Abstract
Secreted-frizzled related proteins (SFRPs) are modulators of the Wnt signaling pathway that is closely involved in normal and malignant hematopoiesis. Epigenetic deregulation of Wnt modulators leading to aberrant signaling has been reported in adult patients with acute myeloid leukemia (AML), but its occurrence in childhood patients with AML and the role of individual modulators are unclear. In this study, we examined SFRP1, SFRP2, SFRP4, and SFRP5 promoter methylation in 83 patients with AML (59 children and 24 adults) and found preferential SFRP1 methylation and mRNA down-regulation in the prognostically favorable subgroup of AML with t(8;21) translocation. Among the 4 genes, SFRP1 methylation independently predicted prolonged event-free and relapse-free survivals in childhood patients with nonacute promyelocytic leukemia with nonadverse cytogenetics. Mechanistically, we further demonstrated that RUNX1-ETO, the t(8;21) fusion product, specifically bound the SFRP1 promoter and repressed its transcription via a consensus RUNX binding site. In t(8;21)–leukemia cells, SFRP1 selectively inhibited canonical Wnt signaling and cellular proliferation that were associated with concomitant down-regulation of Wnt/β-catenin target genes, including CCND1 and MYC. Taken together, we identified SFRP1 as a transcriptional repression target of the t(8;21) fusion protein and demonstrated a novel mechanism of Wnt activation in a specific subtype of AML.
Introduction
The Wnt signaling pathway is an important regulator of hematopoietic cell fate and growth.1,2 Deregulation of this pathway has been implicated in human leukemogenesis.3 Wnt signaling comprises the canonical pathway with β-catenin as a central mediator and noncanonical pathways primarily involving Ca2+ ions and planar cell polarity.4 Signal transduction by Wnts is tightly regulated by several families of extracellular modulators, among which secreted frizzled-related proteins (SFRPs) are the largest family.4 SFRPs are generally regarded as Wnt antagonists. However, they can also activate Wnt signaling and interfere with other signaling pathways through Wnt-independent mechanisms.4 In humans, 5 SFRP genes (SFRP1, SFRP2, SFRP3, SFRP4, and SFRP5) have been identified, and 4 of these genes except SFRP3 contain dense CpG islands around their promoter regions. Interestingly, previous studies reported that SFRP1, SFRP2, SFRP4, SFRP5, and other Wnt modulators were silenced by promoter hypermethylation in different cancers, including acute myeloid leukemia (AML), and this epigenetic silencing was associated with aberrant Wnt activation.5-8 Moreover, in AML, the Wnt signaling can also be activated by mutant fms-like tyrosine kinase 3 (FLT3) and common translocation-associated fusion proteins.9,10 Collectively, these findings indicate that Wnt signaling aberration involves multiple mechanisms and is a common molecular feature in different biologic subtypes of AML.
t(8;21)(q22;q22) is the most frequent karyotypic abnormality in AML, representing ∼ 15% of total cases, and is associated with the French-American-British (FAB) M2 subtype.11 t(8;21) fuses the N-terminal portion of RUNX1, including its Runt domain, to most of the eight-twenty-one (ETO)/MTG8 protein. The resulting RUNX1-ETO protein retains the ability of RUNX1 to bind the consensus sequence TGT/cGGT on target gene promoters. RUNX1-ETO has been postulated as a dominant RUNX1 inhibitor because it represses genes that are activated by RUNX1.12,13 Moreover, RUNX1-ETO can recruit multiple corepressors and chromatin remodeling proteins, including histone deacetylases (HDACs) and DNA methyltransferases (DNMTs) to induce heterochromatic gene silencing,14,15 rendering the fusion protein a potent transcriptional repressor. However, RUNX1-ETO can also activate gene transcription.16,17 In particular, the fusion protein has been reported to induce plakoglobin expression and to activate the Wnt/β-catenin pathway in hematopoietic cells.10 These findings provided evidence for a causal link between t(8;21) and aberrant Wnt signaling in AML pathogenesis.
Previous studies have reported that SFRP promoter methylation might carry prognostic implications in adult patients with AML.6,7,18 Nonetheless, because there is growing evidence to indicate biologic heterogeneity between adult and childhood patients with AML,19 whether SFRP methylation also occurs in childhood patients with AML and carries prognostic effects is unknown. Moreover, the role of individual SFRP genes in AML still remains unclear. In this study, we found specific association of SFRP methylation with distinct cytogenetic subtypes in childhood patients with AML and that SFRP1 methylation was an independent prognostic factor for survivals in a subset of these patients. Further mechanistic studies indicated that SFRP1 was a transcriptional repression target of the RUNX1-ETO protein, and this repression promoted canonical Wnt signaling and cell proliferation of t(8;21)–leukemia cells. These findings uncover an additional mechanism by which RUNX1-ETO perturbs the Wnt pathway and contributes to t(8;21) leukemogenesis.
