Abstract
Idiopathic aplastic anemia (AA) is a common cause of acquired BM failure. Although autoimmunity to hematopoietic progenitors is thought to be responsible for its pathogenesis, little is known about the molecular basis of this autoimmunity. Here we show that a substantial proportion of AA patients harbor clonal hematopoiesis characterized by the presence of acquired copy number-neutral loss of heterozygosity (CNN-LOH) of the 6p arms (6pLOH). The 6pLOH commonly involved the HLA locus, leading to loss of one HLA haplotype. Loss of HLA-A expression from multiple lineages of leukocytes was confirmed by flow cytometry in all 6pLOH(+) cases. Surprisingly, the missing HLA-alleles in 6pLOH(+) clones were conspicuously biased to particular alleles, including HLA-A*02:01, A*02:06, A*31:01, and B*40:02. A large-scale epidemiologic study on the HLA alleles of patients with various hematologic diseases revealed that the 4 HLA alleles were over-represented in the germline of AA patients. These findings indicate that the 6pLOH(+) hematopoiesis found in AA represents “escapes” hematopoiesis from the autoimmunity, which is mediated by cytotoxic T cells that target the relevant auto-antigens presented on hematopoietic progenitors through these class I HLAs. Our results provide a novel insight into the genetic basis of the pathogenesis of AA.
Introduction
Acquired aplastic anemia (AA) is a rare condition associated with BM failure and pancytopenia.1 A series of classic observations and experiments have unequivocally supported that the autoimmunity to hematopoietic stem/progenitor cells (HSPCs) critically underlies the pathogenesis of the BM failure in the majority of AA cases. According to the widely accepted model of immune-mediated BM failure, activated cytotoxic T cells (CTLs) that recognize an auto-antigen(s) presented on HSPCs through their class I HLA molecules have a major role in initiating the autoimmune reactions.2-4 However, no definitive evidence exists that supports this model or the presence of such CTL repertoires. Moreover, little information is available about their target antigens or about the way by which they are recognized by effector T cells.
Another long-standing issue on AA is its close relationship with clonal hematopoiesis.5,6 It was first suspected from an apparent overlap between AA and paroxysmal nocturnal hemoglobinuria (PNH)7,8 and was also implicated by the frequent development of late clonal disorders in AA, such as myelodysplastic syndromes, PNH, or even acute myeloid leukemia (AML).9-11 Clonal hematopoiesis can be explicitly demonstrated by conventional clonality assays at presentation in a substantial proportion of newly diagnosed typical AA cases.12 Although it has been expected that the inciting autoimmune insult somehow confers selective pressures on the evolution of clonal hematopoiesis,5 the exact mechanism for such immunologic selection or escape is still unclear.
The objectives of this study, therefore, were to characterize the clonal nature of the hematopoiesis that is maintained even under the severe autoimmune insult in AA, and to explore the genetic/immunologic mechanism that could underlie the pathogenesis of AA. To achieve these aims, we performed single nucleotide polymorphism (SNP) array-based analysis of genomic copy numbers and/or allelic imbalances in peripheral blood (PB) specimens obtained from 306 patients with AA. Initially, we found that AA patients frequently showed clonal/oligoclonal hematopoiesis that lost specific HLA alleles as a result of copy number-neutral loss of heterozygosity (CNN-LOH) of the 6p arms, which led us to further analyses of the contribution of 6pLOH(+) clones to residual hematopoiesis and a large-scale epidemiologic study on the HLA alleles that are over-represented in AA, involving a total of 6,613 transplants registered in the Japan Marrow Donor Program (JMDP).
Methods
Subjects
PB specimens from a total of 306 patients with AA were analyzed for the presence of genetic alterations using SNP arrays (see Figure 1). The clinical characteristics of these patients are summarized in Table 1 and supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Among the 306 patients, 107 were newly diagnosed and 199 were previously treated. Ninety-six patients received allogeneic BM transplantation from unrelated donors through the JMDP, and their HLA information was available from the JMDP. The other 210 were newly genotyped for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles as described elsewhere.13 A total of 103 patients had been treated with anti–thymocyte globulin plus cyclosporine, cyclosporine alone, or anabolic steroids at the time of sampling. All patients and healthy persons provided their informed consent before sampling in accordance with the Declaration of Helsinki. The study protocol was approved by the ethics committee of the Graduate School of Medical Science, Kanazawa University and also by that of the Graduate School of Medicine, University of Tokyo.
Patient characteristics
| . | Newly diagnosed (n = 107) . | Previously treated (n = 199) . |
|---|---|---|
| Median age at diagnosis, mo (range) | 64 (9-88) | 24 (2-80) |
| Sex, male/female, no. | 58/49 | 110/89 |
| Severity of AA at onset, no. (%) of patients | ||
| Severe | 79 (74) | 185 (93) |
| Nonsevere | 28 (26) | 14 (7) |
| History, mo, median (range) | 19 (0.1-251) | 51 (0.1-372) |
| Past treatment, no. (%) of patients | ||
| ATG + CsA | — | 39 (20) |
| CsA alone | — | 51 (26) |
| Anabolic steroid alone | — | 13 (7) |
| Unknown* | — | 96 (48) |
| . | Newly diagnosed (n = 107) . | Previously treated (n = 199) . |
|---|---|---|
| Median age at diagnosis, mo (range) | 64 (9-88) | 24 (2-80) |
| Sex, male/female, no. | 58/49 | 110/89 |
| Severity of AA at onset, no. (%) of patients | ||
| Severe | 79 (74) | 185 (93) |
| Nonsevere | 28 (26) | 14 (7) |
| History, mo, median (range) | 19 (0.1-251) | 51 (0.1-372) |
| Past treatment, no. (%) of patients | ||
| ATG + CsA | — | 39 (20) |
| CsA alone | — | 51 (26) |
| Anabolic steroid alone | — | 13 (7) |
| Unknown* | — | 96 (48) |
ATG indicates antithymocyte globulin; CsA, cyclosporine A; and —, not applicable.
Information regarding previous therapies of 96 cases (from Japan Marrow Donor Program) was unavailable.
