Abstract
In addition to adaptive T cells, the thymus supports the development of unconventional T cells such as natural killer T (NKT) and CD8αα intraepithelial lymphocytes (IELs), which have innate functional properties, particular antigenic specificities, and tissue localization. Both conventional and innate T cells are believed to develop from common precursors undergoing instructive, TCR-mediated lineage fate decisions, but innate T cells are proposed to undergo positive instead of negative selection in response to agonistic TCR signals. In the present study, we show that, in contrast to conventional αβT cells, innate αβT cells are not selected against functional TCRγ rearrangements and express TCRγ mRNA. Likewise, in contrast to the majority of γδT cells, thymic innate γδT cells are not efficiently selected against functional TCRβ chains. In precursors of conventional T cells, autonomous TCR signals emanating from the pre-TCR or γδTCR in the absence of ligand mediate selection against the TCR of the opposite isotype and αβ/γδ lineage commitment. Our data suggest that developing innate T cells ignore such signals and rely solely on agonistic TCR interactions. Consistently, most innate T cells reacted strongly against autologous thymocytes. These results suggest that innate and adaptive T-cell lineages do not develop from the same pool of precursors and potentially diverge before αβ/γδ lineage commitment.
Introduction
Adaptive immune reactions developed relatively late in evolution to complement more ancient innate responses in fighting infections. The αβT and B lymphocytes involved in adaptive immunity express highly variable, randomly generated, clonally distributed antigen receptors and require antigen priming for efficient responses. Natural killer (NK) and natural killer T (NKT) lymphocytes, regarded as innate or innate-like immune cells, carry invariant or semi-invariant receptors that recognize self-antigens and do not require priming to perform their function. The γδT cells function at the interface of innate and adaptive immunity and contain subsets with diverse and constrained TCR repertoires.1 Whereas the functions of these different lymphocytes are relatively well characterized, the developmental relationships between them are not so clear.2-4
Thymic lymphocyte progenitors traverse well-described developmental stages characterized as: cKit+CD44+25− (DN1), cKit+CD44+25+ (DN2), cKit−CD44−25+ (DN3), and cKit−CD44−25− (DN4). Commitment to the T-cell lineage takes place at the DN2 stage, when TCRγ, TCRδ, and TCRβ begin to rearrange. At the DN3 stage, precursors of αβT cells receive signals from the pre-TCR, which trigger proliferation, induce silencing of TCRγ genes (considered a hallmark of αβ lineage commitment), and direct development into CD4+8+ (DP) cells.5 Silencing of TCRγ during pre-TCR–induced proliferation6 prevents the expression of isotypically mismatched TCR complexes such as pre-TCRαγ or TCRαγ.7,8 No ligand for the pre-TCR has been found, and all of its functions are attributed to the autonomous signals derived from the assembled complex between the pre-Tα and TCRβ chains.9 At the DP stage, pre-Tα is replaced by the rearranged TCRα, and cells highly reactive to self-antigens are removed by negative selection. The γδT-cell precursors can be found in the DN2, DN3, and DN4 stages, but their precise maturation pathway is still under investigation.10 The majority of published results suggest that the αβ and the γδ TCRs play an important role in the process of αβ/γδ lineage commitment and selection against functional TCR of the opposite isotype. Accordingly, it was shown that mouse αβT cells are selected against functional TCRγ and TCRδ chains11-14 Interestingly, however, no selection against functional Vδ6 was found,15 possibly reflecting its very restricted pairing with TCRγ.16 The extent of selection against functional TCRβ rearrangements in γδT cells is less clear. Experiments assessing the status of the TCRβ locus in mouse γδT cells gave no consistent results, ranging from 33% to 70% of functional rearrangements.12,17-19 Nevertheless, γδT cells from pTα-deficient mice are enriched for in-frame TCRβ rearrangements.17 These results indicate that the expression of the γδTCR efficiently directs precursors away from the αβ lineage, whereas the expression of the pre-TCR directs precursors with functional TCRβ chains away from the γδ lineage, albeit with a lower efficacy. Recently, a novel model of αβ/γδ T-cell lineage commitment was proposed, which suggested that TCR signal strength rather than the TCR isotype regulates lineage commitment.20,21
Along with conventional adaptive T cells, the thymus supports the development of innate T cells, such as NKT cells and various intraepithelial lymphocytes (IELs).22-24 NKT cells are CD4−8− and CD4+8−, express characteristically low levels of the TCR and often the NK lineage marker NK1.1. Most αβNKT cells, named invariant NKT (iNKT) cell, express a semi-invariant αβTCR composed of Vα14Jα18 and Vβ8, Vβ7, or Vβ2 chains, and are thought to separate from the conventional αβT cells at the DP stage.25,26 However, in contrast to conventional T cells, their positive selection is mediated by the interaction with CD1d molecules expressed on thymocytes27-29 and seems to require agonistic interactions with self-ligands.30 Nevertheless, the precise nature of the positively selecting ligands is still unclear. Using CD1d tetramers, several stages of iNKT-cell development were defined, including CD1dtet+CD44−HSA+ (stage 1), CD1dtet+CD44−HSA− (stage 2, naive), and CD1dtet+CD44+HSA− (stage 3/4 memory/NK).22,23 In addition to iNKT cells, variant αβNKT cells and NKT cells carrying a γδTCR have been found.31 Little is known about γδNKT cells, but their TCR repertoire is biased for Vγ1Vδ6 and they express PLZF, a transcription factor highly expressed in iNKT cells.32 Two models of iNKT-cell commitment have been proposed22 : the TCR instructive model, supported by the increased frequencies of iNKT cells in Vα14-transgenic mice, and the precommitted precursor model, proposed on the basis of the presence of Vα14 transcripts at day 9.5 of gestation33 and the expression of Vα14 mRNA at the DN4 stage.34 αβΙELs can be divided into different subsets according to the CD4/CD8 coreceptor expression.35 The CD4 and CD8αβ subsets are thought to belong to conventional T cells and home to the gut upon antigenic stimulation in the periphery, whereas the CD8αα subset shares many innate characteristics with iNKT cells.36 Both iNKT and αβCD8ααIELs are proposed to undergo agonist-mediated positive selection (“agonist-selection”37 ). Interestingly, the development of both iNKT and αβCD8ααIELs seems to be more dependent on pre-Tα than on the development of adaptive αβT cells.38,39 However, it is still under debate whether αβCD8ααIEL progenitors leave the thymus as immature DN precursors40 and mature in the gut,39 or if they develop through the DP stage in the thymus, as suggested by lineage-tracing experiments.41-43
Various innate T cells develop in the thymus, but their precise maturation pathways and the role of TCR signals in their lineage commitment are poorly understood. Based on the results of the present study, we propose the existence of separate innate and conventional T-cell lineages, which diverge before αβ/γδ lineage commitment and respond differently to autonomous and agonist-induced TCR signals.
