Abstract
Posttranscriptional mechanisms are now widely acknowledged to play a central role in orchestrating gene-regulatory networks in hematopoietic cell growth, differentiation, and tumorigenesis. Although much attention has focused on microRNAs as regulators of mRNA stability/translation, recent data have highlighted the role of several diverse classes of AU-rich RNA-binding protein in the regulation of mRNA decay/stabilization. AU-rich elements are found in the 3′-untranslated region of many mRNAs that encode regulators of cell growth and survival, such as cytokines and onco/tumor-suppressor proteins. These are targeted by a burgeoning number of different RNA-binding proteins. Three distinct types of AU-rich RNA binding protein (ARE poly-U–binding degradation factor-1/AUF1, Hu antigen/HuR/HuA/ELAVL1, and the tristetraprolin/ZFP36 family of proteins) are essential for normal hematopoiesis. Together with 2 further AU-rich RNA-binding proteins, nucleolin and KHSRP/KSRP, the functions of these proteins are intimately associated with pathways that are dysregulated in various hematopoietic malignancies. Significantly, all of these AU-rich RNA-binding proteins function via an interconnected network that is integrated with microRNA functions. Studies of these diverse types of RNA binding protein are providing novel insight into gene-regulatory mechanisms in hematopoiesis in addition to offering new opportunities for developing mechanism-based targeted therapeutics in leukemia and lymphoma.
Introduction
Approximately two-thirds of protein abundance variation of mammalian cells can be accounted for by posttranscriptional mechanisms.1 Although the role of micro-RNAs (miRNA) in regulating mRNA stability and translation is well established, an expanding number of proteins that bind AU-rich elements (ARE) in the 3′-untranslated regions (UTRs) of mRNA have been discovered to mediate mRNA decay/stabilization or translational control.2 Of the dozen or so AU-rich RNA binding proteins (AUBPs) that have been characterized in detail,3 most bind a multimeric pentamer sequence AUUUA, typically located within a 50- to 150-nucleotide adenine- and uridine-rich 3′-UTR element3,4 (and references therein). An estimated 8% of mammalian mRNA transcripts are potentially targeted by AUBPs, and many well-characterized AUBP mRNA targets encode key regulatory proteins, such as growth factors, cytokines, chemokines, and oncoproteins4 (and references therein). The mechanisms through which AUBPs mediate posttranscriptional regulation, particularly in the context of immune regulation and the inflammatory response, have been reviewed extensively elsewhere.2-8 Most function as accessory proteins to recruit mRNAs and to regulate their fate in various subcellular compartments, such as the exosome mediating 3′ to 5′ decay, processing bodies for 5′ to 3′ decay, and stress granules for translational arrest.3,7 Some function cooperatively or antagonistically with each other or with argonaut endoribonucleases within RNA-induced silencing complexes mediating miRNA-dependent decay.7,8 In addition, most AUBPs display some degree of molecular promiscuity in their ability to modulate gene expression through ARE-independent mechanisms involving either DNA-binding or protein interaction.
Accumulating data implicate 5 AUBP types as key regulators of normal and malignant hematopoiesis. These are: ARE poly-U–binding degradation factor-1 (AUF1), HUR (ELAV-like family 1), KH-type splicing regulatory protein (KSRP/KHSRP), nucleolin, and the members of the ZFP36 (Tis11) family, ZFP36, ZFP36L1, and ZFP36L2. In this review, we summarize the relevant experimental evidence for this and discuss the complex regulatory interactions that exist between these AUBPs and their mRNA targets and how their functions are integrated with miRNA gene-regulatory pathways in hematopoietic cells.