Analysis of genomic copy numbers and detection of 6pLOH
Genomic copy numbers, as well as allele-specific copy numbers, were analyzed by using GeneChip 500K arrays (Affymetrix) as previously described.14,15 Briefly, genomic DNA from AA patients and normal controls were analyzed on GeneChip 500K arrays separately. After adjusting several biases introduced during experiments, signal ratios of the corresponding probes between test (patient) and controls were calculated across the genome to obtain genome-wide copy numbers. Genetic lesions, including copy number gains and losses, as well as CNN-LOHs, were first detected using a hidden Markov model-based algorithm implemented in the CNAG software.14,15 Known copy number variations were carefully excluded by referring to the Database of Genomic Variants (www.projects.tcag.ca/variation). CNN-LOH in 6p involving the HLA locus was more specifically and sensitively detected by statistically evaluating the mean differences in allele-specific copy numbers between heterozygous SNPs on 6p (N = ∼ 1400) that were telomeric from the 5′-end of the HLA-A locus (rs1655927) and all non-6p heterozygous SNPs (N = ∼ 105 000) using the Mann-Whitney U test with the R package (www.r-project.org). Possible false-positive findings arising from multiple testing involving the 306 samples were evaluated by maintaining the false discovery rate under 0.01 as previously described,16 where the microarray data of 1000 JMDP donor specimens obtained from an ongoing whole genome association study (unpublished data) were used to calculate an empiric null distribution.17,18
Determination of the missing HLA alleles in 6pLOH(+) clones in patients with AA
The 500K SNP data of the 1800 JMDP donor-recipient pairs (JMDP dataset), together with their HLA genotyping information, was used to generate an HLA SNP haplotype table on the GeneChip 500K platform, which contains the consensus SNPs of the 3 major haplotypes (P1, P2, and P3) in Japanese subjects18 and the SNP sequences of all observed HLA haplotypes complementary to P1 to P3 within the JMDP set (N = 1576; data not shown). To determine the missing HLA haplotype in each 6pLOH(+) patient, those “HLA” haplotypes were first selected from the aforementioned HLA haplotype table that were compatible with the observed HLA genotypes of that patient. Among these, a candidate haplotype was selected such that it contained the minimum number of SNPs that were incompatible with the patient's genotype. For each candidate haplotype, genomic copy numbers were inferred at the heterozygous SNPs along that haplotype using the circular binary segmentation algorithm,19,20 which divided the haplotype into one or more discrete segments with different mean copy numbers. Finally, each copy number segment was thought to be “missing,” when the alternative hypothesis (Ha: Si ≠ Si, for ∀i) was supported against the null hypothesis (H0: Si = Si, for ∀i) using the Wilcoxon signed rank test with a significance level of .05, where Si represents the allele-specific copy number at the ith heterozygous SNP site within the segment of the candidate haplotype with Si being the corresponding value for the complementary haplotype (supplemental Figure 1). Finally, for those HLA types that appeared more than 8 times among 6pLOH(+) cases, their contribution to the observed allelic loss of HLA haplotypes was evaluated by multivariate logistic regression analysis with stepwise backward selection
Flow cytometry
Heparinized PB and BM were collected from the patients at diagnosis and/or after treatment. HLA-A expression on granulocytes, monocytes, B and T cells, and BM CD34+ cells was analyzed by flow cytometry using a FACSCanto II instrument (BD Biosciences) with the FlowJo 7.6.1 program (TreeStar). The monoclonal antibodies used for this study are provided in supplemental Table 2.
Human androgen receptor assay
The human androgen receptor gene was amplified from genomic DNA of 23 female patients, including 3 6pLOH(+) patients, as described by Ishiyama et al21 with some modifications. Clonality was assessed using an “S value” as a marker of skewing in granulocytes and T lymphocytes.
Association of HLA types with AA
A total of 6613 patients who had received allogeneic BM transplantation through the JMDP between 1992 and 2008 were investigated to see whether the HLA alleles frequently missing in CNN-LOH in 6p with the development of AA could represent risk alleles for the development of AA. Thus, the frequencies of patients with each of the candidate risk alleles (HLA-A*31:01, B*40:02, A*02:01, and A*02:06) and those having none of these alleles were compared between 407 patients with AA and those with other hematopoietic disorders (1827 with AML, 1606 with acute lymphocytic leukemia, 1014 with chronic myeloid leukemia, 825 with myelodysplastic syndrome, 566 with non-Hodgkin lymphoma, and 368 with other hematopoietic neoplasms; supplemental Table 3) by calculating the Fisher P values in the corresponding 2 × 2 contingency tables.
Results
Genetic lesions in AA detected by SNP array analysis
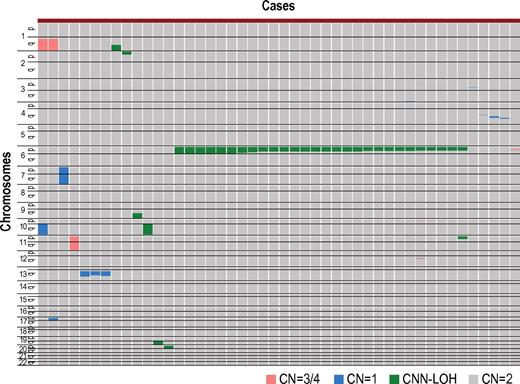
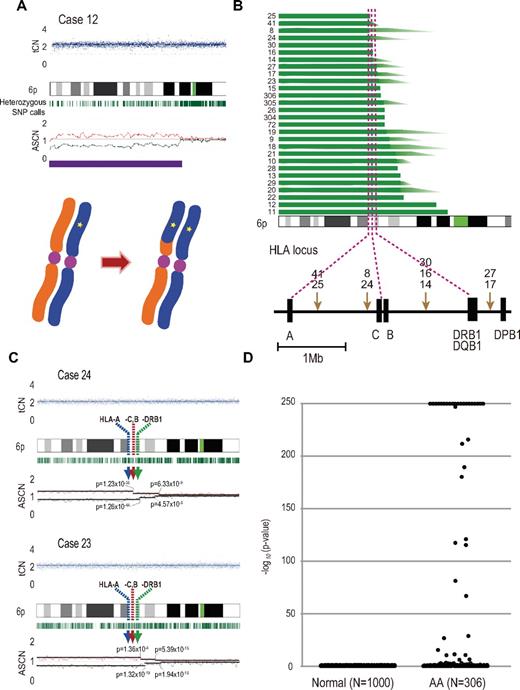
After excluding known or suspected copy number variations, a total of 50 genetic lesions were identified in 46 of the 306 (15%) PB specimens of our AA case series (Table 1; Figure 1). Among these by far, the most conspicuous was the recurrent CNN-LOH involving the 6p arm, which was detected in 28 cases as a significant dissociation of allele-specific copy number graphs in 6p regions using a hidden Markov model–based algorithm implemented in the CNAG software2,14,15 (Figure 2A-2B). Of particular interest was that all CNN-LOH in 6p commonly affected the HLA locus, causing a haploid loss of HLA alleles and uniparental HLA expression. In some cases, the breakpoint of the 6pLOH was predicted to fall within the HLA locus (Figure 2B). These findings strongly indicated that the HLA locus was the genetic target of these 6pLOHs. Also supporting this was the finding that, in half of the cases, the dissociations in the allele-specific copy number graphs were gradually attenuated to the baseline over several mega base pair regions rather than showing a discrete breakpoint, indicating the presence of multiple 6pLOH(+) clones within a single case that had different breakpoints but still shared the same missing HLA alleles (Figure 2C). Moreover, the 6pUPDs existing only in a minor population were more sensitively detected by statistically evaluating the size of dissociation of allele-specific copy numbers in the 6p arm. With this improved statistical test, CNN-LOH in 6p was found in a total of 40 cases (13%; Figure 2D; supplemental Figure 2), where the false discovery rate was maintained at 0.01 to avoid too many false positive findings. In all 6pLOH(+) cases, substantial numbers of heterozygous SNP calls were retained within the affected regions, thus indicating that the CNN-LOHs in 6p were not constitutional but represented acquired genetic events only found in the affected subclones (Figure 1). Indeed, all 6pLOH(+) cases were shown to have “heterozygous” HLA alleles in high-resolution HLA typing of their PB (Table 2). Moreover, 6pLOH was not detected in the CD3-positive T cells in selected cases (cases 25 and 26, supplemental Figure 3). By quantitatively comparing the observed differences in allele-specific copy numbers in the 6pLOH segments with what were expected assuming 100% LOH(+) components, the 6pLOH(+) clones were estimated to account for 0.2% to 53.9% of the PB leukocytes (Table 2). The trend of the lower percentages of the 6pLOH(+) fraction in newly diagnosed patients compared with those in patients at remission was thought to reflect the fact that the former patients tended to have lower counts of granulocytes and monocytes, which were the predominant targets of 6pLOH (see supplemental Table 1).