Methods
Mice
C57BL/6, CD1d deficient (CD1d−/−), CD1d−/−TCRδ−/−, and TCRδ−/− mice were bred and maintained under specific pathogen-free conditions at BioSupport or were purchased from Charles River Laboratories. For experiments, age- and sex-matched mice at the age of 3-16 weeks were used. The study was approved by the local ethics committee of Kantonales Veterinaramt in Zurich, Switzerland.
Abs and tetramers
The following mAbs were used: anti-CD4(RM45), anti-CD8(53.6.7), anti-TCRβ(H57-597), anti-TCRγδ(GL3), anti-TCRVγ4(UC3-10A6), anti-NK1.1(PK136), anti-CD44 (IM7), anti-CD62L(MEL-14), and anti-HSA(M1/69). All mAb and streptavidin conjugates were purchased from BD Pharmingen or eBioscience. PE- and APC-labeled CD1d tetramers loaded with PBS5744 were provided by the National Institutes of Health Tetramer Facility.
RT-PCR analysis
Total RNA was extracted from sorted cells using TRIzol reagent (Invitrogen). cDNA was generated using Superscript III (Invitrogen) according to the manufacturer's instructions. Real-time PCR was done with the MyiQ Real-Time PCR Detection System (Bio-Rad) using SYBR Green (Stratagene) and the following gene-specific primers: Vγ4 5′-ATCCTAGTGCTTCTACATCG-3′; Jγ1 5′-GGAATTACTATGAGCTTAGT-3′; Vγ1 5′-AACTTCTACCTCAACCTTGA-3′; and Jγ4 5′-GAATTACTACGAGCTTTGTC-3′. Thermal cycling conditions were 8 minutes at 95°C, followed by 45 cycles of 95°C for 20 seconds and 60°C for 1 minute. If not indicated otherwise, gene expression was normalized to β-actin and expressed in arbitrary units.
Two-stage RT-PCR
For the analysis of TCRVγ4 mRNA expression in early iNKT cell developmental stages, we could only recover ± 500 (> 50% purity) cells from stage 1 (CD1dtet+CD44−HSA+FCSlo) and ± 1000 cells from stage 2 (CD1dtet+CD44−HSA−FCShi) from several thymi of 3- to 4-week-old CD1d−/− mice. We therefore sorted similar numbers of the stage 3/4 (CD1dtet+CD44+HSA−) cells and the remaining CD1dtet− DN, DP, CD4+, and CD8+ SP thymocytes and generated cDNA as described in “RT-PCR analysis.” To detect a robust signal in the real-time PCR, a pre-amplification step of 15 cycles by conventional PCR was necessary. This was done with the same oligonucleotides that were used for real-time PCR, and 25% of the reaction was used as a template for the real-time PCR analysis. Expression was normalized to cell number and the experiment was performed 2 times with similar results.
Analysis of TCRVγ4 rearrangements
Genomic DNA was extracted from sorted subsets by digestion with proteinase K, followed by phenol/chlorophorm extraction and isopropanol precipitation. The TCRVγ4 rearrangements were amplified by PCR using the same oligonucleotides used for the real-time PCR, cloned into the p123T vector, and sequenced. Only unique sequences were used to calculate percentages of productive rearrangements.
Cloning of TCRδ EGFP bicistronic constructs and retroviral transduction of sorted thymocytes
Two different full-length TCRδ chains (encoding TCRVδ4) were amplified by RT-PCR and cloned into pMYiresGFP retroviral vector. Retrovirus-containing supernatant was produced in the Ecotropic Phoenix packaging cell line and used to infect sorted NK1.1+TCRβlow and NK1.1−CD4+TCRβhi thymocytes, which were stimulated for 2.5 days in tissue culture plates coated with anti–CD3ϵ(145.2C11) and anti–CD28(37.51) mAbs (2 μg/mL each) plus 100 U/mL of IL-2. As an IL-2 source, supernatant from a X63 line producing recombinant IL-2 was used. Cells were transferred into uncoated tissue culture dishes and analyzed 48-72 hours after infection.
Hybridoma generation
The TCRα−β− BW5147 NFAT-EGFP (short BW NFAT-EGFP) fusion partner carrying 4 copies of the minimal human IL-2 promoter, each containing 3 NFAT-binding sites (ACGCCTTCTGTATGAAACAGTTTTTCCTCC) inserted upstream of the EGFP coding sequence, was kindly provided by D. van Essen (MPI, Freiburg, Germany). Sorted thymocytes were activated with plastic-bound anti-CD3ϵ and anti-CD28 Abs in the presence of mouse IL-2 for 2-3 days. Equal numbers of activated T cells and the BW NFAT-GFP fusion partner were then fused using PEG-1500 and plated at limiting dilution in the presence of 100mM hypoxanthine, 400nM aminopterin, and 16mM thymidine (HAT). Autoreactivity was measured as the percentage of GFP+ cells relative to the response observed with αCD3αCD28 stimulation (maximal response).
Measuring hybridoma autoreactivity
Freshly generated hybridomas were grown in 96-well plates. Hybridoma cells were then mixed with at least 5-fold excess of freshly isolated total thymocytes from wild-type (WT) B6 mice and cocultured in IMDM, 2% FCS, and 0.03% Primaton RL/LF (Quest International) for 7-8 hours in V-bottom, 96-well plates to allow maximal contact. To determine the maximal response, a fraction of each hybridoma was stimulated with plate-bound αCD3 and αCD28 for 8 hours. The expression of EGFP was measured by flow cytometry on a FACSCalibur (BD Biosciences) and the results were analyzed with FlowJo Version 8 software (TreeStar). Cell size and granularity differences measured by forward and side scatter allowed us to distinguish hybridoma cells from thymocytes and the determination of the percentage of EGFP+ hybridoma cells. The nonfused BW NFAT-EGFP fusion partner cocultured with thymocytes or cultured on anti-CD3/anti-CD28 did not show any EGFP expression (data not shown)
LCMV infection
Mice were infected intravenously with 200 PFU of LCMV WE. Splenocytes were isolated and sorted on a FACSAria at day 7 after infection. The LCMV WE strain was originally provided by Dr R. M. Zinkernagel and Dr A. Oxenius (ETH, Zurich, Switzerland), and was propagated at low multiplicity on L929 cells. Aliquots were stored at −80°C.