Methods
Patient samples and cell culture
Diagnostic BM from 59 childhood patients with AML (33 males and 26 females) collected in Prince of Wales Hospital since May 1998 were recruited in this study, with institutional review board approval of the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The median age of the patients was 10 years old (range, 0.25-17 years). The patient cohort consisted of 1 M0, 7 M1, 16 M2, 7 M3, 7 M4, 10 M5, 1 M6, 9 M7, and 1 unclassified. Cytogenetic risk groups were classified into favorable [t(8;21), t(15;17), or inv(16)], intermediate (neither favorable nor adverse), and adverse [12p and 5q abnormalities, t(6;9), or −7] according to Harrison et al.19 Patients were treated with the modified United Kingdom Medical Research Council AML 12 protocol, according to cytogenetics and their response to induction chemotherapy.20,21 Essentially, patients were scheduled to receive 2 courses (course 1: 10 + 3 + 5 and course 2: 8 + 3 + 5) of induction therapy with cytarabine, daunorubicin, and etoposide. After achievement of complete remission (CR), a third course of MACE (amsacrine, cytarabine, etoposide) consolidation therapy was given. Patients with favorable cytogenetics received one further course of mitozantrone and cytarabine. Patients with nonfavorable cytogenetics and remission achieved after course 1 chemotherapy received the same 4 courses of chemotherapy or allogeneic transplantation if a matched donor was available. However, patients with nonfavorable cytogenetics who achieved remission after course 2 chemotherapy received amsacrine, cytarabine, and etoposide and then an additional course of chemotherapy with cytarabine and asparaginase followed by mitozantrone and cytarabine as the fifth course, or an allogeneic transplantation if a matched donor was available. For patients with acute promyelocytic leukemia (APL), all-trans retinoic acid was given in addition to the chemotherapy described.
Diagnostic BM from 24 adult patients with AML (12 males and 12 females), including 14 with CBF-AML, were also included for SFRP1 methylation and expression studies. The median age of the patients was 46 years old (range, 18-66 years).
BM samples of all patients were subjected to Ficoll-Hypaque centrifugation, and mononuclear cells were stored at −80°C before use.
Suspension cell lines (Kasumi-1, THP-1, MV4-11, CMK, NB4, ME-1, and U937) were obtained from DSMZ. HeLa and A549 lines were kindly provided by Dr K. W. Lo (Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong). The inducible U937T and U937T-AE lines were generously provided by Prof D. E. Zhang (Department of Pathology, University of California, San Diego). Cells were cultured in RPMI-1640 medium containing 10% FBS (Invitrogen). For U937T and U937T-AE, the medium contained 1 μg/mL tetracycline (Invitrogen) for RUNX1-ETO repression.
Sodium bisulfite modification and MSP
Bisulfite-modified DNA was prepared with the EZ DNA Methylation Kit (Zymo Research). SFRP1, SFRP2, SFRP4, and SFRP5 methylation was examined by methylation-specific PCR (MSP) with the use of primers described before.22,23 For SFRP4, primers were designed with MethPrimer.24 Primer sequences and PCR conditions are provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
RNA extraction and real-time RT-PCR
Total RNA was extracted with TRIZOL (Invitrogen) and treated with DNaseI (USB). Real-time RT-PCR analysis of gene expression was performed with Power SYBR Green PCR Master Mix (ABI) and normalized to GAPDH. Samples were amplified for 40 cycles of 30 seconds at 95°C and 30 seconds at 62°C. Relative expression levels were determined by the 2−ΔΔCt method unless otherwise stated. Primer sequences were listed in supplemental Table 1.
Detection of RUNX1-ETO fusion transcripts was done as described.25 Expression profiling of Wnt signaling genes was performed with the Human Wnt Signaling Pathway RT2 Profile PCR Array (SABiosciences, QIAGEN). PCR products were analyzed by 2% agarose gels.
Detection of FLT3, KIT, NPM1, and CEBPA mutations
FLT3 (internal tandem duplications and D835/I836), KIT (exon 8 and 17), NPM1 C-terminus and CEBPA mutations were analyzed as previously described.25-27
5′-AZA treatment
MV4-11 cells (5 × 106) were treated with 1μM 5-aza-2′-deoxycytidine (5′-AZA; Sigma-Aldrich) for 4 days. Drugs were replenished at day 2 of the treatment. SFRP expression was examined by semiquantitative RT-PCR with the use of primers listed in supplemental Table 1.
