomegaly (67% vs 36% in patients without these mutations, respectively; P = .14) as well as in separate analyses of CMML-1/2 cases (83% vs 37%, P = .075) (supplemental Table 3). In general, increased marrow blasts were seen in RAS and DNMT3A mutants (P = .019 and P = .019, respectively), whereas higher total leukocytes were seen in DNMT3A-positive cases only (P = .016) (supplemental Table 3). When CMML-1/2 cases were analyzed separately (N = 52), the association with increased marrow blasts was seen only in RAS mutant cases (P = .035; supplemental Table 3). We also observed that in the CMML-1/2 group, patients with TET2 mutations were more probable to have normal karyotype by MC compared with patients without mutations (32% vs 61%, respectively; P = .048).
Survival analyses
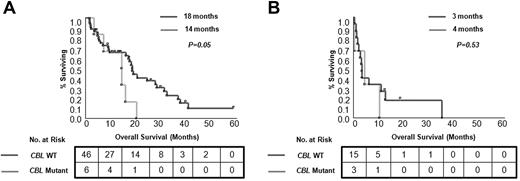
Univariate analyses of clinical and laboratory factors for OS were performed. Patients with CMML with cytogenetic abnormalities detected by SNP-A karyotyping show a shorter OS (Figure 1). In the whole cohort, blast count and disease stage as well as DNMT3A mutational status had a significant effect on patient survival (P = .01, P = .001, and P = .04, respectively). However, in multivariate analyses, which included age and other conventional prognostic factors, only disease stage (CMML-1/2 vs sAML) remained an independent prognostic factor (P = .05). When CMML cases were analyzed separately (without sAML derived from CMML), disease category (CMML-1 vs CMML-2) and the presence of CBL mutations were independent prognostic factors (P = .04 and P = .03, respectively; supplemental Table 4). In general, no differences in OS were observed for patients divided into subgroups on the basis of the presence of individual mutations, except for CBL mutants (Figure 5; supplemental Figure 2). Inferior outcomes were observed for CBL mutant cases compared with patients without mutations (OS, 9 months vs 16 months; P = .04), and this relationship was preserved within the CMML-1/2 subgroup (OS, 14 months vs 18 months; P = .05). In addition, patients with an accumulation of several mutations showed a trend toward inferior outcomes versus patients with single mutations and patients without mutations (in the CMML-1/2 subgroup ≥ 3 mutations vs 1 mutation vs none: OS, 14 months vs 20 months vs 18 months; and in the CMML-derived AML subgroup: OS, 1 month vs 3 months vs 7 months, respectively; supplemental Figure 3).
Kaplan-Meier survival curves estimated according to presence of CBL mutations. Differences in OS for patients with CMML (A) and with CMML-derived AML (B) are shown. For each group median months and number of analyzed cases are presented.
Kaplan-Meier survival curves estimated according to presence of CBL mutations. Differences in OS for patients with CMML (A) and with CMML-derived AML (B) are shown. For each group median months and number of analyzed cases are presented.
Although no significant difference was observed in OS between patients with or without TET2 mutations, the analysis of 5hmC levels (which could be affected by various pathogenic processes, including TET2, IDH1/2,28,40,41 and possibly other undiscovered mutations) showed a difference in OS. When patients were divided into groups on the basis of high or low levels of 5hmC as previously described,28 a trend toward a better OS was noticed in cases with lower levels of 5hmC (OS, 19 months vs 7 months, respectively; P = .06), with significant differences for cases with normal karyotype compared with others (OS, 26 months vs 3 months, respectively; P < .0001). In the high-risk CMML group, patients with lower 5hmC levels showed better OS than patients with higher levels (OS, 20 months vs 4 months, respectively; P = .03).