Cell preparation, flow cytometry, and cell sorting
Single-cell suspensions from mouse organs were made by pressing through a nylon mesh in PBS containing 2% FCS. All of the subpopulations were sorted on FACSAria cell sorter to > 95% purity unless indicated otherwise. DN, DP, CD4+, and CD8+ thymocyte subsets were sorted from a CD4, CD8, TCRβ, and TCRγδ 4-color staining. To obtain αβNKT, TCRVγ4+, and TCRVγ4− γδ thymocyte subsets, total thymocytes were stained for NK1.1, TCRβ, TCRγδ, and TCRVγ4 and sorted according to the following phenotypes: NK1.1+TCRβlowTCRγδ−TCRVγ4−(NKT), NK1.1−TCRβ−TCRγδ+TCRVγ4+ (TCRVγ4+), and NK1.1−TCRβ−TCRγδ+TCRVγ4− (TCRVγ4−). CD4+CD25+ regulatory T cells (Tregs) were sorted from naive mouse thymocytes stained with CD4, CD8, TCRβ, and CD25. To isolate IELs, Peyer patches were removed and the small intestine was cut into small pieces and incubated for 30 minutes under agitation at 37°C in HBS 2% FCS (Ca/Mg free) containing 5mM EDTA and 2mM DTT. Subsequently, single cells were separated from the tissue by vortexing thoroughly and filtering through 40-μm pore size strainers (BD Biosciences). The procedure was repeated 2 times more without DTT, all isolated cells were pooled and stained for TCRβ, TCRγδ, CD8α, and CD8β in the presence of Fc-blocking mAb (2.4G2). αβCD8ααIELs and αβCD8αβIELs cells were sorted according to the following phenotypes: αβCD8ααIELs: TCRγδ−TCRβ+CD8α+CD8β− and αβCD8αβIELs: TCRγδ−TCRβ+CD8α+CD8β+. For the analysis and sorting of iNKT cells, CD16/CD32 Fc receptors were blocked on thymocytes before staining with PBS57-loaded, PE-labeled CD1d tetramers and MACS enrichment using anti-PE beads (Miltenyi Biotec), followed by staining of ΤCRβ, CD44, and HSA. Expression of markers was measured by flow cytometry on a FACSCalibur instrument (BD Biosciences), and the results were analyzed with FlowJo Version 8 software (TreeStar). Cells were cultured in IMDM, 2% FCS, and 0.03% Primaton RL/LF (Quest International).
Expression of intracellular TCRβ in γδ T cells
Thymocytes were depleted of CD4+ and CD8+ cells by MACS and stained for surface TCRβ, TCRγδ, and TCRVδ6 in the presence of Fc receptor blocking Ab. After fixation and permeabilization, intracellular TCRβ was stained. For analysis, TCRγδ+ surface-TCRβ− cells were gated and then the Vδ6+ and Vδ6− subsets of γδ T cells were analyzed for intracellular TCRβ expression.
Statistical analysis
Two-tailed, paired, and unpaired t tests and the χ2 test were done using Prism Version 4.0 software (GraphPad). P < .05 was considered significant.
Results
TCRγ expression in αβNKT cells
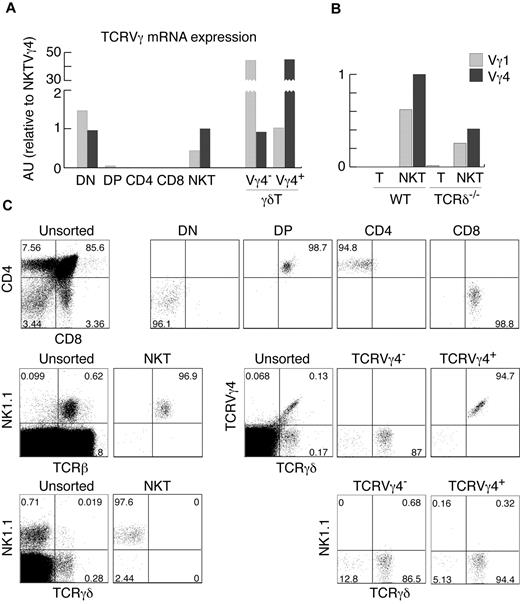
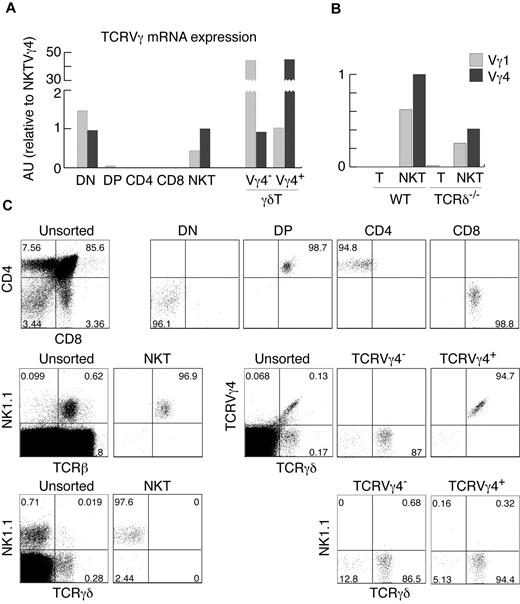
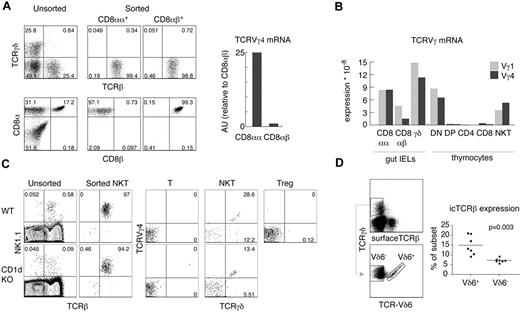
The analysis of αβ and γδ TCR gene expression and rearrangement status in cells from different lymphocyte lineages can provide useful information about the developmental origin of these cells. For example, detection of TCRγ rearrangements and mRNA expression in NK cells defined their minor, thymus-dependent developmental route.2 We therefore set out to define the status of TCRγ in various αβT-cell subsets. In agreement with previously published results,6 we did not find any significant amount of TCRγ mRNA in purified DP or CD4+ and CD8+ thymocytes using real-time PCR (Figure 1A-B). Surprisingly, we found substantial expression of TCRγ genes in highly purified αβNKT cells (Figure 1A-C). Thymic αβNKT cells showed relatively high expression of TCRVγ4 and TCRVγ1 mRNAs. The amounts were comparable to the TCRVγ4 mRNA in surface TCRVγ4− γδT cells and TCRVγ1 mRNA in surface TCRVγ4+ γδT cells (Figure 1A), suggesting a much closer developmental relationship of αβNKT and γδT cells than previously thought.