DNA constructs, transient transfection, and reporter gene assays
Human SFRP1, SFRP2, and SFRP3 promoter sequences were cloned into pGL3-Basic (Promega). pCMV-RUNX1-ETO and pCMV-CBFβ-MYH11 expression plasmids were kindly provided by Prof. S.W. Hiebert (Department of Biochemistry, Vanderbilt University School of Medicine). pCMV-RUNX1-ETOΔ469 and mutant SFRP1 promoter construct were prepared as previously described.25 Human SFRP1 cDNA expression plasmid (pCMV-SFRP1) was obtained from Origene. T-cell factor/lymphoid enhancer-binding factor (TCF/LEF), activator protein-1 (AP-1) and nuclear factor of activated T cells (NFAT) firefly luciferase reporters were obtained from SABiosciences.
Transient transfection was performed with Lipofectamine 2000 (Invitrogen) as previously described.25 For HeLa, cells were transfected with 0.5 μg of SFRP promoter constructs, 1 μg of pCMV expression plasmids, and 6.25 ng of Renilla luciferase plasmid pRL-CMV (Promega). For U937, U937T, and U937T-AE, cells were transfected with 1.5 μg of SFRP1 promoter constructs and 1 μg of pRL-CMV in the presence or absence of 1.5 μg of pCMV expression plasmids. For TCF/LEF, AP-1, and NFAT reporter studies, cells were transfected with 1 μg of luciferase reporters and 0.5 μg of pCMV expression plasmids. Luciferase activity was measured 24 hours after transfection unless otherwise stated.
To examine the effect of RUNX1-ETO on endogenous SFRP1 expression, we transiently transfected A549 lung cancer cells with 10 μg of pCMV-RUNX1-ETO with the use of Lipofectamine 2000 as previously described.25 SFRP mRNA levels were examined by real-time RT-PCR 48 hours after transfection.
ChIP assays
ChIP assays were performed following the manufacturer's protocol (Upstate Biotechnology) with 10 μg of anti-RUNX1 N-terminus (Calbiochem), anti-ETO (Calbiochem), anti-HDAC1 (Millipore), anti-MeCP2 (Abcam), anti-DNMT1 (New England Biolabs), or anti-DNMT3b (Abcam) Ab. Immunoprecipitation with anti-IgG was done as control. Primers for SFRP1 detection were as follows: oligo1 forward, 5′-GGACTGCGCCTTTTGTCC-3′, and oligo1 reverse, 5′-CTCTGCGCCCTGTTCTCC-3′ (103 bp); oligo2 forward, 5′-TAGATGCAGGAGGCTCAGGT-3′, and oligo2 reverse 5′-AGGAAACGGGAATTTCATCC-3′ (127 bp). Primers for β-globin detection were HBB forward, 5-′CAGTGCCAGAAGAGCCAAG-3′, and HBB reverse, 5-′CGGCAGACTTCTCCTCAG-3′ (237 bp).
Western blot analysis
Nuclear and whole-cell lysates were prepared as previously described.25 β-catenin, MYC, and cyclin D1 expression were detected by rabbit monoclonal Abs against the respective proteins (Cell Signaling Technology). A β-actin Ab (Abcam) served as loading control. Immunostained proteins were visualized by the Luminata chemiluminescent horseradish peroxidase detection reagent (Millipore).
Measurement of phospho-JNK and phospho-p38 MAPK
Flow cytometric analysis of phospho-JNK and phospho-p38 MAPK was performed as previously described.28 Briefly, cells were fixed in 1.5% formaldehyde and permeabilized with methanol. Cells were then incubated with rabbit monoclonal phospho-JNK and phospho-p38 MAPK Abs (Cell Signaling Technology), followed by goat anti–rabbit IgG Alexa Fluor 488–labeled secondary Ab (Invitrogen). Stained cells were analyzed by a FACSCalibur flow cytometer (BD Biosciences).
Measurement of cell proliferation
Cells were treated with the indicated concentrations of recombinant SFRP1 or SFRP4 proteins (R&D Systems) for the indicated time period. Cell proliferation was measured with the WST-1 Cell Proliferation Reagent (Roche Applied Science).