AUF1
The AUF1 protein occurs in 4 different isoforms (p37, p40, p42, and p45) and regulates gene expression through multiple mechanisms. It directly regulates a number of mRNAs that play key roles in hematopoietic cells, including BCL2, TNFα, IL3, IL6, COX2, cyclin D1, IL8, cFOS, and cJUN.9-14 It can also modulate mRNA translation and gene transcription. For example, although AUF1 mediates decay of cMYC proto-oncogene mRNA in cell-free systems,15 it induces MYC translation in an ARE-dependent manner in K562 erythroleukemia and THP-1 myelomonocytic cell line models.10 AUF1 associates with the AUBP nucleolin to form LR1,16 a heterodimeric protein complex that binds to the GGNCNAG(G/C)CTG(G/A) consensus sequence present in the MYC P1 promoter17 to induce transcription. LRI binds a similar consensus sequence present in the EBV EBNA-1 promoter and immunoglobulin heavy chain S regions (G-rich regions) involved in class switch recombination.16,18 By contrast, AUF1 (p42 and p45 isoforms) exhibits phosphorylation-dependent sequence-specific binding to and transcriptional repression of the CD21 promoter in several B-cell lines and in primary B cells.19
AUF1 function is essential for normal lymphopoiesis (Figure 1). Knockout mice that are deficient in all 4 AUF1 isoforms display decreased numbers of splenic T and B cells (especially follicular B cells) together with a modest expansion of the splenic marginal zone B-cell compartment20 (Figure 1). The reduced numbers of follicular B cells is attributable to increased turnover and apoptosis, most probably arising as a consequence of decreased levels of prosurvival proteins, Bcl2, BclXl, and Bfl1/A1.20 AUF1-deficient mice also display reduced IgG serum levels in response to T-independent antigen despite no impairment in germinal center formation and class switch recombination.20
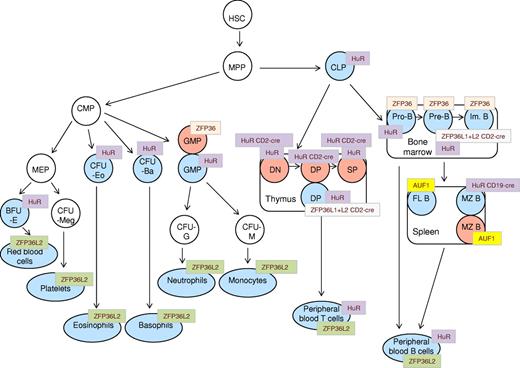
Schematic representation of phenotypic abnormalities in hematopoietic compartments of AUBP-deficient mice. A generalized scheme of hematopoietic cell differentiation is shown, onto which are “mapped” the various compartments that are reported to be affected in AUF1, HUR, HUR-CD19cre, HUR-CD2cre, ZFP36, ZFP36L2, and ZFP36L1+L2-CD2cre knockout mice. Blue represents hematopoietic cell types with diminished cell numbers; and red, elevated cell numbers. HSC indicates hematopoietic stem cells; MPP, multipotent progenitor cells; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN, double-negative (CD4−/CD8−) thymocytes; DP, double-positive (CD4+/CD8+) thymocytes; SP, single-positive (CD4+, CD8+) thymocytes; Im. B, immature B cells; FL B, follicular B cells; MZ B, marginal zone B cells; PB T, peripheral blood T cells; PB B, peripheral blood B cells; GMP, granulocyte-macrophage progenitor; CFU-G, colony factor unit granulocytes; CFU-M, colony-forming unit macrophage; MEP, megakaryocyte erythroid progenitor; CFU-Meg, colony-forming unit megakaryocytes; CFU-Eo, colony-forming unit eosinophil; CFU-Ba, colony-forming unit basophil; and BFU-E, burst-forming unit-erythroid.
Schematic representation of phenotypic abnormalities in hematopoietic compartments of AUBP-deficient mice. A generalized scheme of hematopoietic cell differentiation is shown, onto which are “mapped” the various compartments that are reported to be affected in AUF1, HUR, HUR-CD19cre, HUR-CD2cre, ZFP36, ZFP36L2, and ZFP36L1+L2-CD2cre knockout mice. Blue represents hematopoietic cell types with diminished cell numbers; and red, elevated cell numbers. HSC indicates hematopoietic stem cells; MPP, multipotent progenitor cells; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN, double-negative (CD4−/CD8−) thymocytes; DP, double-positive (CD4+/CD8+) thymocytes; SP, single-positive (CD4+, CD8+) thymocytes; Im. B, immature B cells; FL B, follicular B cells; MZ B, marginal zone B cells; PB T, peripheral blood T cells; PB B, peripheral blood B cells; GMP, granulocyte-macrophage progenitor; CFU-G, colony factor unit granulocytes; CFU-M, colony-forming unit macrophage; MEP, megakaryocyte erythroid progenitor; CFU-Meg, colony-forming unit megakaryocytes; CFU-Eo, colony-forming unit eosinophil; CFU-Ba, colony-forming unit basophil; and BFU-E, burst-forming unit-erythroid.
AUF1 associates with the nucleophosmin-anaplastic lymphoma kinase (NMP-ALK) fusion protein that results from the most common translocation, t(2;5)(p23;q35), found in anaplastic large cell lymphoma (ALCL) and some other lymphomas.21 It colocalizes with NMP-ALK–containing granules where it is targeted for phosphorylation. In its phosphorylated state, AUF1 has been suggested to result in stabilization of mRNA targets that would otherwise be targeted for decay. The expression of several AUF1 targets, such as cyclin A, cyclin D, MYC,21 and JUN,22 correlates with ALCL disease (Table 1).