αβNKT cells express TCRγ mRNA. (A-B) Expression of TCRVγ genes normalized to β-actin in the indicated thymocyte subpopulations, measured by real-time RT-PCR in arbitrary units relative to NKT Vγ4 (AU). (C) Numbers in dot plots show percentages and indicate the purity of thymocyte subsets, sorted as indicated in “Cell preparation, flow cytometry, and cell sorting,” from which mRNA was isolated. Sorted NKT cells were not contaminated with γδT cells. Proportions of given subpopulations in unsorted thymocytes are also shown. Data are representative of at least 2 independent experiments.
αβNKT cells express TCRγ mRNA. (A-B) Expression of TCRVγ genes normalized to β-actin in the indicated thymocyte subpopulations, measured by real-time RT-PCR in arbitrary units relative to NKT Vγ4 (AU). (C) Numbers in dot plots show percentages and indicate the purity of thymocyte subsets, sorted as indicated in “Cell preparation, flow cytometry, and cell sorting,” from which mRNA was isolated. Sorted NKT cells were not contaminated with γδT cells. Proportions of given subpopulations in unsorted thymocytes are also shown. Data are representative of at least 2 independent experiments.
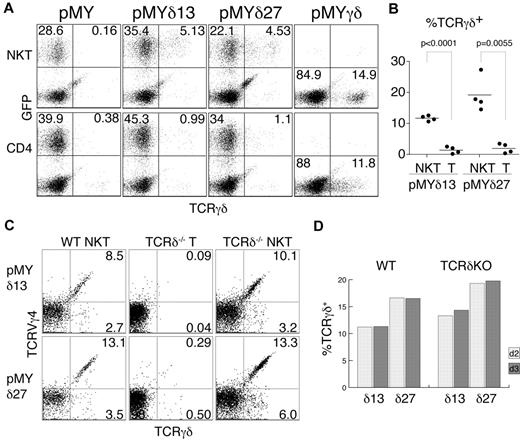
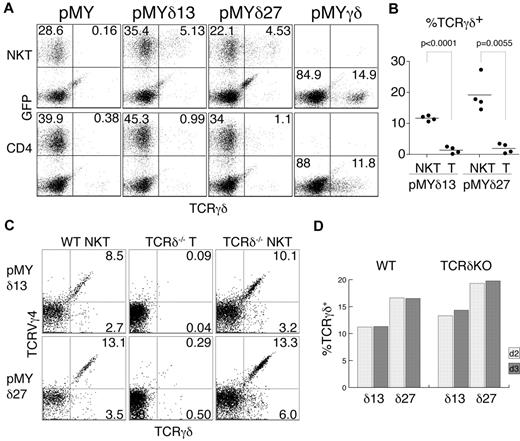
Intrigued by this finding, we wanted to test whether TCRγ proteins are produced in αβNKT cells, but surface and intracellular FACS stainings showed no signal (data not shown). However, TCRγ chains may be unstable in the absence of the TCRδ chains, precluding easy identification by intracellular staining. To circumvent this problem, we decided to provide TCRδ chains by retroviral transduction. To this end, we cloned 2 TCRVδ4 chains into the pMYiresGFP vector. Sorted NK1.1+TCRβ+ and NK1.1−CD4+TCRβhi thymocytes were then transduced with the generated constructs (pMYδ13iresGFP and pMYδ27iresGFP). GFP expression was used to distinguish the infected from the noninfected cells, and surface staining for the TCRγδ receptor allowed the determination of the fraction of cells expressing functional TCRγ chains (Figure 2A). Interestingly, as depicted in Figure 2B, a substantial fraction of the transduced NK1.1+TCRβ+ (NKT) cells expressed TCRγδ on the surface (11.7% ± 0.4% TCRγδ13+ and 19.2% ± 2.8% TCRγδ27+ of GFP+ cells), whereas the majority of NK1.1−CD4+TCRβhi (CD4) thymocytes did not express TCRγδ (1.4% ± 0.6% TCRγδ13+ and 2.0% ± 0.8% TCRγδ27+ cells of GFP+ cells). Importantly, cells infected with an empty vector (pMYiresGFP) showed no TCRγδ on the surface. To show that the lack of TCRγδ expression on CD4+ cells was not because of their inherent inability to display the TCRγδ on the surface, we infected the sorted cells with a vector containing both functional TCRγ and TCRδ chains (pMYγδ). As shown in Figure 2A, both populations (αβNKT and αβT cells) could express the TCRγδ on the surface, albeit at different levels, likely resulting from competition for the CD3 complex with the endogenous αβTCRs.
αβNKT cells are not selected against functional TCRγ chains. Sorted NKT and CD4+ thymocytes were retrovirally transduced with the indicated constructs and stained with anti–γδTCR Ab. (A) Representative results of the FACS analysis used to determine the percentage of γδTCR+ cells among the GFP+ cells (B). (B) Summarized results from 4 independent experiments. The P values of a paired t test are shown. (C-D) Percentages of TCRVγ4+TCRγδ+ and TCRVγ4−TCRγδ+ among GFP+ cells from WT or TCRδ-deficient (KO) mice measured on day 2 (C) and day 3 after infection.
αβNKT cells are not selected against functional TCRγ chains. Sorted NKT and CD4+ thymocytes were retrovirally transduced with the indicated constructs and stained with anti–γδTCR Ab. (A) Representative results of the FACS analysis used to determine the percentage of γδTCR+ cells among the GFP+ cells (B). (B) Summarized results from 4 independent experiments. The P values of a paired t test are shown. (C-D) Percentages of TCRVγ4+TCRγδ+ and TCRVγ4−TCRγδ+ among GFP+ cells from WT or TCRδ-deficient (KO) mice measured on day 2 (C) and day 3 after infection.
The sorted subpopulations used for this analysis were very pure (Figure 1C). Nevertheless, to definitively exclude contamination with γδT cells, we analyzed cells from mice lacking γδT cells because of TCRδ deficiency. As shown in Figures 1B and 2C, αβNKT cells from these mice expressed functional TCRγ chains of both the Vγ4+ and Vγ4− isotypes. Furthermore, we detected the same fraction of TCRγ+ αβNKT cells when measured 2 or 3 days after retroviral TCRδ reconstitution, indicating that cells expressing TCRγ chains were not enriched in culture. Therefore, the measured percentages represent the actual percentages in the freshly isolated NKT cells (Figure 2D). Interestingly, we observed no difference in TCRγ expression between WT and TCRδ-deficient αβNKT cells, suggesting little selection against functional TCRγ in WT αβNKT cells. These results show that, in contrast to conventional αβT cells, a substantial fraction of αβNKT cells expresses functional TCRγ chains.