Copy number changes and allelic imbalances in 46 of the 306 AA cases. The copy number changes and allelic imbalances (or CNN-LOHs) in each case are summarized in the chromosomal order vertically for 46 AA cases with copy number abnormalities. Gains and losses, as well as CNN-LOHs, are shown in the indicated colors.
Copy number changes and allelic imbalances in 46 of the 306 AA cases. The copy number changes and allelic imbalances (or CNN-LOHs) in each case are summarized in the chromosomal order vertically for 46 AA cases with copy number abnormalities. Gains and losses, as well as CNN-LOHs, are shown in the indicated colors.
Acquired 6pLOHs in AA patients that target the HLA locus. (A) Typical CNAG outputs in SNP array analysis showing CNN-LOH (purple line) that appears as significant dissociation in allele-specific copy number graphs (red and green lines) from the baseline with normal total copy numbers (tCN; top panel). As a result of an allelic conversion, the affected segment causes LOH (* indicates 1; bottom panel). The “acquired” origin of these lesions is indicated by the retention of substantial numbers of heterozygous SNP calls (green bars below the chromatogram) that would otherwise mostly disappear. (B) The breakpoints of 6pLOHs found in a total of 28 AA cases, all involving the HLA locus in common. In more than half of cases (indicated by arrowheads in panel B), the exact location of the breakpoint was difficult to uniquely determine, where dissociation of the allele-specific copy number graphs continuously tapered along the 6p arm, indicating the presence of multiple 6pLOH(+) clones with common missing alleles (C). Indeed, the breakpoint containing regions are separated into multiple segments having significantly different copy numbers in the circular binary segmentation model, as indicated by solid lines with P values. Note that the most telomeric breakpoint is located within (case 24) or centromeric to (case 23) the HLA locus in each case. (D) A skewed distribution of the logarithm of P values in AA cases compared with normal persons. The P values were calculated in the Mann-Whitney U test, with which the difference in the mean allele-specific copy numbers between 6p and other chromosomal regions was evaluated (see “Analysis of genomic copy numbers and detection of 6pLOH”). A total of > 250 values are plotted as 250.
Acquired 6pLOHs in AA patients that target the HLA locus. (A) Typical CNAG outputs in SNP array analysis showing CNN-LOH (purple line) that appears as significant dissociation in allele-specific copy number graphs (red and green lines) from the baseline with normal total copy numbers (tCN; top panel). As a result of an allelic conversion, the affected segment causes LOH (* indicates 1; bottom panel). The “acquired” origin of these lesions is indicated by the retention of substantial numbers of heterozygous SNP calls (green bars below the chromatogram) that would otherwise mostly disappear. (B) The breakpoints of 6pLOHs found in a total of 28 AA cases, all involving the HLA locus in common. In more than half of cases (indicated by arrowheads in panel B), the exact location of the breakpoint was difficult to uniquely determine, where dissociation of the allele-specific copy number graphs continuously tapered along the 6p arm, indicating the presence of multiple 6pLOH(+) clones with common missing alleles (C). Indeed, the breakpoint containing regions are separated into multiple segments having significantly different copy numbers in the circular binary segmentation model, as indicated by solid lines with P values. Note that the most telomeric breakpoint is located within (case 24) or centromeric to (case 23) the HLA locus in each case. (D) A skewed distribution of the logarithm of P values in AA cases compared with normal persons. The P values were calculated in the Mann-Whitney U test, with which the difference in the mean allele-specific copy numbers between 6p and other chromosomal regions was evaluated (see “Analysis of genomic copy numbers and detection of 6pLOH”). A total of > 250 values are plotted as 250.
6pLOH(+) AA cases and imputed allelic status of HLA alleles
| UID . | 6pUPD(+) fraction,* % . | Missing alleles . | Retained alleles . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A . | B . | C . | DRB1 . | DQB1 . | DPB1 . | A . | B . | C . | DRB1 . | DQB1 . | DPB1 . | ||
| 19 | 53.9 | 31:01†‡ | 40:02† | 03:04† | 12:01 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 05:01 |
| 12 | 51.8 | 02:01†‡ | 40:02† | 03:03 | 15:01 | 06:02 | 05:01 | 26:02 | 40:06 | 08:01 | 09:01 | 03:03 | 05:01 |
| 17 | 51.6 | 24:02 | 13:01 | 03:04† | 12:02 | 03:01 | 04:02 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 304 | 49.3 | 31:01†‡ | 55:02 | 01:02 | 12:02 | 03:01 | 41:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 04:02 |