HUR/ELAV1
The ubiquitously expressed HUR/ELAV1 member of the ELAV family of AUBP acts both to stabilize and to modulate the translational efficiency of various mRNAs involved in normal and malignant hematopoiesis. It stabilizes mRNAs encoding BCL2, MCL1, cyclin A, cyclin B1, cyclin D1, lymphotoxin-α, GM-CSF, IL4, VEGF, CD3ζ, CD95L, GATA-3, XIAP, and survivin.23-30 HUR has also been reported to destabilize mRNAs for AML1/RUNX1, CD2, VAV1, NFκBIE, CD3ϵ, TNFα, and STAT3.27 HUR enhances the translational efficiency of mRNAs encoding p53, cytochrome c, XIAP, and BCL2 while suppressing translation of p27, MYC, and WNT5α.26,29,31 The mechanism of HuR-mediated translational repression of MYC has been studied in some detail and requires recruitment of let-7 miRNA in an Ago2-dependent manner.32 Let-7 miRNA functions as translational repressor through recognition of the m7G cap on MYC mRNA leading to either impaired recruitment of elF4E or to inhibition of elF4E-elF4G association.33 HUR itself is subject to miRNA-mediated regulation through translational repression by several miRNAs (Figure 2), including miR-16 and miR-125α,34,35 miR-519,36 and miR-34α.37
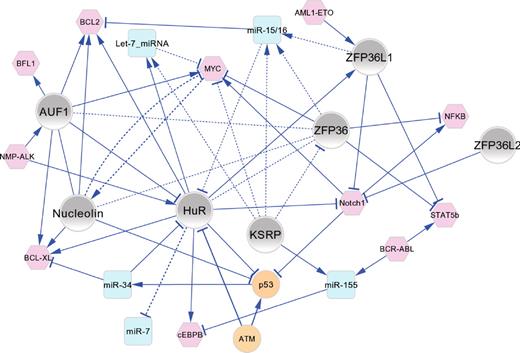
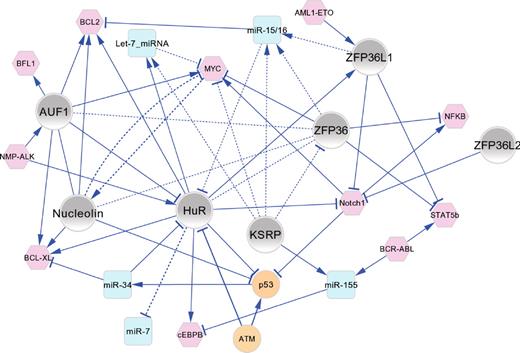
Functional connectivity map of AUBPs in hematopoiesis and leukemogenesis. The major regulatory interactions between AUBPs (gray nodes), miRNAs (blue nodes), and oncoproteins/tumor suppressor proteins (pink/orange nodes) are shown as a network graph in which edges represent experimentally validated functional connections that have been manually curated from the literature and are described in the text. In most cases, the directionality of positive (→) or negative (⊣) regulatory interactions is known. Solid line edges represent interactions documented in normal/malignant hematopoietic cells; and dashed-line edges, interactions identified in other cell types that are probable to occur in hematopoietic cells. The network graph shows an organic layout view constructed using Cytoscape Version 2.8.0.
Functional connectivity map of AUBPs in hematopoiesis and leukemogenesis. The major regulatory interactions between AUBPs (gray nodes), miRNAs (blue nodes), and oncoproteins/tumor suppressor proteins (pink/orange nodes) are shown as a network graph in which edges represent experimentally validated functional connections that have been manually curated from the literature and are described in the text. In most cases, the directionality of positive (→) or negative (⊣) regulatory interactions is known. Solid line edges represent interactions documented in normal/malignant hematopoietic cells; and dashed-line edges, interactions identified in other cell types that are probable to occur in hematopoietic cells. The network graph shows an organic layout view constructed using Cytoscape Version 2.8.0.