Lack of selection against productive Vγ4 rearrangements in αβNKT cells
Considering that the transduced TCRδ chains may be unable to form stable heterodimers with all of the TCRγ chains, the frequency of TCRγ+ cells established in the TCRδ reconstitution experiments might have been underestimated. However, precise determination of that frequency was necessary to establish whether TCRγ rearrangements found in NKT cells are random, selected for, or selected against productive junctions. We therefore cloned and sequenced the V-J junctions of Vγ4 rearrangements from sorted NK1.1+TCRβ+ and NK1.1−TCRβhi thymocytes. Table 1 shows a summary of the results from sequencing 66 Vγ4 junctions from NK1.1+TCRβ+ and 91 Vγ4 junctions from NK1.1−TCRβhi cells. Twenty-three percent of the Vγ4 rearrangements in NKT cells were productive, which was in good agreement with the TCRδ transduction experiments. As expected, a significantly lower proportion (10%) was productive in conventional T cells. Furthermore, only 35% of the in-frame TCRγ rearrangements in αβNKT cells contained STOP codons, whereas this fraction was substantially higher in αβT cells (63%). In the absence of selection, the fraction of productive Vγ4 rearrangements is expected to be lower than 33% because of an in-frame TAA STOP codon that has to be removed during the recombination process. This fraction has been estimated experimentally to be 16%-18%,11,13 indicating that our result is compatible with lack of selection. We therefore conclude that, whereas αβT cells are selected against functional TCRVγ4 rearrangements, there is no such selection among αβNKT cells.
Selection against functional TCR chains of the opposite isotype is a common characteristic of conventional but not innate T cells
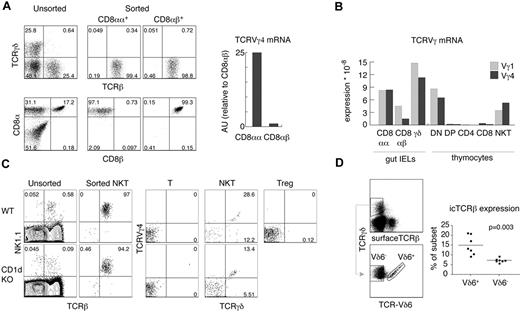
To determine whether other αβT-cell subsets expressed productive TCRγ, we analyzed gut IELs and found substantial expression of TCRVγ4 mRNA in sorted CD8αα+TCRβ+TCRγδ− IELs, but not in conventional CD8αβ+TCRβ+TCRγδ− IELs (Figure 3A-B). This expression was comparable to the levels found in αβNKT cells. Interestingly, for TCRVγ1 mRNA, the difference was smaller, indicating that silencing of TCRVγ1 locus was less stringent in αβIELs than in αβT cells (Figure 3B). Unfortunately, stimulation with αCD3 and αCD28 led to a substantial death of IELs, precluding TCRδ reconstitution. However, as shown in Table 1, 27% of the TCRVγ4 rearrangements found in CD8αα+IELs were productive, whereas only 11.5% of the TCRVγ4 rearrangements found in CD8αβ+IELs were productive. We conclude that, in contrast to CD8αβ+IELs, CD8αα+IELs are not depleted of functional TCRγ chains. We then compared TCRδ-reconstituted αβNKT cells from WT and CD1d-deficient mice. As shown in Figure 3C, αβNKT cells selected by ligands different from CD1d expressed amounts of surface TCRγδ similar to that of WT αβNKT cells.
αβ and γδ innate T cells are not efficiently selected against functional TCR chains of the opposite isotype. (A) Expression of TCRVγ4 as determined by real time RT-PCR on mRNA derived from small bowel TCRβ+TCRγδ− IELs sorted according to the expression of CD8α and CD8β. (B) Direct comparison of the expression of TCRVγ1 and TCRVγ4 mRNA in various T-cell subsets by real-time RT-PCR relative to β-actin. (C) Sorted NKT cells, T cells, and Tregs from WT and CD1d KO mice were retrovirally transduced with pMYδ27 and stained as indicated. Numbers in dot plots show percentages and indicate the purity of the sorted NKT cells (left panels) and percentages of TCRVγ4+TCRδ+ and TCRVγ4−TCRδ+ among GFP+ NKT cells, T cells, and Tregs (right panels). Data are representative of 2 independent experiments. (D) Percentages of intracellular (ic) TCRβ+ cells among TCRVδ6+ and TCRVδ6− γδ thymocytes analyzed in individual WT mice as shown in the dot plots; data are representative of 3 independent experiments. P value of a paired t test is shown
αβ and γδ innate T cells are not efficiently selected against functional TCR chains of the opposite isotype. (A) Expression of TCRVγ4 as determined by real time RT-PCR on mRNA derived from small bowel TCRβ+TCRγδ− IELs sorted according to the expression of CD8α and CD8β. (B) Direct comparison of the expression of TCRVγ1 and TCRVγ4 mRNA in various T-cell subsets by real-time RT-PCR relative to β-actin. (C) Sorted NKT cells, T cells, and Tregs from WT and CD1d KO mice were retrovirally transduced with pMYδ27 and stained as indicated. Numbers in dot plots show percentages and indicate the purity of the sorted NKT cells (left panels) and percentages of TCRVγ4+TCRδ+ and TCRVγ4−TCRδ+ among GFP+ NKT cells, T cells, and Tregs (right panels). Data are representative of 2 independent experiments. (D) Percentages of intracellular (ic) TCRβ+ cells among TCRVδ6+ and TCRVδ6− γδ thymocytes analyzed in individual WT mice as shown in the dot plots; data are representative of 3 independent experiments. P value of a paired t test is shown
We conclude that depletion of productive TCRVγ4 rearrangements and permanent TCRγ locus silencing are common features of adaptive but not innate αβT cells. Interestingly, a similar trend could be observed in the γδT-cell lineage: thymic (Vδ6+) γδNKT cells expressed intracellular TCRβ chains more often (15% ± 1.8%) than conventional γδT cells (7.3% ± 0.4%; Figure 3D).
Our data show that innate T cells are not efficiently selected against functional TCRs of the opposite isotype, indicating that their precursors diverge at an early developmental stage before commitment to the αβ or γδ T-cell lineage.