| 11 | 48.0 | 02:06†‡ | 40:02† | 03:04† | 15:01 | 06:02 | ND‖ | 11:01 | 67:01 | 07:02 | 16:02 | 05:02 | ND‖ |
| 21 | 46.2 | 31:01ठ| 51:01 | 14:02 | 14:05 | 05:03 | 03:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 04:02 |
| 24 | 44.9 | 31:01† | 40:02† | 03:04† | 11:01 | 03:01 | 02:01 | 24:02 | 40:06 | 08:01 | 09:01 | 03:03 | 05:01 |
| 26 | 44.3 | 31:01†‡§ | 40:01 | 03:04† | 04:05 | 04:01 | 03:01 | 26:03 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 27 | 43.5 | 02:06† | 40:02† | 03:04† | 04:10 | 04:02 | 02:01 | 11:01 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 10 | 42.1 | 31:01† | 40:02† | 03:04† | 08:03 | 06:01 | 02:01 | 24:02 | 51:01 | 14:02 | 09:01 | 03:03 | 02:01 |
| 8 | 40.8 | 02:06†‡ | 40:02† | 03:03 | 12:01 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 04:02 |
| 23 | 35.2 | 02:01† | 40:02† | 03:04† | 09:01 | 03:03 | 02:01 | 24:02 | 54:01 | 01:02 | 04:05 | 04:01 | 04:02 |
| 25 | 32.1 | 02:06†‡ | No LOH | 01:01 | No LOH | ||||||||
| 9 | 23.5 | 02:06†‡ | 39:01 | 07:02 | 08:02 | 04:02 | 02:01 | 24:02 | 15:18 | 07:04 | 04:01 | 03:01 | 14:01 |
| 20 | 21.7 | 26:01‡ | 40:02† | 03:03 | 15:01 | 06:02 | 05:01 | 02:18 | 46:01 | 01:02 | 08:03 | 06:01 | 05:01 |
| 14 | 21.7 | 31:01†‡ | 51:01 | 14:02 | 09:01 | 03:03 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 22 | 20.6 | 02:01† | 39:01 | 07:02 | 08:03 | 06:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 18 | 17.6 | 02:01†‡ | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 | 24:02 | 35:01 | 03:03 | 15:01 | 06:02 | 04:02 |
| 15 | 17.4 | 02:06† | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 02:01 |
| 41 | 15.2† | 31:01†‡ | 35:01 | 03:03 | 09:01 | 03:03 | 03:01 | 26:01 | 39:01 | 07:02 | 08:03 | 06:01 | 05:01 |
| 28 | 12.8 | 24:02 | 54:01 | 01:02 | 01:01 | 05:01 | 04:02 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 29 | 11.7 | 31:01† | 40:02† | 03:04† | 15:01 | 06:02 | 02:01 | 24:02 | 54:01 | 01:02 | 04:05 | 04:01 | 05:01 |
| 305 | 10.3 | 02:06†‡ | 40:02† | 15:02 | 15:02 | 06:01 | 04:01 | 24:02 | 51:01 | 14:02 | 09:01 | 03:03 | 02:01 |
| 13 | 9.6 | 24:02‡ | 40:02† | 03:04† | 15:01 | 06:02 | 02:01 | 02:01‡ | 35:01 | 08:01 | 09:01 | 03:03 | 02:01 |
| 306 | 8.5 | 24:02‡ | 40:02† | 03:04† | 09:01 | 03:03 | 02:01 | 26:02 | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 |
| 16 | 8.1 | 11:01 | 40:06 | 08:01 | No LOH | 24:02 | 46:01 | 01:02 | No LOH | ||||
| 30 | 8.0 | 02:06† | 39:01 | 07:02 | No LOH | 24:02 | 40:06 | 08:01 | No LOH | ||||
| 72 | 5.6 | 02:01† | 40:02† | 03:04† | 09:01 | 03:03 | 05:01 | 02:07 | 46:01 | 01:02 | 08:03 | 06:01 | 02:02 |
| 36 | 4.0 | 02:01†‡ | ND¶ | ND# | 15:02 | 06:01 | 09:01 | 24:02 | ND¶ | ND# | 15:02 | 06:01 | 09:01 |
| 124 | 3.5 | 24:02 | 40:02† | 03:04† | 12:01 | 03:01 | 02:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 223 | 2.8 | 31:01†‡ | 48:01 | 03:04† | 09:01 | 03:03 | 05:01 | 02:06† | 39:01 | 07:02 | 15:01 | 06:02 | 02:01 |
| 215 | 2.8 | 31:01† | 51:01 | 14:02 | 08:02 | 04:02 | 04:02 | 03:01 | 44:02 | 05:01 | 13:01 | 06:03 | 05:01 |
| 181 | 1.3 | 02:06† | 13:01 | 03:04† | 12:02 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 97 | 1.0 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 05:01 | 02:01† | 39:01 | 07:02 | 15:01 | 06:02 | 02:01 |
| 252 | 0.9 | ND** | 40:02† | 03:04† | 09:01 | 03:03 | 05:01 | ND** | 46:01 | 01:02 | 04:05 | 04:01 | 05:01 |
| 118 | 0.9 | 02:06‡§ | 40:02† | 03:04† | 08:02 | 03:02 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 298 | 0.8 | 24:02 | 40:02† | 03:04† | 15:01 | 06:02 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 188 | 0.7 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 | 02:01† | 52:01 | 12:02 | 11:01 | 03:01 | 05:01 |
| 291 | 0.7 | 31:01† | 51:01 | 14:02 | 15:01 | 06:02 | 02:01 | 24:02 | 40:01 | 03:04† | 11:01 | 03:01 | 05:01 |
| 196 | 0.2 | ND†† (A*02:06/24:02, B*35:01/51:01, C*03:03/15:02, DRB1*04:03/15:01, DQB1*03:02/06:02, DPB1*0:201/02:01) | |||||||||||
| UID . | 6pUPD(+) fraction,* % . | Missing alleles . | Retained alleles . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A . | B . | C . | DRB1 . | DQB1 . | DPB1 . | A . | B . | C . | DRB1 . | DQB1 . | DPB1 . | ||
| 19 | 53.9 | 31:01†‡ | 40:02† | 03:04† | 12:01 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 05:01 |
| 12 | 51.8 | 02:01†‡ | 40:02† | 03:03 | 15:01 | 06:02 | 05:01 | 26:02 | 40:06 | 08:01 | 09:01 | 03:03 | 05:01 |
| 17 | 51.6 | 24:02 | 13:01 | 03:04† | 12:02 | 03:01 | 04:02 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 304 | 49.3 | 31:01†‡ | 55:02 | 01:02 | 12:02 | 03:01 | 41:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 04:02 |
| 11 | 48.0 | 02:06†‡ | 40:02† | 03:04† | 15:01 | 06:02 | ND‖ | 11:01 | 67:01 | 07:02 | 16:02 | 05:02 | ND‖ |
| 21 | 46.2 | 31:01ठ| 51:01 | 14:02 | 14:05 | 05:03 | 03:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 04:02 |