Recent transcriptome-wide screening has identified many thousands of additional mRNAs that are direct and functional targets of HUR in human cells.38,39 HUR binding sites that overlap with or are adjacent to miRNA binding sites facilitate combinatorial regulation by HUR and miRNAs, most probably through alleviation of miRNA-mediated repression by HUR.39 Significantly, many HuR binding sites occur in introns and are often associated with splice sites to regulate alternative splicing. RNA processing mechanisms also account for HUR-mediated repression of the miR-7 miRNA that functions as a potent tumor suppressor in many human cancer types.38
Hur is required for normal mouse embryonic development.40 The postnatal function of Hur was investigated in a tamoxifen-dependent cre recombinase deletion mouse model.40 After tamoxifen-induced Hur deletion, mice die within 10 days. An analysis of the hematopoietic system within the first 4 days of tamoxifen treatment revealed that these mice are characterized by possession of atrophic spleens, lymph nodes, and thymus accompanied by reduced numbers of lymphoid, myeloid, and erythroid progenitor cells in the bone marrow40 (Figure 1). The peripheral blood lymphocyte count is markedly diminished while granulocyte numbers are modestly increased; red blood cell and platelet counts are unaffected.40 Pro-B and pre-B cells, but not mature, late pre-B cells, are characterized by widespread apoptosis and necrosis in HuR-deficient mice that can be explained by increased levels of proapoptotic p53, NOXA, PUMA, p21, and caspase-9 mRNA levels and decreased levels of pro-survival Bcl2 and BclXl mRNA levels in bone marrow of Hur-deficient mice.40 The disparity found between immature and mature B cells may be because Hur is normally expressed at higher levels in immature B compared with mature B cells. There is also a shift in subcellular distribution of Hur protein from both nuclear and cytoplasmic in immature pre-B cells to exclusively cytoplasmic in mature B cells.40
The same Hur-deficient mice also display decreased numbers of double-positive (CD4+ CD8+, DP) T cells but with an increase in the number of double-negative (CD4−CD8−, DN) and single-positive cells (CD4+ or CD8+, SP) thymic T cells.40 However, when deletion of HuR was specifically targeted to thymocytes,27 there were increased numbers of thymic DN, SP, and DP cells in thymus (Figure 1). In the latter study, Hur was found to affect the proliferation of DN cells, the process of positive selection, and the egress of mature T cells to the periphery.27 Increased proliferation of the DN cells was attributed to decreased p53 protein levels.27 Decreased positive selection was attributed to decreased phosphorylation of ZAP-70, Lck, and PKCθ, whereas reduced egress of T cells was attributed to reduced chemotaxis in response to the CCR7 ligand CCL21 and the CXCR4 ligand SDF1.27
Finally, when deletion of Hur was targeted to B cells (CD19-Cre), the mice were characterized by reduced numbers of marginal zone B cells in the spleen and reduced number of B1 cells in the peritoneal cavity, whereas follicular B cells remained unchanged41 (Figure 1). In addition, these mice displayed an impaired germinal center formation and IgG1 secretion in response to a T cell–dependent antigen.41
In common with AUF1, HUR colocalizes with NMP-ALK in NMP-ALK granules in ALCL and is a substrate for tyrosine phosphorylation by this kinase.42 The level of cytoplasmic HUR is not affected by the presence of NMP-ALK, which instead promotes its localization to polysomes and binding to cEBPβ mRNA42 This results in stabilization and increased translation of cEBPβ mRNA.42 Increased expression of c-EBPβ is a unique feature of ALCL compared with several other leukemias and lymphomas.42 The c-EBPβ protein regulates apoptosis in ALK-positive cell lines as well as tumor growth in mice in vivo.43
HUR may also play a wider role in leukemogenesis (Table 1). It is overexpressed in M4 acute myeloid leukemia (AML) and correlated with high levels of elF4E.44 It is also overexpressed in acute phase and blast crisis in chronic myeloid leukemia (CML) compared with chronic phase disease with expression increasing progressively during transit from the chronic phase to the blast crisis.45 In chronic lymphocytic leukemia (B-CLL), HUR mRNA is differentially expressed between cases with high and low levels of miR-16/miR-1546 consistent with its mRNA being targeting by miR-16.34
One intriguing mRNA target for HUR in leukemogenesis is the mRNA encoding the prosurvival protein survivin (Table 1). Overexpression of HUR results in inhibition of p53-dependent transcription and down-regulation of expression of the survivin protein.23 HUR stabilizes p53 mRNA leading to increased levels of p53 protein, which in turn negatively regulates survivin gene transcription. However, when p53 is silenced, HUR overexpression actually increases levels of survivin by stabilizing its mRNA. This suggests that the outcome of HUR expression on survivin levels may depend on the p53 mutational status of leukemic cells.23
Finally, HuR have been implicated in posttranscriptional genotoxic/oxidative stress pathways in mammalian cells,47 and recent data havew shown that it functions as a key mediator of the checkpoint kinase, ataxia telangiectasia mutated (ATM) in B lymphocyte cell lines.48 In response to double-stranded DNA breaks, the ATM kinase phosphorylates a cascade of downstream effectors, including the checkpoint kinase CHK2, which in turn phosphorylates HUR to modulate its mRNA-binding and translational control functions. Ionizing radiation elicits a dramatic change in the profile of mRNAs that are associated with HUR in an ATM-dependent manner. Of these, the mRNAs encoding cancer-associated proteins p21, FOXO3, MEK1, MEK2, DUSP10, and (interestingly) the AUBP, ZFP36L1 were experimentally validated as ATM-regulated HUR targets. In each case, radiation-responsive up-regulation of their encoded proteins was mediated by translational control rather than by changes in mRNA levels.48
Persons with the autosomal recessive ataxia telangiectasia (AT) condition who inherit defective ATM function are predisposed to develop lymphomas and lymphoid leukemias at high frequency. The ATM gene is also commonly mutated in sporadic lymphoid malignancies. Thus, HUR probably plays a key role in mediating the effects of loss of function of the ATM checkpoint kinase tumor suppressor in sporadic lymphoid malignancies as well as in AT patients48 (Table 1).
