Abstract
We have investigated the prognostic significance of isocitrate dehydrogenase 2 (IDH2) mutations in 1473 younger adult acute myeloid leukemia patients treated in 2 United Kingdom Medical Research Council trials. An IDH2 mutation was present in 148 cases (10%), 80% at R140 and 20% at R172. Patient characteristics and outcome differed markedly between the 2 mutations. IDH2R140 significantly correlated with nucleophosmin mutations (NPM1MUT), whereas IDH2R172 cases generally lacked other molecular mutations. An IDH2R140 mutation was an independent favorable prognostic factor for relapse (P = .004) and overall survival (P = .008), and there was no significant heterogeneity with regard to NPM1 or FLT3 internal tandem duplication (FLT3/ITD) genotype. Relapse in FLT3/ITDWTNPM1MUTIDH2R140 patients was lower than in favorable-risk cytogenetics patients in the same cohort (20% and 38% at 5 years, respectively). The presence of an IDH2R172 mutation was associated with a significantly worse outcome than IDH2R140, and relapse in FLT3/ITDWTNPM1WTIDH2R172 patients was comparable with adverse-risk cytogenetics patients (76% and 72%, respectively).
Introduction
Recurrent mutations in the genes coding for isocitrate dehydrogenase isoforms 1 and 2 (IDH1 and IDH2) have recently been described in acute myeloid leukemia (AML) patients.1-3 Both enzymes convert isocitrate to α-ketoglutarate in an NAD phosphate+-dependent manner, but they have different subcellular locations: IDH1 in the cytoplasm and IDH2 in mitochondria. Heterozygous mutations at IDH1R132, the functionally equivalent IDH2R172, and also IDH2R140 were found to cause loss of normal enzymatic function and accumulation of 2-hydroxyglutarate (2-HG) through neomorphic enzyme activity, with a substrate switch from isocitrate to α-ketoglutarate, which is then converted to 2-HG.4 IDH2 mutations have been reported in 9% to 19% of AML cases, predominantly IDH2R140 rather than IDH2R172 alterations.5-8 However, the prognostic significance of these mutations remains unclear with either no impact on outcome,5-7 or an adverse effect when IDH1 and IDH2 mutant patients are analyzed together in the subgroup of normal karyotype patients with a nucleophosmin 1 mutation (NPM1MUT) but not an internal tandem duplication in the fms-like tyrosine kinase gene (FLT3/ITDWT).8 In addition, there is some evidence that IDH2R172 mutations confer an extremely poor prognosis, although this is based on very small numbers of patients, many of them elderly.5,9
We recently demonstrated that the prognostic impact of an IDH1 mutation is dependent on FLT3/ITD genotype.10 To determine whether IDH2 mutations have a similar effect on outcome and to investigate whether IDH2R140 and IDH2R172 mutations differ in their impact, we analyzed IDH2 mutant status and clinical outcome in 1473 adult nonacute promyelocytic leukemia AML patients, mostly 60 years of age or younger, treated on the United Kingdom Medical Research Council AML10 and 12 trials.
Methods
Details of the study cohort and samples, patient therapy, clinical end points, and statistical methods are in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Amplicons of IDH2 exon 4 were screened by denaturing high-performance liquid chromatography and mutations confirmed by restriction enzyme digestion or sequencing (supplemental Methods). Ethical approval for the trials and tissue collection for research was obtained from the Multi-Centre Research Committee of Wales.
Results and discussion
An IDH2 mutation was detected in 148 of 1473 patients (10%): 119 (80%) altered IDH2R140 (113 R140Q, 3 R140L, 2 R140W, and 1 R140G) and 29 (20%) affected IDH2R172 (all R172K). Patient details are in supplemental Table 1. Overall, IDH2 mutant-positive patients (IDH2MUT) were significantly older than IDH2-wild type (IDH2WT) patients (median, 47 vs 43 years, P = .001), but there was no difference according to sex, type of leukemia (de novo/secondary), or presenting white blood cell count. Most IDH2MUT patients (92%) were in the cytogenetic intermediate-risk group using the Medical Research Council classification11 : 64% had a normal karyotype and 28% an intermediate-risk abnormal karyotype. In addition, 3% had adverse-risk and 5% favorable-risk cytogenetics (supplemental Table 1). There was no correlation between presence of an IDH2 mutation and FLT3/ITD, FLT3/TKD, or CEBPA mutations, but a highly significant positive correlation with presence of an NPM1 mutation; 61% of IDH2MUT patients were NPM1MUT (P < .0001). There was also a negative correlation with IDH1 mutations (P = .05).