| 24 | 44.9 | 31:01† | 40:02† | 03:04† | 11:01 | 03:01 | 02:01 | 24:02 | 40:06 | 08:01 | 09:01 | 03:03 | 05:01 |
| 26 | 44.3 | 31:01†‡§ | 40:01 | 03:04† | 04:05 | 04:01 | 03:01 | 26:03 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 27 | 43.5 | 02:06† | 40:02† | 03:04† | 04:10 | 04:02 | 02:01 | 11:01 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 10 | 42.1 | 31:01† | 40:02† | 03:04† | 08:03 | 06:01 | 02:01 | 24:02 | 51:01 | 14:02 | 09:01 | 03:03 | 02:01 |
| 8 | 40.8 | 02:06†‡ | 40:02† | 03:03 | 12:01 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 04:02 |
| 23 | 35.2 | 02:01† | 40:02† | 03:04† | 09:01 | 03:03 | 02:01 | 24:02 | 54:01 | 01:02 | 04:05 | 04:01 | 04:02 |
| 25 | 32.1 | 02:06†‡ | No LOH | 01:01 | No LOH | ||||||||
| 9 | 23.5 | 02:06†‡ | 39:01 | 07:02 | 08:02 | 04:02 | 02:01 | 24:02 | 15:18 | 07:04 | 04:01 | 03:01 | 14:01 |
| 20 | 21.7 | 26:01‡ | 40:02† | 03:03 | 15:01 | 06:02 | 05:01 | 02:18 | 46:01 | 01:02 | 08:03 | 06:01 | 05:01 |
| 14 | 21.7 | 31:01†‡ | 51:01 | 14:02 | 09:01 | 03:03 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 22 | 20.6 | 02:01† | 39:01 | 07:02 | 08:03 | 06:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 18 | 17.6 | 02:01†‡ | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 | 24:02 | 35:01 | 03:03 | 15:01 | 06:02 | 04:02 |
| 15 | 17.4 | 02:06† | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 02:01 |
| 41 | 15.2† | 31:01†‡ | 35:01 | 03:03 | 09:01 | 03:03 | 03:01 | 26:01 | 39:01 | 07:02 | 08:03 | 06:01 | 05:01 |
| 28 | 12.8 | 24:02 | 54:01 | 01:02 | 01:01 | 05:01 | 04:02 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 29 | 11.7 | 31:01† | 40:02† | 03:04† | 15:01 | 06:02 | 02:01 | 24:02 | 54:01 | 01:02 | 04:05 | 04:01 | 05:01 |
| 305 | 10.3 | 02:06†‡ | 40:02† | 15:02 | 15:02 | 06:01 | 04:01 | 24:02 | 51:01 | 14:02 | 09:01 | 03:03 | 02:01 |
| 13 | 9.6 | 24:02‡ | 40:02† | 03:04† | 15:01 | 06:02 | 02:01 | 02:01‡ | 35:01 | 08:01 | 09:01 | 03:03 | 02:01 |
| 306 | 8.5 | 24:02‡ | 40:02† | 03:04† | 09:01 | 03:03 | 02:01 | 26:02 | 40:06 | 08:01 | 09:01 | 03:03 | 02:01 |
| 16 | 8.1 | 11:01 | 40:06 | 08:01 | No LOH | 24:02 | 46:01 | 01:02 | No LOH | ||||
| 30 | 8.0 | 02:06† | 39:01 | 07:02 | No LOH | 24:02 | 40:06 | 08:01 | No LOH | ||||
| 72 | 5.6 | 02:01† | 40:02† | 03:04† | 09:01 | 03:03 | 05:01 | 02:07 | 46:01 | 01:02 | 08:03 | 06:01 | 02:02 |
| 36 | 4.0 | 02:01†‡ | ND¶ | ND# | 15:02 | 06:01 | 09:01 | 24:02 | ND¶ | ND# | 15:02 | 06:01 | 09:01 |
| 124 | 3.5 | 24:02 | 40:02† | 03:04† | 12:01 | 03:01 | 02:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 223 | 2.8 | 31:01†‡ | 48:01 | 03:04† | 09:01 | 03:03 | 05:01 | 02:06† | 39:01 | 07:02 | 15:01 | 06:02 | 02:01 |
| 215 | 2.8 | 31:01† | 51:01 | 14:02 | 08:02 | 04:02 | 04:02 | 03:01 | 44:02 | 05:01 | 13:01 | 06:03 | 05:01 |
| 181 | 1.3 | 02:06† | 13:01 | 03:04† | 12:02 | 03:01 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 97 | 1.0 | 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 05:01 | 02:01† | 39:01 | 07:02 | 15:01 | 06:02 | 02:01 |
| 252 | 0.9 | ND** | 40:02† | 03:04† | 09:01 | 03:03 | 05:01 | ND** | 46:01 | 01:02 | 04:05 | 04:01 | 05:01 |
| 118 | 0.9 | 02:06‡§ | 40:02† | 03:04† | 08:02 | 03:02 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 298 | 0.8 | 24:02 | 40:02† | 03:04† | 15:01 | 06:02 | 05:01 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 |
| 188 | 0.7 | 24:02 | 52:01 | 12:02 | 15:02 | 06:01 | 09:01 | 02:01† | 52:01 | 12:02 | 11:01 | 03:01 | 05:01 |
| 291 | 0.7 | 31:01† | 51:01 | 14:02 | 15:01 | 06:02 | 02:01 | 24:02 | 40:01 | 03:04† | 11:01 | 03:01 | 05:01 |
| 196 | 0.2 | ND†† (A*02:06/24:02, B*35:01/51:01, C*03:03/15:02, DRB1*04:03/15:01, DQB1*03:02/06:02, DPB1*0:201/02:01) | |||||||||||
UID indicates unique ID.
The percentage of 6pUPD(+) fraction is derived from total peripheral blood leukocytes that include lymphoid as well as myeloid element.
HLA types significantly deviated to missing alleles.
The allelic loss was confirmed by flow cytometry.
The missing haplotype was determined by flow cytometry.
DPB1*04:02/05:01.
B*15:18/52:01.
C*08:01/12:02.
A*02:01/02:07.
Missing allele was not determined because copy number changes in these segments were not statistically significant.
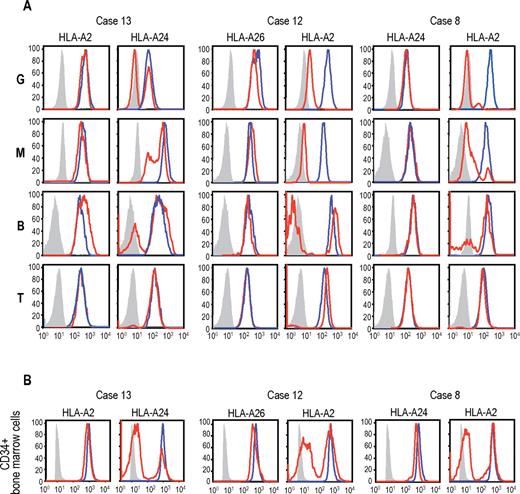
Uniparental expression of HLA in AA cases with CNN-LOH in 6p. Allele-specific expression of HLA-A antigens in AA specimens was examined by flow cytometry using monoclonal antibodies that specifically recognize the indicated HLA types (red lines), where leukocytes from healthy persons were used as a control (blue lines). (A-B) The uniparental expression of HLA-A antigens in PB leukocytes and BM CD34+ cells obtained from 3 AA cases with CNN-LOH in 6p. Different leukocyte compartments were separately examined, including granulocytes (G), monocytes (M), B-lymphocytes (B), and T-lymphocytes (T).