KSRP
KSRP is a multifunctional protein involved in various processes, including transcription, alternative pre-mRNA splicing, and mRNA localization in addition to its role as an AUBP mediating mRNA decay. Some key targets of KSRP that are relevant to hematopoiesis include ZFP36, NOXA, PRDM1 variant 1 (Blimp-1 variant 1), BACH2, cIAP2, BMP2, BMP6, CCL20, cyclin D3, CCR1, CCR3, CCR7, CXCL2, CXCL3, CXCL10, CXCL11, ID2, IL6, SOCS2, E selectin, and TLR4.49 In addition, KSRP is involved in the maturation of several miRNAs that are dysregulated in leukemias and lymphomas, as summarized in Table 1. KSRP has been found to immunoprecipitate with Drosha and DGCR8 proteins and to regulate the maturation of let7a, miR-15b, miR-16, miR-20, miR-21, miR-26b, miR-27b, miR-98, miR-106a, miR-125b, miR-196a, miR-199a, miR-301, and miR-595.50,51 KSRP seems to promote the association of Drosha with pri-miRNA and of Dicer with pre-miRNA at least for let-7a and miR-2151 and as thus to promote their maturation. Another report found that KSRP is involved in the maturation of miR-155 from pri- and pre-miR-155 after LPS stimulation in macrophages.52
The KSRP protein also targets ID3 inhibition of E2A-regulated transcription of the Notch1 gene.53 Deregulated Notch1 expression arising either through mutational mechanisms or the t(7;9) translocation has been causally associated with the development of human T-acute lymphoblastic leukemia (T-ALL).53 Although one published study has reported high expression of KSRP in CML acute phase/blast crisis compared with chronic phase disease45 (Table 1), it remains to be determined whether altered expression/function of KSRP plays a causal role in leukemogenesis via miRNA maturation or other pathways.
Nucleolin
Nucleolin is found in the nucleolus, nucleoplasm, and the cell membrane54 and possesses a diverse set of functions, such as regulation of PolI- and PolII-mediated transcription, nucleo-cytoplasmic transport, the formation of nucleosomes, and chromatin remodeling, in addition to its role in the stabilization of certain mRNAs through binding to AREs.54 Among the mRNAs that are stabilized by nucleolin are several that encode proteins with established roles in normal and malignant hematopoiesis, including IL2, BCL2, BCLXl, and CD40L (CD154).55-57 Independently of its ability to bind BCL2 mRNA, nucleolin can also interact with phosphorylated BCL2 protein in the nucleus during the transition from prophase to anaphase in mitosis.58 This may affect spindle formation as well as chromosome segregation.58 In hematopoietic cells, nucleolin regulates transcription of the CD34 gene by direct binding to the CD34 promoter,59 a process that is negatively regulated by hypophosphorylated Rb.60 As mentioned previously, nucleolin, together with AUF1, forms the transcriptional regulatory LR1 complex that regulates multiple genes in lymphocytes.16,17 In addition, nucleolin binds to and stabilizes G-quadruplex DNA in the Myc promoter to inhibit Myc transcription in vivo.61 Because nucleolin transcription is itself activated by MYC,62 it may function as part of a negative regulatory feedback loop with MYC. The functions of nucleolin are also integrated with p53-dependent pathways. It binds to the 5′-UTR of p53 mRNA and causes inhibition of translation under basal conditions and after irradiation.63 Down-regulation of nucleolin results in induction of apoptosis and inhibition of proliferation through induction of p53.64 However, nucleolin can also interact with the Hdm-2 protein resulting in stabilization of p53.65
A unique feature of nucleolin compared with the other AUBPs is that it can undergo N-glycosylation–dependent localization to the plasma membrane66 to modulate key signaling pathways via protein interaction mechanisms. Membrane bound nucleolin interacts with and induces accumulation of the KRAS proto-oncogene and subsequent activation of the MAPK/ERK pathway.67 In response to CD21 activation, it also becomes tyrosine phosphorylated and associates with the unphosphorylated p85 subunit of the PI3K pathway.68 Finally, through RNA-dependent association with DGCR8, nucleolin may serve an adaptor function in transferring the pri-miRNAs to the nucleolus and in the processing-cleavage of pri-miRNAs to pre-miRNA within the nucleolus by a Drosha-DGCR8-nucleolin complex.69
As summarized in Table 1, deregulated expression of nucleolin is a consistent feature of several types of leukemia. In B-CLL, nucleolin expression is correlated with high levels of BCL2 protein,55 reflecting the function of nucleolin in stabilizing BCL2 mRNA. Similarly, in AML, high levels of nucleolin70 may act to maintain transcription of the CD34 gene, particularly because AML cells typically lack Rb function, which acts to negatively regulate the transcriptional regulatory functions of nucleolin.60 Nucleolin is also overexpressed in pediatric ALL, irrespective of disease subtype.71 In a more recent study, expression of nucleolin, together with nucleophosmin, was found to be deregulated in de novo primary ALL and AML patients as well as in refractory and relapsed leukemia patients.72 Expression of these 2 proteins positively correlated with higher relapse rates and negatively correlated with overall survival and relapse-free survival72 (Table 1).
ZFP36 family
The 3 ZFP36 family members, ZFP36, ZFP6L1, and ZFP36L2, function primarily by targeting an extensive, overlapping repertoire of mRNAs for degradation via the exosome or via Xrn1 exonuclease.3,5 One exception to this is ZFP36, which has recently been shown to also target the NF-κB pathway via ARE-independent mechanisms involving recruitment of HDAC1 or HDAC3 to the NF-κB p65 subunit73 and through inhibition of p65 nuclear import.74
Several studies in cell line models and primary cells have implicated all 3 members of the ZFP36 family of proteins in both proapoptotic functions and as regulators of cell differentiation through a variety of mechanisms.5 The ZFP36L1 protein, for example, is required for rituximab-mediated apoptosis of B-CLL cells.75 It also functions as a negative regulator of plasma cell differentiation in the mouse BCL1 cell line model by targeting mRNA encoding the plasma cell transcription factor, PRDM1/BLIMP1 (A. Nasir, J.D.N., M. Aldrovandi, A. Zekarati, M.B., M. Bijmakers, S. Thompson, J.J.M., unpublished observations, October 2011). Because PRDM1/BLIMP1 acts to repress transcription of the ZFP36L1 gene,76 these 2 regulatory proteins may function as a bi-stable switch mechanism during the transition from mature B to plasma cells. Overexpression of ZFP36L1 and ZFP36 in human CD34+ cord blood cells inhibits erythroid differentiation and proliferation by targeting degradation of STAT5b mRNA, which in turn leads to repression of STAT5b-dependent transcription of GATA-1.77
Definitive insight into the distinctive role of each ZFP36 family member in hematopoiesis has been provided by gene targeting studies in mice (Figure 1). Loss of ZFP36 function results in an increase in the numbers of granulocytes in the spleen and bone marrow accompanied by a decrease in the numbers of B cells in the bone marrow.78 This is attributable to increased proliferation of short-term hematopoietic stem cells and multipotent progenitor cells arising from increased levels of GCSF, IL1β, IL6, and TNFα found in the plasma of ZFP36 knockout mice, and these effects are noncell-autonomous.78 The defective hematopoiesis may also arise from dysregulation of the transcription factor E47, the mRNA of which is normally targeted for degradation by ZFP36.79 The defective hematopoietic phenotype seen in ZFP36L2-deficient mice (Figure 1) is associated with elevated Cxc11 expression and diminished levels of Cxcl4, Cxcl7, integrin a2b (CD41), integrin b3 (CD61), CD59α, SLAMF1, and PBX1.80 Conditional knockout mice in which loss of ZFP36L1/L2 function is targeted to lymphocytes (Figure 1) display widespread defects in lymphopoiesis. Notably, all ZFP36L1/L2 double-knockout mice developed thymic tumors, and the majority also displayed other abnormalities, including splenomegaly and lymphadenopathy.81 Interestingly, in these same mice, there was a block in B-cell development from the CD25+B220+ stage (Figure 1), although presumably this was not B-cell intrinsic as deletion of ZFP36L1/L2 in these mice was under control of CD2-cre and CD2 is not expressed in B cells.82
Consistent with their proapoptotic functions, members of the ZFP36 family have been implicated as tumor suppressors in solid tumors.83 A similar functional role for these proteins is emerging in leukemogenesis. Compelling evidence for this is provided by the recent finding that concomitant targeted deletion of ZFP36L1 and ZFP36L2 in mouse thymocytes results in T-ALL as early as3 months of age, whereas single deletion has no effect.81 The CD8+ tumors are oligoclonal in nature, and their development is attributable to deregulated expression of Notch1 mRNA, which is normally targeted for degradation by ZFP36L1.81 As mentioned previously, deregulated Notch1 expression has been causally associated with the development of human T-ALL.53
At a mechanistic level, loss of function of different ZFP36 family members probably contributes to leukemogenesis through multiple mechanisms (Table 1). In adult T cell lymphoma (ATL), ZFP36, but not ZFP36L1, has been found to physically associate with the TAX protein and to inhibit its transactivation function.84 The TAX-ZFP36 complex is targeted to the proteasome resulting in increased degradation of TAX.84 In addition, ZFP36 may recruit HDAC1 and HDAC3 to Tax with concomitant inhibition of p300/CBP recruitment as shown for the p65 NF-κB subunit.73 Indeed, TAX protein has been found to associate with p300/CBP and HDAC1 or HDAC3; the presence of HDAC1 inhibits the binding of p300/CBP to TAX that is required for TAX transactivation function.85 The physical association between ZFP36 and TAX also appears to titrate out ZFP36 function because TAX inhibits degradation of TNFα mRNA by ZFP36 in ATL.84 A similar mechanism may contribute to the deregulated expression of other cytokines in ATL, such as IL2, IL6, IL10, and GM-CSF, which are induced by HTLV-1 virus and that are targets for ZFP36.84 Consistent with a role for loss of ZFP36 function in disease progression of ATL, ZFP36 mRNA is down-regulated in acute-phase ATL compared with chronic-phase disease86 (Table 1).
The transcription factor NF-κB functions in one of the most frequently deregulated pathways in leukemias and lymphomas87 and is a key target for mRNA decay and transcriptional repression by the ZFP36 protein as mentioned previously.73,74 ZFP36, together with ZFP36L1, also functions in miRNA pathways in leukemogenesis. mRNA targets for miR-16/miR-15 are enriched in AREs motifs,46 and miR-16 requires the presence of ZFP36 and ZFP36L1 to bind to ARE-containing mRNA.88 miR-16/miR-15 are commonly down-regulated in B-CLL; and indeed, miR-16/miR-15 knockout mice develop B-CLL.89 One intriguing possibility is that in B-CLL cases, in which miR-16/miR-15 expression is not down-regulated, the absence of ZFP36 family members may promote the development of CLL indirectly by attenuating the function of miR-16/miR-15. ZFP36L1 is expressed at low levels in most B-CLLs.75 In some cases, this may be attributable to an interstitial deletion of the ZFP36L1 locus at 14q24.90 Interestingly, ZFP36L2 mRNA is differentially expressed between B-CLL cases with high and low levels of miR-16/miR-15,46 consistent with its mRNA being targeting by miR-16.34 Other genetic/epigenetic mechanisms are also probably involved in down-regulating expression of ZFP36 family members. In B-cell lymphomas, for example, the promoters of the ZFP36 and ZFP36L1 genes are targeted for transcriptional repression by the BCL6 oncogene.91,92
In myeloid leukemias, the expression of ZFP36 family members is correlated with clinicopathologic features of disease (Table 1). In 2 independent studies of CML, ZFP36 expression displays a progressive decrease during transit from chronic phase to blast crisis.45,93 ZFP36 is known to target STAT5b mRNA for degradation in CD34+ hematopoietic cells.77 Because proliferation and survival of BCR-ABL–transformed cells requires STAT5 function,94 loss of ZFP36 function would be expected to enhance the malignant properties of CML cells. ZFP36L1, by contrast, is reportedly overexpressed in M2 AML carrying the AML1-ETO translocation, and this is correlated with induction of cell proliferation and inhibition of differentiation.95 Similarly, ZFP36L2 mRNA was found to be overexpressed in resistant/relapsed AML patients.96
Most established tumor suppressor genes are found in mutant form in at least a subset of tumors in which they are involved. Recent data indicate that at least for ZFP36L1 and ZFP36L2 this criterion may be fulfilled (Table 1). High throughput sequencing of primary multiple myeloma cases has identified deletions within a regulatory intron of the ZFP36L1 gene in a significant minority of cases.97 Although the functional significance of this remains to be established, the same study also found that the Rrp44 gene (encoding exosome component 11) is mutated within its coding region in 11% of multiple myeloma patients.97 The resulting predicted loss of exosome function would probably lead to widespread impairment of cellular mRNA degradation machinery in multiple myeloma. Mutations in the ZFP36L2 gene have also recently been reported in various subtypes of AML and ALL98 ; one of these mutants was shown to encode a protein with impaired antiproliferative activity compared with wild-type ZFP36L2.98