Lack of permanent TCRγ silencing in innate T cells
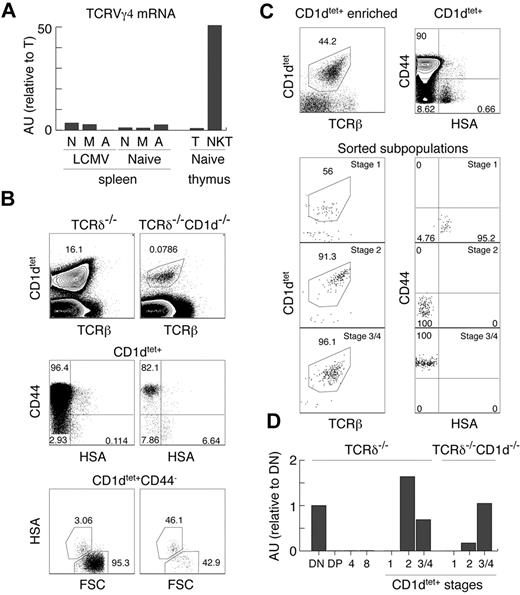
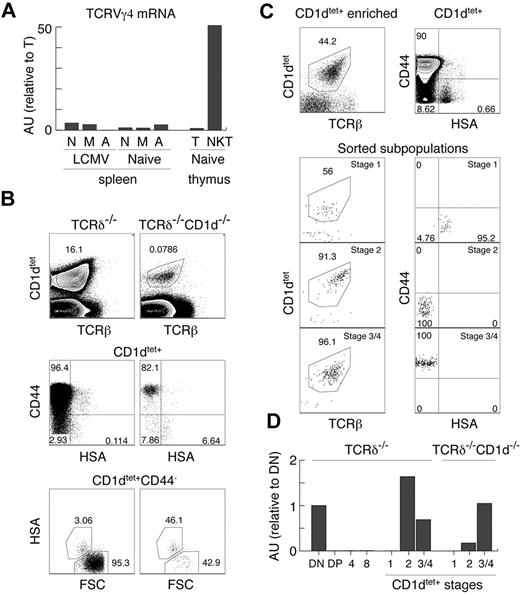
Our experiments clearly show that the TCRγ locus is open in innate T cells, which is in strong contrast to the majority of αβT cells, which silence TCRγ before the DP stage of thymic development. iNKT cells, CD8αα+IELs, and Tregs are thought to be selected by high-affinity ligands and belong to the so-called “agonist-selected” T cells.37 In support of this, Tregs and iNKT cells have been shown recently to receive strong TCR stimulation during positive selection.45 To test whether this particular mode of selection or strong TCR stimulation reopen the initially silenced TCRγ locus, we performed several experiments. First, we reconstituted TCRδ in Tregs (Figure 3C) and could find no TCRγδ expression, indicating that “agonist-selection” per se does not lead to TCRγ opening. Second, we sorted naive, memory, and activated αβT cells from the spleens of naive and LCMV-infected mice. As shown in Figure 4A, these cells also expressed only very small amounts of TCRγ mRNA compared with αβNKT cells, indicating that strong TCR stimulation does not up-regulate TCRγ mRNA. This result also demonstrates that memory or activated T cells, which can up-regulate NK1.146 and could have contaminated sorted αβNKT cells (especially from the CD1d-knockout mice), are not the source of the TCRγ found in these cells.
Lack of permanent TCRγ silencing in innate T cells. (A) Expression of TCRVγ4 mRNA normalized to β-actin in the indicated subpopulations, measured by real time RT-PCR in arbitrary units relative to mature thymocytes (AU). N indicates naive splenocytes TCRγδ−TCRβ+CD44−CD62L+; M, memory splenocytes TCRγδ−TCRβ+CD44+CD62L+; A, activated splenocytes TCRγδ−TCRβ+CD44+CD62L−; T, mature thymocytes TCRγδ−TCRβhi; and NKT, thymic NKT TCRγδ−TCRβloNK1.1+. (B-C) Analysis of MACS enriched, CD1dtet+ iNKT cells from the thymus of TCRδ−/− and CD1d−/−TCRδ−/− (B) and the sorted CD1dtet+ T-cell subsets from TCRδ−/− mice. (C) Numbers in dot plots show percentages. (D) Expression of TCRVγ4 mRNA normalized to cell number in the indicated subpopulations sorted from TCRδ−/− and CD1d−/−TCRδ−/− mice measured by real-time RT-PCR (see “Two-stage RT-PCR”) in arbitrary units relative to DN thymocytes (AU).
Lack of permanent TCRγ silencing in innate T cells. (A) Expression of TCRVγ4 mRNA normalized to β-actin in the indicated subpopulations, measured by real time RT-PCR in arbitrary units relative to mature thymocytes (AU). N indicates naive splenocytes TCRγδ−TCRβ+CD44−CD62L+; M, memory splenocytes TCRγδ−TCRβ+CD44+CD62L+; A, activated splenocytes TCRγδ−TCRβ+CD44+CD62L−; T, mature thymocytes TCRγδ−TCRβhi; and NKT, thymic NKT TCRγδ−TCRβloNK1.1+. (B-C) Analysis of MACS enriched, CD1dtet+ iNKT cells from the thymus of TCRδ−/− and CD1d−/−TCRδ−/− (B) and the sorted CD1dtet+ T-cell subsets from TCRδ−/− mice. (C) Numbers in dot plots show percentages. (D) Expression of TCRVγ4 mRNA normalized to cell number in the indicated subpopulations sorted from TCRδ−/− and CD1d−/−TCRδ−/− mice measured by real-time RT-PCR (see “Two-stage RT-PCR”) in arbitrary units relative to DN thymocytes (AU).
These results suggest that the subset of precursors selected toward the innate lineage might not silence TCRγ. However, they do not definitely exclude an instructive mechanism in which specific signals received by innate precursors during positive selection reopen the TCRγ locus. To resolve this issue, one would have to identify precursors of innate T cells before positive selection, which is not possible at this time. Analyzing CD1dtet+ cells from CD1d−/− mice does not help, because TCRVα14Jα18 rearrangements do not happen exclusively in innate precursors, and therefore CD1dtet-binding does not distinguish innate from adaptive precursors at the immature HSA+ stage. Consistently, as in total DP cells, we detected no TCRVγ4 mRNA in HSA+CD44−CD1dtet+ cells (Figure 4D). Unexpectedly, however, we found low numbers of mature HSA−CD1dtet+ cells in CD1d−/− mice (Figure 4B). Whereas these cells are selected by a different ligand, they still expressed TCRVγ4 mRNA (Figure 4D). Interestingly, HSA−CD44+CD1dtet+ cells expressed the same levels as cells developing in the presence of CD1d, but HSA−CD44−CD1dtet+ cells expressed approximately 5 times less (Figure 4D). Nevertheless, these results do not definitely resolve the issue of “no-silencing/reopening” of the TCRγ locus, because they could indicate CD1d-triggered TCRVγ4 locus opening or that in CD1d−/− mice, HSA−CD44−CD1dtet+ cells contain adaptive T cells with silenced TCRγ. Supporting the latter possibility, HSA−CD44− cells were relatively enriched among CD1dtet+ cells in CD1d−/− mice. Because we used TCRδ−/− and TCRδ−/−CD1d−/− mice for these experiments, the detected TCRγ mRNA was derived exclusively from αβNKT cells.
Innate αβ and γδT cells are highly autoreactive.