Uniparental expression of HLA in AA cases with CNN-LOH in 6p. Allele-specific expression of HLA-A antigens in AA specimens was examined by flow cytometry using monoclonal antibodies that specifically recognize the indicated HLA types (red lines), where leukocytes from healthy persons were used as a control (blue lines). (A-B) The uniparental expression of HLA-A antigens in PB leukocytes and BM CD34+ cells obtained from 3 AA cases with CNN-LOH in 6p. Different leukocyte compartments were separately examined, including granulocytes (G), monocytes (M), B-lymphocytes (B), and T-lymphocytes (T).
The disease status of the 40 patients at the sampling was before treatment in 16 cases, during remission for 1 to 16 years after therapies in 15, and before BM transplantation for refractory disease in 9. All evaluable 6pLOH(+) AA cases responded to immunosuppressive therapy (IST) (23 of 23), whereas 101 of 126 evaluable cases with 6pLOH(−) responded (P = .014; Table 3).
Response rate (CR + PR) according to the Camitta criteria
| . | Newly diagnosed (n = 107) . | Previously treated (n = 103) . | ||
|---|---|---|---|---|
| 6pLOH(−) (n = 91), no. (%) . | 6pLOH(+) (n = 16), no. (%) . | 6pLOH(−) (n = 88), no. (%) . | 6pLOH(+) (n = 15), no. (%) . | |
| Immunosuppressive therapies (all) | 36/49 (73) | 11/11 (100) | 65/77 (84) | 12/12 (100) |
| ATG + CsA | 14/19 (74) | 7/7 (100) | 27/33 (82) | 5/5 (100) |
| CsA alone | 22/30 (73) | 4/4 (100) | 38/44 (86) | 7/7 (100) |
| Anabolic steroid alone | 0/0 (0) | 0/0 (0) | 7/11 (64) | 2/2 (100) |
| Unknown/not evaluable | 42 | 5 | 0 | 1 |
| . | Newly diagnosed (n = 107) . | Previously treated (n = 103) . | ||
|---|---|---|---|---|
| 6pLOH(−) (n = 91), no. (%) . | 6pLOH(+) (n = 16), no. (%) . | 6pLOH(−) (n = 88), no. (%) . | 6pLOH(+) (n = 15), no. (%) . | |
| Immunosuppressive therapies (all) | 36/49 (73) | 11/11 (100) | 65/77 (84) | 12/12 (100) |
| ATG + CsA | 14/19 (74) | 7/7 (100) | 27/33 (82) | 5/5 (100) |
| CsA alone | 22/30 (73) | 4/4 (100) | 38/44 (86) | 7/7 (100) |
| Anabolic steroid alone | 0/0 (0) | 0/0 (0) | 7/11 (64) | 2/2 (100) |
| Unknown/not evaluable | 42 | 5 | 0 | 1 |
CR indicates complete remission; PR, partial remission; ATG, antithymocyte globulin; and CsA, cyclosporine A.
Uniparental expression of HLA-A in multilineage hematopoietic cells
The genetic loss of one HLA haplotype in SNP array analysis was further confirmed by expression analysis of HLA-A in PB leukocytes using flow cytometry in 19 eligible cases with 6pLOH(+), in which the HLA-A alleles were heterozygous and fresh PB samples were available. Loss of expression of one HLA-A antigen was confirmed in all 19 6pLOH(+) cases (Figure 3A; supplemental Figure 4). The HLA-A missing cells in the PB were shown to have appeared shortly after the onset or before the initiation of treatments in 2 cases, and were confirmed to persist for 1 to 16 months (median, 6 months) in 14 patients (supplemental Table 1; supplemental Figure 5). The percentage of granulocytes lacking HLA-A antigens in the 2 patients who were responsive to IST remained almost the same during the convalescent period of 2 to 3 months (supplemental Figure 6). Importantly, uniparental expression of HLA-A alleles was detected in multiple cell lineages, including granulocytes, monocytes, B cells, and, to a lesser extent, in T cells. Moreover, uniparental HLA-A expression was demonstrated in BM CD34+ cells in 5 patients whose BM samples were available for flow cytometry. All 5 patients possessed various proportions of BM CD34+ cells (49.7%-71.3%), which had lost the expression of one HLA-A antigen; and in each case, the missing HLA-A allele was identical to that in the PB leukocytes (Figure 3B). The uniparental expression of HLA-A in case 13 was also observed in the CD34+ compartment of the archived BM specimen obtained 2 years before analysis (supplemental Figure 7). Together, these findings suggested that the 6pLOH involved early HSPCs and that the 6pLOH occurred at the level of long-term repopulating stem cells.
Clonality of the HLA-missing granulocytes
The human androgen receptor-based clonality assays in granulocytes were performed in 3 6pLOH(+) and 20 6pLOH(−) patients, in which all 3 6pLOH(+) and 4 (20%) of the 6pLOH(−) patients showed evidence of clonality in granulocyte populations (supplemental Figure 8).
Missing HLA alleles in 6pLOH
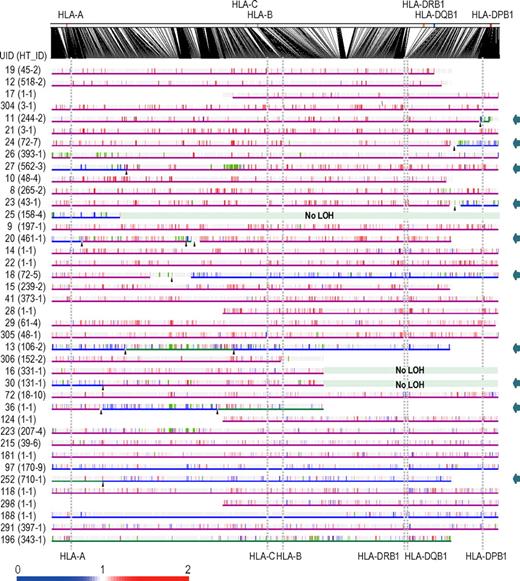
Given that the HLA is the genetic target of 6pLOH in AA, the missing HLA alleles in 6pLOH are of particular interest because in this context they are thought to be directly involved in the presentation of the target auto-antigens to CTLs and, therefore, to be critically important in the pathogenesis of AA. We determined the missing HLA alleles in each 6pLOH(+) AA patient by the haplotype imputation of HLA alleles based on the large data of HLA haplotypes observed in the JMDP set, followed by statistical evaluation of allele-specific copy numbers along the imputed haplotypes (Figure 4). The imputed haplotypes were confirmed in 4 cases by the family studies on the HLA. The allelic status was imputed at least partially in 39 of the 40 6pLOH(+) cases. The imputed results were consistent with the patterns of uniparental expression of HLA-A in flow cytometry in 18 cases with 6pLOH (Table 2; Figure 4), except for those in case 26, in which no valid SNP haplotype around the HLA-A locus was identified and the status of HLA-A was determined by flow cytometry. The missing HLA alleles in 6pLOH(+) AA showed a conspicuous deviation to some selected HLA alleles, including HLA-A*31:01, B*40:02, C*03:04, and, to a lesser extent, HLA-A*02:01 and A*02:06. After the effects of linkage disequilibrium between individual HLA alleles were taken into consideration by multivariate analysis, 4 HLA alleles were shown to remain as the principal determinants of the missing haplotypes, HLA-A*31:01, B*40:02, A*02:01, and A*02:06 (supplemental Table 4).