Data analysis
Unpaired t and Fisher exact tests were used to analyze the relation with continuous and categorical variables between groups. Complete survival data were available from 58 childhood patients with AML. Patients with APL (n = 7) were excluded for survival analysis. Event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) were analyzed using the Kaplan-Meier method, and survival curves were compared by the log-rank test. EFS was measured from the date of diagnosis until failure to achieve CR, relapse, or death from any cause (whichever occurred first), censoring for those alive and event-free at last follow-up. RFS was measured from the date of achievement of CR until relapse or death from any cause (whichever occurred first), censoring for those alive and relapse-free at last follow-up. OS was measured from the date of diagnosis until death from any cause, censoring for those alive at last follow-up. Cox regression analysis was performed to test the significance of SFRP1 methylation after controlling for other potential prognostic factors, including age, sex, presentation white blood cell count, FAB subtype (M7 and non-M7), FLT3 mutations, KIT mutations, NPM1 mutations, CEBPA double mutations, and treatment received (4 courses, 5 courses, and allogeneic transplantation). Two-sided P < .05 were considered statistically significant. Survival analysis was performed with SPSS 13.0 (SPSS).
Results
Methylation of SFRP promoters in childhood patients with AML
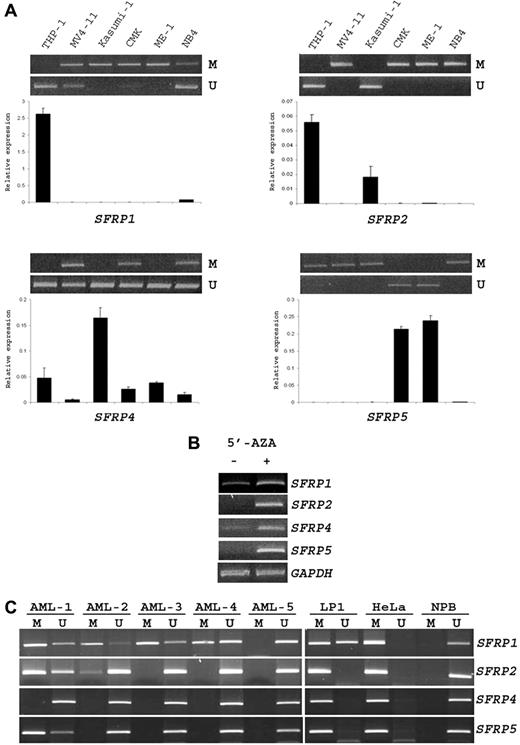
To verify our MSP platform, we first used a multiple myeloma (LP1) and cervical cancer (HeLa) cell line as positive controls and PBMCs from a healthy donor as negative control. Concordant with the published results,6,29,30 SFRP1, SFRP2, SFRP4, and SFRP5 promoters were methylated in LP1 and HeLa cells but unmethylated in the normal peripheral blood sample. The 4 SFRP promoters were also unmethylated in 5 normal BM samples examined (data not shown). In addition, SFRP methylation was found to correlate with a dramatic down-regulation of respective SFRP mRNA levels in 6 AML cell lines (Figure 1A). Treatment with the DNA demethylating agent 5′-AZA increased or reactivated the expression of the 4 SFRP genes in MV4-11 cells (Figure 1B).
Methylation of SFRP promoters in AML cell lines and childhood patients with AML. (A) MSP (top) and real-time RT-PCR (bottom) analysis of SFRP promoter methylation and expression in 6 AML cell lines. M and U represent PCR products with primers specific for the methylated and unmethylated sequences, respectively. SFRP mRNA levels were normalized to GAPDH and compared with the mean expression levels of 3 normal BM samples. Results are presented as mean ± SE from triplicate assays. Note that SFRP promoter methylation was associated with mRNA down-regulation in these cell lines. (B) Semiquantitative RT-PCR analysis of SFRP expression in MV4-11 cells after treatment with the DNA demethylating agent 5′-AZA. Amplification of GAPDH cDNA served as internal control. (C) Representative MSP analysis of the 4 SFRP promoters in 5 childhood patients with AML (AML-1 to -5). The results of positive (LP1 and HeLa cell lines) and negative (NPB, normal peripheral blood) controls are also shown.
Methylation of SFRP promoters in AML cell lines and childhood patients with AML. (A) MSP (top) and real-time RT-PCR (bottom) analysis of SFRP promoter methylation and expression in 6 AML cell lines. M and U represent PCR products with primers specific for the methylated and unmethylated sequences, respectively. SFRP mRNA levels were normalized to GAPDH and compared with the mean expression levels of 3 normal BM samples. Results are presented as mean ± SE from triplicate assays. