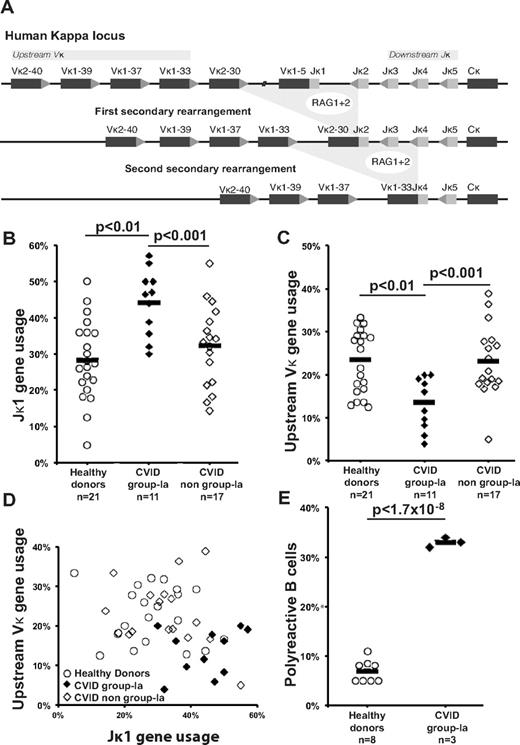
The letter by Rakhmanov et al1 suggested that normal κ/λ ratios in CD21+ naive B cells and increased Igλ-chain usage by CD21−/lo B cells in common variable immunodeficiency disease (CVID) patients with expanded CD21−/lo B-cell populations (CVID group-Ia2 ) were evidence of normal or increased receptor editing during central selection. Although increased Igλ-chain usage could correlate with extensive secondary recombination because Igλ genes normally rearrange after Igκ genes, the best evidence of receptor editing and central tolerance defects is found in proximal B-cell populations, specifically immature B cells in marrow or new emigrant/transitional B cells in peripheral blood. These subpopulations, similar in antibody repertoire and reactivity, offer an accurate historical record of central events uninfluenced by proliferation.3,4 Accordingly, the analysis by Warnatz and colleagues1 of Igλ-chain usage in CD21+ naive B cells and CD21−/lo B cells from 4 CVID group-Ia patients does not inform central events but implies peripheral selection in 2 B-cell subpopulations with well-documented histories of homeostatic expansion.4,5 Therefore, we assessed if receptor editing is functional in CVID group-Ia patients by analyzing the antibody repertoire and reactivity of new emigrant/transitional B cells from these patients.
We report here the Igκ repertoire from 1226 single CD19+CD10++IgMhiCD27− new emigrant/transitional B cells from 49 individuals including 21 healthy controls, 11 CVID group-Ia patients and 17 CVID non group-Ia patients with CD21−/lo B cells < 20% of total B cells.2 Secondary recombination mediating receptor editing replaces previous Igκ gene rearrangements with variable (Vκ)-joining (Jκ) gene segment joints resulting in increased Vκ genes located upstream of the locus combined to downstream Jκ genes (Figure 1A).6 We found that the Igκ repertoires of new emigrant/transitional B cells from CVID group-Ia patients were biased toward Jκ1, the most upstream Jκ gene segment, suggesting decreased secondary recombination (Figure 1B). CVID group-Ia Igκ repertoires were also characterized by decreased usage of upstream Vκ segments, genes overrepresented in diseases marked by extensive secondary recombination (Figure 1C).7,8 Moreover, bivariate analysis of Jκ1 and upstream Vκ usage further demonstrated CVID group-Ia new emigrant/transitional B cells occupy a distinct Igκ repertoire niche reflecting a dearth of secondary recombination (Figure 1D). The molecular basis of secondary recombination defects in these patients remains unknown and its correlation with the presence of peripheral CD21−/lo B cells is not understood. One clue about the regulation of secondary recombination came from BTK-deficient patients who display impaired B-cell receptor (BCR) signaling correlating with extensive secondary recombination events potentially through a failure to down-regulate RAG gene expression.7 Increased BCR signaling might therefore induce the premature RAG gene down-regulation resulting in the diminished secondary recombination activity observed in CVID group-Ia patients. This hypothesis, however, is not supported by BCR-induced calcium flux data demonstrating normal signal strength in CD21+ mature naive B cells from such patients.5,9 Alternatively, decreased recombination events in CVID group-Ia Igκ repertoires may suggest defective V(D)J recombination and/or DNA repair. The presence of such defects may account for radiosensitivity and malignancy susceptibility reported in some CVID patients.10
Diminished secondary recombination and corresponding defective central tolerance in the new emigrant/transitional B cells of CVID group-Ia patients. (A) Receptor editing is mediated by successive rounds of secondary recombination catalyzed by RAG enzymes with replacement of existing Igκ gene rearrangements with newly joined upstream Vκ and downstream Jκ gene segments. Increased Jκ1 (B) combined to decreased upstream Vκ (C) gene segment usage in the new emigrant/transitional B cells of CVID group-Ia patients reveals a unique repertoire niche (D) reflecting a history of decreased secondary recombination compared with healthy controls and non-group-Ia CVID patients. (E) An increased frequency of polyreactive new emigrant B cells in CVID group-Ia patients compared with healthy controls demonstrates a central defect in B-cell tolerance in these patients. Each diamond represents an individual, and the average is shown with a bar.
Diminished secondary recombination and corresponding defective central tolerance in the new emigrant/transitional B cells of CVID group-Ia patients. (A) Receptor editing is mediated by successive rounds of secondary recombination catalyzed by RAG enzymes with replacement of existing Igκ gene rearrangements with newly joined upstream Vκ and downstream Jκ gene segments. Increased Jκ1 (B) combined to decreased upstream Vκ (C) gene segment usage in the new emigrant/transitional B cells of CVID group-Ia patients reveals a unique repertoire niche (D) reflecting a history of decreased secondary recombination compared with healthy controls and non-group-Ia CVID patients. (E) An increased frequency of polyreactive new emigrant B cells in CVID group-Ia patients compared with healthy controls demonstrates a central defect in B-cell tolerance in these patients. Each diamond represents an individual, and the average is shown with a bar.
Finally, using an RT-PCR method to clone and express in vitro antibodies express by single B cells,3 we found that CVID group-Ia patients displayed increased frequencies of polyreactive clones in their new emigrant/transitional B-cell compartment, demonstrating a defective central B-cell tolerance checkpoint in these individuals (Figure 1E). Hence, the altered regulation of secondary recombination/receptor editing in developing B cells from CVID group-Ia patients correlates with the impaired counterselection of autoreactive clones in their bone marrow.
Authorship
N. R. and Y.-S. N. contributed equally to this work.
Acknowledgments: The authors thank Dr S. Rudchenko and S. Semova for cell sorting. This work was supported by Grant Number T32 AI007174 and CIS/Talecris Fellowship Award (to N.R.), AI061093, AI071087 and AI082713 from National Institutes of Health-National Institute of Allergy and Infectious Diseases (to E.M.).
Contribution: N. R., Y.-S. N., and E. M. performed research and analyzed data; C.C.-R. and E. M. designed the study; and N. R. and E. M. wrote the letter.
Conflict-of-interest disclosure: C.C.R. has served on the medical advisory board of Baxter Therapeutics and has received research grants from Baxter Therapeutics and Octapharma. The remaining authors declare no competing financial interests.
Correspondence: Eric Meffre, Yale University School of Medicine, 300 George St, New Haven, CT 06520; e-mail: eric.meffre@yale.edu.
References
National Institutes of Health