Imputation of missing HLA haplotypes. The observed allelic copy numbers at heterozygous SNP sites along each candidate SNP haplotype are color-coded as indicated at the bottom. Green bars showed the SNPs that are incompatible with the patient's genotype. Case IDs and haplotype ID (HT_ID) are indicated on the left. The locations of the 500K SNPs and HLA-A, C, B, DRB1, DQB1, and DPB1 are indicated in the figure. For each allele, genomic copy numbers were imputed using the circular binary segmentation algorithm. This divided each haplotype into one or more segments having discrete mean allelic copy numbers (blue arrows on the right). The positions of breakpoints are indicated by arrowheads. Finally, the mean allelic copy number of each segment was statistically compared with that of the corresponding segment on the other haplotype using the Wilcoxon signed rank test. Missing HLA haplotypes were determined based on the result of the statistic tests. Purple and blue lines indicated the retained and missing segments, respectively, whereas the allelic status was not determined statistically for those segments shown by green lines.
Imputation of missing HLA haplotypes. The observed allelic copy numbers at heterozygous SNP sites along each candidate SNP haplotype are color-coded as indicated at the bottom. Green bars showed the SNPs that are incompatible with the patient's genotype. Case IDs and haplotype ID (HT_ID) are indicated on the left. The locations of the 500K SNPs and HLA-A, C, B, DRB1, DQB1, and DPB1 are indicated in the figure. For each allele, genomic copy numbers were imputed using the circular binary segmentation algorithm. This divided each haplotype into one or more segments having discrete mean allelic copy numbers (blue arrows on the right). The positions of breakpoints are indicated by arrowheads. Finally, the mean allelic copy number of each segment was statistically compared with that of the corresponding segment on the other haplotype using the Wilcoxon signed rank test. Missing HLA haplotypes were determined based on the result of the statistic tests. Purple and blue lines indicated the retained and missing segments, respectively, whereas the allelic status was not determined statistically for those segments shown by green lines.
Over-representation of frequently missing HLAs in AA populations
Because these missing HLA alleles in 6pLOH could be involved in the pathogenesis of AA, we next tested whether these relevant HLA alleles are associated with the risk of the development of AA among the 6,613 JMDP registrants. As shown in Table 4, the 4 major missing HLA alleles, HLA-A*31:01, B*40:02, A*02:01, and A*02:06, were more frequently observed in AA cases compared with nonsignificant HLA alleles (ie, all HLA alleles other than these 4 alleles), where the odds ratios for the risk of the development of AA between each of these alleles and nonsignificant alleles were 1.87 (95% confidence interval [CI], 1.43-2.43) for A*02:01, 2.22 (95% CI, 1.70-2.90) for A*02:06, 1.37 (95% CI, 1.00-1.88) for A*31:01, and 1.95 (1.48-2.58) for B*40:02 (Table 4). The combined relative risk for all these alleles was 1.75 (1.42-2.17; P = 1.3 × 10−7).
Association of missing HLA alleles with AA in Japanese patients
| Risk allele . | AA (N = 407) . | Other diseases (N = 6206) . | Total (N = 6613) . | P (χ2 test) . | Odds ratio (95% CI) (vs no risk alleles) . |
|---|---|---|---|---|---|
| A*02:01 | 103 | 1173 | 1276 | 2.5 × 10−6 | 1.87 (1.43−2.43) |
| A*02:06 | 100 | 957 | 1057 | < 1.0 × 10−7 | 2.22 (1.70−2.90) |
| A*31:01 | 58 | 899 | 957 | 0.048 | 1.37 (1.00−1.88) |
| B*40:02 | 86 | 938 | 1024 | 1.8 × 10−6 | 1.95 (1.48−2.58) |
| All risk alleles | 268 | 3250 | 3518 | 1.3 × 10−7 | 1.75 (1.42−2.17) |
| No risk alleles | 139 | 2956 | 3095 | — | — |
| Risk allele . | AA (N = 407) . | Other diseases (N = 6206) . | Total (N = 6613) . | P (χ2 test) . | Odds ratio (95% CI) (vs no risk alleles) . |
|---|---|---|---|---|---|
| A*02:01 | 103 | 1173 | 1276 | 2.5 × 10−6 | 1.87 (1.43−2.43) |
| A*02:06 | 100 | 957 | 1057 | < 1.0 × 10−7 | 2.22 (1.70−2.90) |
| A*31:01 | 58 | 899 | 957 | 0.048 | 1.37 (1.00−1.88) |
| B*40:02 | 86 | 938 | 1024 | 1.8 × 10−6 | 1.95 (1.48−2.58) |
| All risk alleles | 268 | 3250 | 3518 | 1.3 × 10−7 | 1.75 (1.42−2.17) |
| No risk alleles | 139 | 2956 | 3095 | — | — |
— indicates not applicable.
Discussion
The origin of clonal hematopoiesis in AA is a focus of long-standing disputes, in which a profoundly reduced hematopoietic stem cell pool and/or escape from the autoimmune insults have been implicated in the evolution of the clonal hematopoiesis in AA.5,22,23 Our findings on 6pLOH in AA provide an intriguing insight not only into the underlying mechanism of the clonal hematopoiesis in AA but also into the origin of the autoimmunity that is responsible for the pathogenesis of AA. A recent study from the United States also reported 3 cases with 6pLOH.24 With a sensitive detection algorithm, the presence of the 6pLOH(+) components was demonstrated in as many as 13% of typical cases with AA, and the evidence from the subsequent studies strongly indicated that the HLA genes are the genetic targets of 6pLOH in AA patients. First, the HLA locus was commonly and critically involved in all 6pLOHs found in AA. Second, some AA patients carried multiple 6pLOH(+) subclones with different breakpoints, but in all cases, the 6pLOH involved the HLA locus and occurred in a manner that targeted the same parental HLA allele. Moreover, particular class I HLA alleles were over-represented among 6pLOH(+) cases and consistently found in the missing haplotypes. Finally, many of these HLA alleles were shown to be tightly associated with the development of AA in Japanese patients in case-control studies using the large JMDP registry.