There were striking differences in the characteristics of patients with the different IDH2 mutations (supplemental Table 1). IDH2R172 cases had significantly lower presenting white blood cell counts than IDH2R140 cases (3.6 vs 30.9 × 109/L, P < .0001) and were more likely to have an intermediate-risk abnormal karyotype than a normal karyotype, 60% and 36%, respectively, of those with known cytogenetics compared with 20% and 71% for IDH2R140 cases (P = .0001). Furthermore, they were less likely to have other molecular mutations, the incidence of FLT3/ITDs was 7% versus 24%, respectively (P = .05), and NPM1 mutations 3% versus 75% (P < .0001). None of the IDH2R172 cases had FLT3/TKD, biallelic CEBPA, or IDH1 mutations.
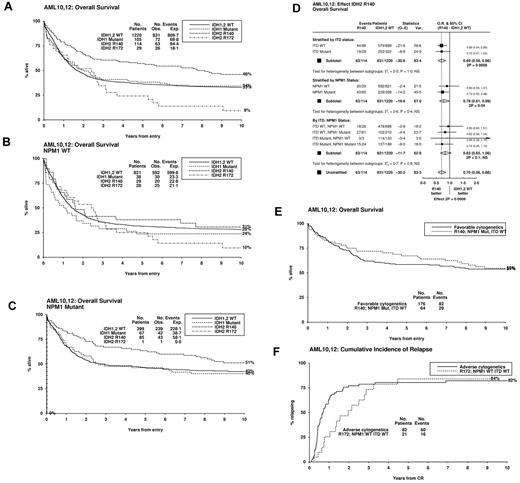
For outcome analysis, patients were separated into 4 groups, IDH2R140 and IDH2R172 mutated, IDH1MUT, and IDH1/2WT. Five patients with both IDH1 and IDH2 mutations were excluded. IDH2R140 patients had a favorable response to therapy, whereas IDH2R172 patients had a poor response, complete remission rates were 88%, 48%, 81%, and 75% for the 4 groups, respectively (Table 1). The differences were reduced if patients remitting without complete hematologic recovery were included (92%, 79%, 81%, and 82%, respectively), and IDH2R140 or IDH2R172 status was not a significant factor in multivariable analysis for complete remission (supplemental Table 2). Long-term outcome was significantly different between IDH2R140 and IDH2R172 cases (P = .0001 for cumulative incidence of relapse [CIR], P = .0002 for overall survival [OS]). IDH2R140 patients had a reduced CIR (supplemental Figure 1) and improved OS (Figure 1A), and the significance was maintained in multivariable analysis (P = .004 and P = .008 respectively; supplemental Table 2). IDH2R172 patients had an increased CIR (supplemental Figure 1) and reduced OS (Figure 1A), although this did not reach significance (P = .09 for OS, IDH2R172 vs IDH1/2WT). Results were similar if analyses were censored at allogeneic transplantation in first complete remission.
Impact of IDH2 mutant status on outcome. (A) Kaplan-Meier curves for OS stratified by IDH2 and IDH1 mutant status. OS in (B) NPM1WT and (C) NPM1MUT subgroups. (D) Mantel-Byar analysis for the effect on overall survival of an IDH2R140 mutation stratified by NPM1 and FLT3/ITD mutant status. (E) OS in FLT3/ITDWTNPM1MUTIDH2R140 patients compared with those with favorable-risk cytogenetics. (F) Relapse in FLT3/ITDWTNPM1WTIDH2R172 patients compared with those with adverse-risk cytogenetics.
Impact of IDH2 mutant status on outcome. (A) Kaplan-Meier curves for OS stratified by IDH2 and IDH1 mutant status. OS in (B) NPM1WT and (C) NPM1MUT subgroups. (D) Mantel-Byar analysis for the effect on overall survival of an IDH2R140 mutation stratified by NPM1 and FLT3/ITD mutant status. (E) OS in FLT3/ITDWTNPM1MUTIDH2R140 patients compared with those with favorable-risk cytogenetics. (F) Relapse in FLT3/ITDWTNPM1WTIDH2R172 patients compared with those with adverse-risk cytogenetics.
When the results were stratified according to NPM1 genotype, relapse was reduced in IDH2R140 cases both with and without an NPM1 mutation (supplemental Figure 2A-B), and there was no significant heterogeneity between the 2 groups (P = .8). For OS, although the beneficial impact of an IDH2R140 mutation appeared larger in NPM1MUT patients, 5-year OS 64% for NPM1MUTIDH2R140 and 47% for NPM1MUTIDH1/2WT patients (P = .03), 38% for NPM1WTIDH2R140, and 32% for NPM1WTIDH1/2WT patients (P = .8; Figure 1B-C), there was no significant heterogeneity between the 2 groups (P = .4). There was also no difference in the benefit of an IDH2R140 mutation according to FLT3/ITD genotype (Figure 1D; supplemental Figure 2C-D). Of note, the outcome for FLT3/ITDWTNPM1MUTIDH2R140 patients was equivalent to or better than that for favorable-risk cytogenetics patients in the same cohort: OS 67% versus 59%, respectively (Figure 1E), CIR 20% versus 38% (supplemental Figure 3A).
As most patients with an IDH2R172 mutation (26 of 29, 90%) lacked either a FLT3/ITD or an NPM1 mutation, we examined the subgroup of 772 FLT3/ITDWTNPM1WT patients. IDH2R172 patients had an extremely poor prognosis with a much higher incidence of relapse than IDH2R140 patients (CIR 89% vs 36% at 5 years; supplemental Figure 3B) and worse 5-year OS (28% vs 38%, respectively); but with the small numbers of IDH2-mutated patients in these subgroups, neither difference reached statistical significance (P = .06 for relapse, P = .14 for OS). Of the 25 IDH2R172 patients with available cytogenetics, only one had an adverse karyotype. Nevertheless, by 5 years after remission, the risk of relapse for FLT3/ITDWTNPM1WTIDH2R172 patients was equivalent to that for adverse-risk cytogenetics patients in this cohort: 76% versus 72%, respectively (Figure 1F). OS was slightly better, 23% versus 11%, respectively (supplemental Figure 3C).
The favorable outcome associated with an IDH2R140 mutation has not been reported previously and was unexpected. Structural modeling suggests that, although IDH2 residue R172 is analogous to the mutated residue in IDH1 (R132), IDH2R140 is also predicted to interact with the β-carboxyl of isocitrate and its mutation to bind and orient α-ketoglutarate in the enzymes' active site in a manner similar to mutated IDH2-R172.3 Indeed, both mutations produce elevated levels of 2-HG rather than isocitrate.2-4 Further work is required to determine whether changes in substrate concentrations, such as the shift in reduced NAD phosphate/NAD phosphate+ levels induced by the switch in enzyme function of the IDH mutants, or the absolute level of 2-HG, affect the ability of the cell to respond to cellular stress and, ultimately, impact on chemosensitivity.12,13 Understanding the underlying biology of the differences in outcome observed for IDH1 and IDH2 mutants will be important for future studies and may lead to the development of novel approaches to therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the clinical investigators who entered and managed patients in these two trials.
This work was supported by Leukaemia & Lymphoma Research United Kingdom and the United Kingdom Medical Research Council. The work was undertaken at University College London Hospitals/University College London, who received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
Authorship
Contribution: R.E.G. and D.C.L. designed the study; C.L.G., C.M.E., and L.Z. performed assays; R.K.H. analyzed the data; A.K.B. was the principal trial coordinator; C.L.G., R.E.G., R.K.H., and D.C.L. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, UCL Cancer Institute, Paul O'Gorman Bldg, 72 Huntley St, London, WC1E 6DD, United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.