Serial studies
Sequential samples of 6 patients with mutated CMML before AML transformation, at the time of AML transformation, or at the time of AML relapse were also studied (supplemental Table 5). Patient no. 12 was initially found to have a TET2 mutation at the time of initial diagnosis of CMML-1, but, on remission induction therapy for AML, the TET2 mutation was not detected. This mutation reappeared when therapy was discontinued because the patient progressed. The patient subsequently underwent a matched related donor nonmyeloablative allogeneic peripheral blood stem cell transplantation that resulted in a complete remission, and no TET2 mutation was detected at that time and for years after. Patient no. 28 was initially classified as having refractory cytopenia with unilineage dysplasia (RCUD). No molecular mutations were noted, but his disease progressed and a diagnosis of CMML-1 was established, at which point a homozygous CBL mutation was detected. Subsequently, the patient transformed to AML, but no new mutations were found. Patient no. 41 was originally diagnosed with CMML-1 and was found to have a DNMT3A mutation. Soon after, the patient progressed to CMML-2, and the same type of mutation was present during disease evolution. Patient no. 65 was found to have TET2, ASXL1, and DNMT3A mutations at the time of initial diagnosis of therapy-related MDS. The same clones persisted at the time of CMML diagnosis and at subsequent disease transformation to AML several years later. Patient no. 49 had a BM examination that did not support the diagnosis of MDS and had normal cytogenetics but was found to have heterozygous TET2 and ASXL1 mutations. Subsequent follow-up showed the diagnosis of CMML-2 with CN-LOH4q spanning region of TET2 gene and TET2 and ASXL1 mutations. Patient no. 2 was found to have homozygous TET2 and EZH2 mutations at initial diagnosis of CMML-1; after allogeneic hematopoietic stem cell transplantation, molecular remission was achieved. On relapse, the original homozygous TET2 and heterozygous EZH2 mutations were found.
Discussion
The prognosis of CMML is generally poor, and effective therapies are limited. The identification of the underlying pathogenesis, including recurrent molecular lesions, is of the utmost importance in developing disease-modifying treatment strategies.
On routine analysis of CMML with MC, clonal chromosomal abnormalities were identified in < 50% of cases, whereas SNP-A increased the potential identification of clonal markers in ≤ 70% of cases; patients with previously cryptic lesions had decreased survival. This finding is consistent with results obtained in MDS.7 Here, we show that targeted mutational analysis of patients with CMML can identify a mutation and clonal marker in ∼ 86% of patients, including those with an otherwise normal karyotype as assessed by routine and array-based karyotyping.
Recently, somatic mutations in genes involved in epigenetic regulation have been described in myeloid malignancies, including ASXL1, EZH2, UTX, and DNMT3A.11,20-22,29,30,35 On the basis of the functional involvement in epigenetic regulation, our study focused on the potential role of UTX, EZH2, and DNMT3A alterations in CMML. We report for the first time UTX mutations in patients with CMML. UTX mutations were present at the C-terminus and the N-terminus of the UTX protein but mostly in the region adjacent to the JmjC domain required for UTX activity. In addition, among identified germ line UTX variants, 4 of 6 were missense substitutions (with a frequency of < 1/400 in male controls35 ), but the functional consequences of these variants remain unclear. Overall, UTX mutations were detected less often than mutations in TET2 or ASXL1 and in some instances were concomitantly present. Consequently, inactivation of UTX because of mutation or haploinsufficiency through LOH may also impair differentiation programs in affected progenitor cells. Recurrent homozygous EZH2 mutations have been previously identified in patients with MDS, MPN, and MDS/MPN,20-22 implicating loss of EZH2 function in the pathogenesis of these conditions. In our cohort, recurrent EZH2 mutations were identified in 5.5% of all patients. Notably, all mutations except one missense variant were predicted to result in premature termination as a consequence of nonsense or frameshift mutations. These mutations were often associated with the presence of CN-LOH encompassing EZH2 but not del(7) or del(7q). The mutations targeted evolutionary highly conserved residues in the SET and adjacent CXC domains, required for histone methyltransferse activity. We have not identified simultaneous occurrence of both EZH2 and UTX mutations, although this observation is based on the small number of identified mutants. With respect to the function of these genes in the regulation of H3K27me3 levels, one could stipulate that corresponding mutations will be associated with alterations in H3K27 methylation patterns. Indeed, in all studied acquired EZH2 mutations, significantly decreased levels of H3K27 trimethylation were found as shown by our previous studies and other investigators.21,22 Most probable, similar, although opposite in action, consequences might occur with the inactivation of UTX. Nevertheless, no significant changes on trimethylation status of H3K27 were found so far in 2 patients with mutated UTX,42 a finding consistent with another report.35
The foregoing mutations in CMML will help to reconcile the existence of a presumptive reduction of H3K27me3 levels because of EZH2 heterozygosity or homozygosity and loss-of-function of the histone H3K27 demethylase UTX. In addition, overexpression of EZH2 has been reported in patients with AML and with MDS,43 most probably resulting in silencing of genes involved in differentiation. Loss of UTX activity would be enzymatically equivalent to a gain of function for EZH2 or simply overexpression of this enzyme. In either situation greater steady state levels of H3K27me3 are induced in malignant cells. H3K27me3 is associated with transcriptionally repressed genes, whereas H3K4me3 turns transcription on.44 A rapid decrease of H3K27me3 occurs during stem cell differentiation.45 Thus, inactivation of UTX in stem cells should lead to the maintenance of H3K27me3 marks on target genes and should prevent differentiation. Indeed, this is parallel to our observations whereby UTX mutations were detected in more-aggressive forms of CMML and AML derived from CMML. Conversely, EZH2 mutations were detected among patients with CMML-1, suggesting an early event in malignant evolution.
On the basis of the recent discovery of DNMT3A mutations, we have also investigated their presence in CMML, detecting mutations in 10% of patients. DNMT3A mutations were present in the methyltransferase domain and mostly affected R882 amino acid, possibly causing changes in the DNA-binding groove or its interaction with DNMT3L.29,30 All of these mutations were heterozygous, except for 2 homozygous variants associated with CN-LOH in the region containing the gene. DNMT3A-recurrent mutations at a single amino acid position suggest a gain-of-function mechanism, although very heterogenous mutations at different positions in this gene generally might suggest loss of function. DNMT3A mutations were enriched in patients with normal karyotype, similarly to those reported in de novo AML.30
Similar to DNMT3A, TET2 and IDH1/2 may affect DNA methylation and thereby convey a clonally restricted epigenetic instability phenotype. Like TET1, TET2 is an α-ketoglutarate–dependent enzyme that converts 5mC to 5hmC,27,28 and loss of TET2 function is associated with altered DNA methylation.28 Neomorphic IDH mutations may indirectly affect the activity of α-ketoglutarate–dependent enzymes, including TET proteins and UTX, through the generation of high levels of 2-hydroxyglutarate and may therefore have consequences analogous to mutations of these genes (eg, decreased 5hmC levels or alteration of H3K27 methylation status).40,41,46
In our study, mutations of TET2 were found at a high frequency in patients with CMML, comparable to frequencies reported by other groups.12,13,23 A similar observation was found for ASXL1; 43% of patients harbored a mutation, including 15% of cases that carried the controversial variant c.1934dupG p.Gly646TrpfsX12.39 CBL mutations were identified in 14%, RAS mutations in 11%, and the JAK2 V617F in only 1% of patients with otherwise typical CMML. IDH1/2 mutations were identified in 4% of patients at more advanced stage of disease, probably contributing as secondary events to their progression.47 Only in 14% of patients did we not find any mutation in all genes analyzed. Most probably, apart from new mutations yet to be identified, they could carry RUNX1 mutations found in patients with CMML.8
Combined analyses of mutational spectrums in CMML shows the molecular heterogeneity of the disease, allows for subclassification of patients on the basis of mutations or the combination of mutations present, and facilitates analyses of outcome studies on the basis of the presence of objective mutational biomarkers. Our study clearly shows the molecular heterogeneity of CMML and that the disease can be associated with multiple mutations that probably accumulated in the course of disease. It is not clear which of these mutations are of ancestral type and which clearly herald progression. Clinical analysis of patients with UTX or EZH2 mutations did not show any distinct features, probably because of the small number of identified cases and overlap with other mutations. With regard to DNMT3A mutations, clinical association has been enriched, and correlation with higher marrow blasts and with blood cells counts was observed, but, again, this preliminary observation has to be confirmed in a larger cohort. Similarly, no specific phenotype associated with ASXL1 or TET2 mutations was identified, with the exception for the association of TET2 and CBL mutations with older age and increased marrow blasts in RAS-mutant cases.
The analysis of clinical outcomes did not show a significant effect of specific mutations; however, there are a number of methodologic issues that may explain this finding. In general, the diagnosis of CMML carries a poor prognosis, with a median survival of 12 months,48 precluding detection of clinical differences within the current cohort size. Nevertheless, CBL mutant cases showed less favorable survival, and a similar trend was seen for DNMT3A mutant cases. Moreover, patients who have accumulated more mutations had inferior outcomes compared with those with single mutations or those without any mutations.
A similar lack of distinctive features has been reported for ASXL1, IDH1/2, and TET2 in CMML.10-13,49 However, a worse prognosis and acute transformation has been linked to ASXL1 mutations.23 In contrast, the reports on the prognostic effect of TET2 mutations were controversial,10,12,13 but our study suggests that they do not convey additional negative effects on prognosis, at least in this very homogenous and well-defined CMML cohort. It is also possible that other mutations may be present in some patients with known lesions, such as those with TET2, indicating that analyzed cohorts are not sufficiently homogeneous. For instance, decreased levels of 5hmC were found in patients without TET2 or IDH1/2 mutations, implying the presence of other functional related molecular defects.28 We therefore also investigated global 5hmC levels as a possible prognostic marker in CMML that would unify patients with a defective common molecular pathway. When we correlated this surrogate marker with survival, a trend toward better outcomes was seen for patients with low 5hmC levels. Although this has to be clarified in a larger study, similar outcomes were reported for TET2 mutants in other studies.12,13
Screening of sequential samples suggests that some of the molecular mutations may precede disease evolution. In particular, ASXL1 and TET2 mutations are detectable early in the clinical course. It is also probable that multiple mutations detected in one patient are a result of a stepwise evolution process. However, only deep sequencing and calculation of clonal sizes could, albeit indirectly, prove this point. Additional mechanisms, such as epigenetics and altered signaling pathways, may also be responsible for disease progression in CMML. The persistence of the same clone at the time of relapse and disease progression also provide helpful insight, because it suggests that future therapies aimed at the original molecular defects may result in disease eradication. Following molecular markers during the disease course may also be helpful in determining the likelihood of successful therapy and, conversely, the possibility of impending disease relapse.
In summary, our study shows that, despite a relatively uniform clinical phenotype and globally poor prognosis, CMML shows tremendous underlying molecular heterogeneity, with some mutations affecting convergent pathways or compensating for each other. For example, CBL mutations, via activation of RTK receptors, could result in activation of the RAS pathway, producing similar effects to RAS mutations and aberrant pSTAT5 activation. Similarly, TET2 and IDH-1/2 mutations may lead to a decrease in 5mC hydroxylation or EZH2 and UTX mutations to altered H3K27me3 and be functionally similar to mutations of ASXL1 which also affect histone methylation. Above and beyond these findings our study shows, for the first time, the detection of UTX and DNMT3A mutations in patients with CMML. In general, we propose that epigenetic machinery may be frequently affected by mutations in myeloid malignancies in particular, as shown in this CMML cohort. However, larger patient cohorts will be needed to determine whether any of these gene mutations are critical in the pathogenesis of CMML. These diverse mutations can be associated with a uniform phenotype not distinguishable on clinical grounds, further showing that molecular diagnostics is needed for selection of targeted therapies for individual patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by in part by grants R01HL098511 (J.P.M), U54 RR019391 (M.A.S. and J.P.M.), K24 HL-077522 (J.P.M.), DOD MPD510343 (M.A.M.), AI44432 (A.R.), RC1 DA028422 (A.R.), and grants from AA&MDS International Foundation and the Robert Duggan Charitable Fund (J.P.M.).
National Institutes of Health
Authorship
Contribution: A.M.J., H.M., and H.S. designed, performed, and analyzed most of the experimental work; Y.S., F.T., V.V., and C.P. helped with patient genotyping; A.M.J., M.A.M., and J.P.M. compiled the manuscript; C.O. performed SNP-A and cytogenetic analyses; R.V.T performed classification of patients and database and performed clinical analyses; Y.H., and A.R. generated the anti-CMS antiserum, developed the quantitative assay for genomic 5-hmC,28 and quantified the levels of 5-hydroxymethylcytosine in genomic DNA; E.D.H. reviewed pathologic specimens, interpreted results, and edited the manuscript; M.A.S. helped in classification and identification of patients and edited the manuscript; A.R. provided vital reagents and contributed to the design and analysis of the study; M.A.M. and A.L. selected some of the patients and contributed to the design and analysis of the study; and J.P.M. conceptualized the project, supervised the experimentation, analyzed the results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, MD, PhD, Taussig Cancer Institute/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.