To explain the apparent lack of functional TCRγ chain depletion in innate αβT cells, we considered the possibility that innate precursors may require agonistic TCR signals for αβ/γδ lineage commitment. Therefore, not all γδTCR+ cells, only those recognizing a ligand, would commit to the innate γδT-cell lineage, resulting in minor, undetectable depletion of functional TCRγ in innate αβT cells (eg, αβNKT). This hypothesis predicts that all innate T cells are autoreactive.
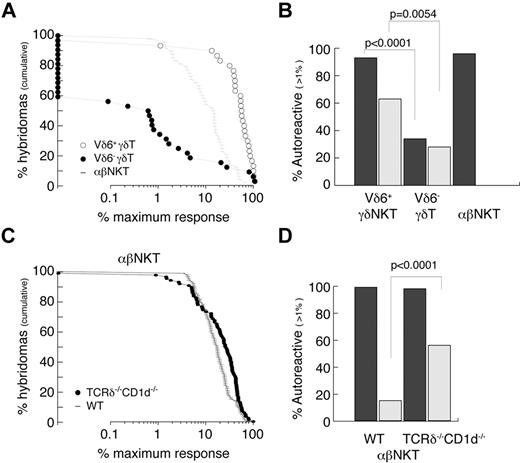
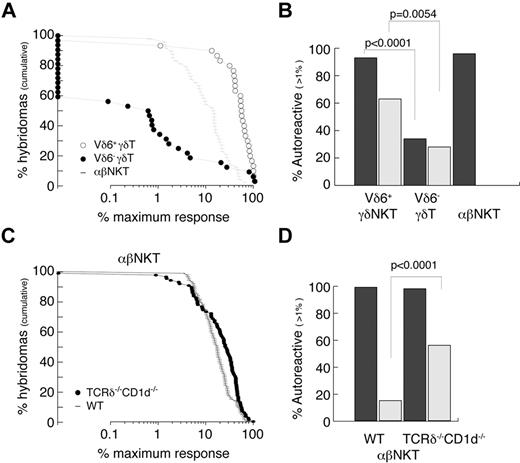
The autoreactivity of γδT cells was always suspected because of their often-activated phenotype, but this was not thoroughly studied. We therefore attempted to quantitatively measure the frequency of autoreactive cells among Vδ6+ (enriched for innate γδ) and Vδ6− (enriched for conventional γδ) thymocytes and compared it with thymic αβNKT cells (Figure 5A-B). The experimental strategy was to generate hybridomas by fusion of sorted thymocytes with a TCR− thymoma BW carrying an NFAT-EGFP reporter. As expected, 96% of αβNKT cell-derived hybrids responded (≥ 1% of the maximum response) when stimulated with freshly isolated autologous thymocytes (Figure 5A-B). Interestingly, 93% of Vδ6+ γδ thymocyte–derived hybridomas but only 34% of Vδ6− γδ thymocyte–derived hybridomas responded in the same assay. In agreement with previousy published results,47 many γδ thymocyte-derived hybridomas expressed NFAT-GFP spontaneously (Figure 5B), indicating that they expressed the auto-antigens to which they were reactive. TCRδ-reconstitution experiments suggested that αβNKT cells developing in the absence of CD1d belong to the innate lineage; therefore, we also tested their reactivity against autologous thymocytes. To avoid γδT-cell contamination during sorting, we again used TCRδ−/−CD1d−/− mice. Figure 5C and D shows that, similarly to the CD1d-selected iNKT cells, they were all autoreactive. Interestingly, in contrast to the CD1d-selected iNKT cells, many showed spontaneous reactivity similar to the γδT-cell–derived hybridomas.
Innate T cells are autoreactive. (A-B) Reactivity of WT TCRVδ6+, TCRVδ6− γδT, and αβNKT-cell–derived hybridomas. (C-D) Reactivity of αβNKT-cell–derived hybridomas from WT and CD1d−/−TCRδ−/− mice. (A,C) Cumulative fraction of hybridomas for which the response exceeded the level indicated on the x-axis. (B,D) Percentage of autoreactive hybridomas determined as percentage of hybridomas for which the response exceeded 1% of maximum. Dark bars represent reactivity toward autologous thymocytes; light bars spontaneous reactivity. P values of χ tests are shown.
Innate T cells are autoreactive. (A-B) Reactivity of WT TCRVδ6+, TCRVδ6− γδT, and αβNKT-cell–derived hybridomas. (C-D) Reactivity of αβNKT-cell–derived hybridomas from WT and CD1d−/−TCRδ−/− mice. (A,C) Cumulative fraction of hybridomas for which the response exceeded the level indicated on the x-axis. (B,D) Percentage of autoreactive hybridomas determined as percentage of hybridomas for which the response exceeded 1% of maximum. Dark bars represent reactivity toward autologous thymocytes; light bars spontaneous reactivity. P values of χ tests are shown.
These results show that most, if not all, innate T cells are autoreactive, and suggest that they undergo “agonist selection.”
Discussion
Parallel development of innate and conventional T cells from separate precursors
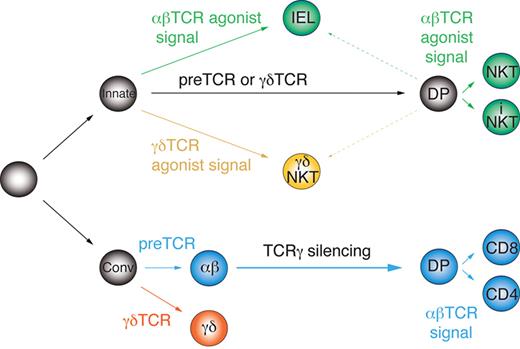
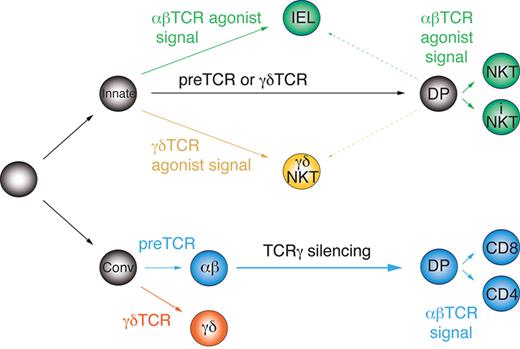
The current scheme of early T-cell development and lineage commitment does not distinguish between innate and conventional T-cell precursors, but instead is based on the idea of a common, equivalent precursor undergoing a series of instructive cell-fate decisions. Accordingly, αβ/γδ lineage commitment occurs in response to the autonomous signals delivered by the pre-TCR or the γδTCR. Later, αβT-cell precursors undergo conventional selection by the MHC class I or II or “agonist selection” by nonclassic MHC molecules such as CD1d and acquire adaptive (CD4 or CD8 T cells) or innate characteristics (eg, iNKT cells), respectively.23 However, the distinct molecular signature of innate and conventional αβT cells reported herein, in particular the different Vγ4 rearrangement status, cannot be explained by this model. Instead, it indicates the existence of separate precursors of innate and conventional T-cell lineages. Therefore, based on our results, we propose a novel model of early thymocyte differentiation. In this model (Figure 6), commitment to the αβ and γδ lineages takes place independently in distinct innate and conventional DN precursors, which react differently to the autonomous (weaker) and agonist-induced (stronger) TCR signals.
Parallel development model of innate and conventional T-cell lineages. Innate and conventional precursors diverge early before αβ/γδ lineage commitment and react differently to autonomous and agonistic TCR signals.
Parallel development model of innate and conventional T-cell lineages. Innate and conventional precursors diverge early before αβ/γδ lineage commitment and react differently to autonomous and agonistic TCR signals.
We propose that innate precursors require strong agonist-induced signals for αβ/γδ lineage commitment/selection, whereas weaker, TCR-autonomous signals induce developmental progression to the DP stage without αβ lineage commitment (ie, without permanent TCRγ silencing) independently of the TCR isotype. These DP cells are compatible with agonist selection and, when receiving strong agonistic signals, give rise to innate T cells; however, weak signals are not enough to save them from death by neglect. Before completion of TCRα rearrangements on both chromosomes, such precursors can still give rise to γδNKT cells upon recognition of agonist ligands. In support of this, we observed a higher fraction of intracellular TCRβ+ cells among thymic γδNKT cells than in conventional γδT cells (Figure 3D), and rare TCRα rearrangements have been found in γδT cells.19 Alternatively, innate T cells may actually represent a third lineage, which merely requires strong agonistic signals for maturation irrespectively of the TCR isotype and does not undergo αβ/γδ lineage commitment sensu stricto. Conversely, conventional T-cell precursors are deleted by strong agonist-induced signals, require autonomous TCR signals for αβ/γδ lineage commitment48 and weak TCR signals for positive selection.
Conceivably, in an attempt to explain our results by resorting to a common precursor, one could postulate that all innate αβT cells derive from precursors with nonfunctional TCRδ rearrangements or expressing TCRVδ6 chains, because these do not pair with TCRVγ4.16 Such cells would display random rearrangements at the TCRVγ4 locus. However, we could not find any mechanism explaining why such precursors would preferentially give rise to innate αβT cells. In addition, the apparent reciprocal enrichment of intracellular TCRβ+ cells in innate γδT cells cannot be explained this way. One could also interpret our data as supportive for a TCR-independent mechanism of commitment to the αβ and γδ lineages. In this case the innate lineage might represent a default pathway from which the other lineages diverge in a TCR-independent manner. However, to explain selection against functional TCR chains of the opposite lineage, one would have to imply that γδTCRs are toxic for αβT cells and vice versa. We could not find any evidence for that in the literature, nor could we observe toxicity in CD4+ thymocytes reconstituted with TCRγδ (Figure 2A last panel). Taken together, whereas we consider our model (Figure 6) most plausible, further experiments are needed to precisely clarify the innate pathway and the nature of the corresponding DN precursors.
The permanent silencing of TCRγ locus in conventional αβT cells was shown to be dependent on pre-TCR–induced proliferation.6 However, DP cells generated by receptors other than pre-TCR do not silence TCRγ loci, showing that it is not an inherent feature of the DP stage. Interestingly, DP cells generated by αCD3 injection into RAG mice do silence TCRγ loci,6 suggesting that proliferation on the way to DP, rather than particular TCR signals, induces TCRγ silencing. The lack of permanent TCRγ silencing in innate αβT cells shown here could indicate that pre-TCR–mediated signals are interpreted differently by innate and conventional precursors, and that the former proliferate less extensively than the latter. As a result, adaptive precursors would give rise to the majority of DP, which are depleted of functional Vγ4 rearrangements and silence the TCRγ loci.
Implications for the TCR signal strength model of αβ/γδ lineage commitment
Our model offers an alternative explanation for the apparent incompatibility of αβ lineage commitment with strong TCR signals, which lies at the ground of the “TCR signal strength model” of lineage commitment. This model is based mainly on 2 studies using TCR-transgenic mice. Interestingly, both of these studies used autoreactive TCRs, one reactive to T22 (KN6)21 and the other presumably to a heat-shock protein,20,49 and showed that changing the TCR signal strength changed the ratio of αβ to mature γδ T cells. The main conclusion from these studies, that attenuation of TCR signaling diverts precursors from the γδ lineage toward the αβ lineage, is based upon 2 assumptions: (1) that all αβ and γδ thymocytes derive from the same, equivalent precursors and (2) that all γδTCRs transduce stronger signals than pre-TCR or αβTCR. However, the experiments presented in the present study indicate the existence of heterogeneous precursors before the αβ/γδ lineage split. In addition, whereas further experiments testing all of the thymic stromal cells are required to fully analyze the reactivity of γδΤ cells, the data suggests that not all γδTCRs transduce equally strong signals. Our results suggest that changing the TCR signal strength may change the relative contributions of innate and conventional precursors to the αβ and γδ lineages, rather than αβ/γδ lineage commitment per se.
In summary, we propose that innate and conventional T-cell lineages diverge early, before the αβ/γδ T-cell commitment, and develop in parallel. Their precursors perceive TCR signals differently, so innate precursors are compatible with agonist selection and can reach the DP stage without commitment to the αβ lineage or may leave the thymus and give rise to gut IELs. Conversely, conventional T-cell precursors undergo negative selection upon agonistic ligand binding and require pre-TCR–mediated signals for αβ lineage commitment, TCRγ silencing, and progression to the DP stage.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank D. van Essen (MPI, Freiburg, Germany) for the BW NFAT-GFP reporter cell line, G. Nolan (Stanford University, Stanford, CA) for Phoenix cells, T. Kitamura (University of Tokyo, Tokyo, Japan) for the pMY-IRES-GFP plasmid, P. Pereira (Institut Pasteur, Paris, France) for the αTCRγδ hybridomas, the National Institutes of Health Tetramer Facility for the CD1d tetramers, M. Kisielow and A. Schütz (ETH, Zürich, Switzerland) for cell sorting, and P. Kisielow (Institute of Immunology and Experimental Therapy, Wroclaw, Poland) for discussions and critical reading of the manuscript.
This project was funded by the Swiss National Science Foundation 310030_124922.
Authorship
Contribution: J.K. designed the research, performed the experiments, and wrote the manuscript; L.T. and J.W. performed the experiments; K.K. analyzed the data and edited the manuscript; and M.K. obtained the funding, analyzed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Kisielow, PhD, Molecular Biomedicine, Swiss Federal Institute of Technology (ETH), Wagistr 27, CH-8952 Zürich-Schlieren, Switzerland; e-mail: jan.kisielow@env.ethz.ch.