In conclusion, given the multitude of important regulators of cell growth, differentiation, and survival that are targeted at the posttranscriptional level by AUBPs, it is perhaps unsurprising that the phenotypes of mice that are deficient in their genes manifest in various defects in hematopoiesis. As discussed previously, multiple AUBPs often function in association within defined subcellular compartments and bind concomitantly to the same mRNA.3,6,7 For example, ZFP36 physically interacts with KSRP, nucleolin, HUR, and AUF1. AUF1 and HUR both target several mRNAs through direct binding to nonoverlapping 3′-UTR sequences.99 A similar phenomenon occurs for nucleolin with ZFP36 and PABP proteins,100 and with ZFP36L1 and ZFP36L2.81 The mRNAs of several AUBPs that contain 3′ ARE elements are themselves subject to auto- and/or trans-regulation, as occurs with ZFP36 family members.101 Examples of trans-regulation include the destabilization of HUR by AUF1102 and the inhibition of translation of ZFP36L1 by HUR.48 The AUBPs discussed in this review are also integrated with miRNA pathways at multiple levels in hematopoietic cells. For example, HUR represses miR-7 expression.38 It is subject to translational repression by several miRNAs34-37 and also functions in argonaut 2-dependent Let-7 repression,32 KSRP is involved in miRNA maturation,51,52 whereas ZFP36 family members function in miRNA-mediated decay.46,88 Finally, these AUBPs together with the miRNAs with which they functionally interact are intimately associated with some of the key oncogene/tumor suppressor pathways that are perturbed in malignant hematopoietic cells. The posttranscriptional “regulome” of hematopoietic cells can therefore be viewed as a highly interconnected regulatory network in which AUBPs represent major hubs. In Figure 2, we have constructed a global functional connectivity map of AUBPs in normal/malignant hematopoiesis based on the experimental data discussed in this review. This graphic view is not exhaustive but provides an appreciation of some of the major regulatory interactions that have been documented for AUBPs in hematopoietic cells. In particular, it highlights the close functional interconnectivity between AUBPs that in many cases mirrors their physical association and illustrates the critical intersection of AUBPs and miRNAs in the pathways that regulate oncogene/tumor suppressor functions in leukemogenesis.
Several lines of evidence implicate individual AUBPs in the pathogenesis of leukemia/lymphoma through diverse mechanisms. However, only the ZFP36 family exhibit characteristics of tumor suppressors where malignancy is associated with loss of function. The use of mouse models of various hematopoietic malignancies should, in the future, prove particularly instructive in elucidating the precise function role and mechanisms of individual AUBPs in specific disease types. Finally, AUBPs and the mechanisms through which they function offer new potential therapeutic targets for leukemia and lymphoma, as exemplified by a recent study in which restoration of ZFP36L1 function to solid tumor cells suppressed angiogenesis and tumorigenesis.103
Acknowledgments
The authors thank several colleagues and investigators in the field for helpful discussions during the preparation of the manuscript.
This work was supported by Cancer Research UK.
Authorship
Contribution: M.B. conducted literature research, wrote the manuscript, and designed Table 1 and Figure 1; J.D.N. conducted literature research, wrote the manuscript, and designed Figures 1 and 2; and J.J.M. conducted literature research, drafted sections of the manuscript, and checked the content of the final manuscript for accuracy.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John D. Norton, Department of Biological Sciences, University of Essex, Wivenhoe Park, Colchester CO4 3SQ, United Kingdom; e-mail: jnorton@essex.ac.uk.