The conspicuous bias of the missing HLA alleles in 6pLOH to particular HLA types and the significant association of AA with those HLA types strongly suggest that the recurrent 6pLOH in AA is a phenomenon tightly related to the pathogenesis of AA rather than mere secondary event during the course of AA. Based on these observations, it is well reasoned that, in 6pLOH(+) AA cases, the autoimmunity to HSPCs is mediated by the CTLs that target the antigens presented via specific class I HLA molecules and that the 6pLOH(+) cells found in AA could be explained as escape hematopoiesis that survives the autoimmune insult by genetically deleting the relevant HLA species that are required for antigen presentation (Figure 5). These scenarios are further supported by the recent reports showing that the CNN-LOH in 6p provides a common mechanism of leukemic relapse after HLA haploidentical stem cell transplantations, in which leukemic cells that lost the mismatched HLA haplotype through CNN-LOH in 6p are thought to escape the immunologic surveillance of the engrafted donor T cells.25,26 Importantly, it was experimentally demonstrated by immunologic assays that the 6pLOH(+) leukemic cells actually escaped GVL by CTLs, whereas 6pLOH(−) leukemic cells were effectively killed by the same CTLs. Although the immunologic targets of CTLs are different between relapse after haploidentical transplants (mismatched HLAs themselves) and AA (still unknown autoantigens presented on missing HLAs), the prominent similarities found in both cases further support that CNN-LOH in 6p confers an escape mechanism from autoreactive CTLs in AA.
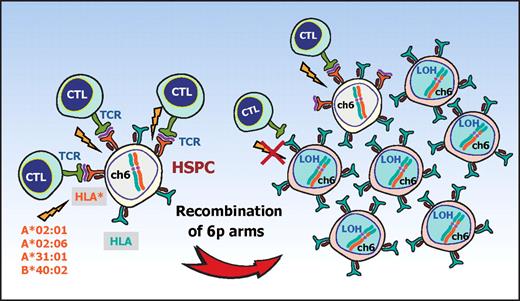
A proposed mechanism for escape hematopoiesis in 6pLOH(+) AA. In AA, the targets of CTLs are the HSPCs that present some auto-antigen through particular class I HLA molecules, including HLA-A*02:01, A*02:06, A*31:01, and B*40:02. In the presence of these autoimmune insults, the HSPCs that lose their expression of the antigen-presenting HLA molecule as a result of CNN-LOH in 6p would acquire a growth advantage over other HSPCs expressing the relevant HLA, leading to clonal outgrowth of the 6pLOH(+) progenies.
A proposed mechanism for escape hematopoiesis in 6pLOH(+) AA. In AA, the targets of CTLs are the HSPCs that present some auto-antigen through particular class I HLA molecules, including HLA-A*02:01, A*02:06, A*31:01, and B*40:02. In the presence of these autoimmune insults, the HSPCs that lose their expression of the antigen-presenting HLA molecule as a result of CNN-LOH in 6p would acquire a growth advantage over other HSPCs expressing the relevant HLA, leading to clonal outgrowth of the 6pLOH(+) progenies.
In light of the above considerations, the chronologic behavior of the 6pLOH(+) components in PB is also interesting and worth discussing. Despite the assumption that 6pLOH is an effective escape mechanism from CTLs, the 6pLOH(+) stem cells were unable to repopulate the BM to cure AA, unless effective IST was applied (supplemental Figure 6). This is most probably explained by the presence of inflammatory cytokines, such as IFN-γ and TNF-α, which have also been shown to play an important role in the BM failure in AA and are thought to be responsible for the continued prevention of the 6pLOH(+) stem cells from fully expanding and reconstituting the BM (supplemental Figure 9A-B).27,28
When the autoimmune insults are removed after IST, no further injury of normal stem cells would occur. However, this does not necessarily mean the surviving normal stem cells can eventually outnumber the 6pLOH(+) stem cells over time. Note that, once the autoimmune insults disappear, nothing could biologically or immunologically discriminate a 6pLOH(+) stem cell from a 6pLOH(−) stem cell (supplemental Figure 9A). In particular, a 6pLOH(+) stem cell and a 6pLOH(−) stem cell will produce the same number of progeny on average and feed the same number of mature blood cells. As a consequence, once established, the predominance of 6pLOH(+) stem cells over 6pLOH(−) stem cells should be maintained, after the severely reduced hematopoietic stem cell pool has been re-expanded with removal of the inciting autoimmunity. It is also of note that the recovery of myeloid components after IST, which are affected more strongly by 6pLOH than lymphoid cells, contributes to an apparent increase in 6pLOH components in the SNP array analysis in PB (supplemental Figure 6A).
One of the most significant findings in the current study is the identification of the HLA alleles that are over-represented in the Japanese AA populations, including HLA-A*31:01, B*40:02, A*02:01, and A*02:06. All of these HLA alleles belong to class I MHCs and thus are thought to be involved in the antigen presentation to CTLs. This provides another prominent example, in which specific HLA types play a critical role in the development of a human disease, and the information about these particular HLA types provides a solid basis on which we can ultimately isolate the relevant antigens responsible for the development of AA. Of particular note, there was a previous report indicating that HLA-B*40:02 and A*02:06 were over-represented in PNH as well as AA, although the study size was much smaller than the current study.29 Combined with our study, these findings support the hypothesis that AA and PNH are the different outcomes of the same immunologic insult5,30 and may also provide the genetic basis of the high prevalence of AA and PNH in East Asia.31,32
In some AA cases, hematopoiesis could be maintained over years by the progenitors that escaped and survived the inciting autoimmune insult by deleting the target HLA through CNN-LOH in 6p. Given that the 6pLOH was detected in only 13% of our series, it is probable that other escape mechanisms may also operate to maintain hematopoiesis in AA. Indeed, clonality was clearly demonstrated in 20% of the 6pLOH(−) cases in the human androgen receptor assay study (supplemental Figure 8). In addition, our SNP array analysis also revealed a variety of clonal abnormalities in AA cases (Figure 1), although it is still open to question whether these abnormalities actually represent the mechanism of escape hematopoiesis or were related to some neoplastic process. Further studies on the genetic basis of the escape mechanisms would contribute to our understanding of the molecular pathogenesis of AA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and donors and their physicians, including K. Kawakami of Suzuka General Hospital and A. Okamoto of Nagoya Daini Red Cross Hospital, for contributing to this study.
This work was supported in part by the Core Research for Evolutional Science and Technology, the Japan Science and Technology Agency, the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aids for Scientific Research), and the Ministry of Health, Labor and Welfare of Japan (Grant-in-Aids).
Authorship
Contribution: S. Ohtake, S. Ogawa, and S.N. developed the concept of the study and supervised the project; T.K., S. Ohtake, and S.N. designed the experiments; T.K., A.S.-O., Y. Sato, Y. Mori, M.K., M.S., K.H., and Y. Sasaki performed the experiments and analyzed the data; K.K. performed high-resolution HLA typing; S.M. and Y. Morishima provided the information of JMDP donor-recipient pairs (JMDP dataset); T.K., A.S.-O., S. Ogawa, and S.N. wrote the paper; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shinji Nakao, Cellular Transplantation Biology, Kanazawa University Graduate School of Medical Science, 13-1 Takaramachi, Kanazawa, Ishikawa 920-8640 Japan; e-mail: snakao@med3.m.kanazawa-u.ac.jp.
References
Author notes
T.K. and A.S.-O. contributed equally to this study.
S. Ogawa and S.N. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal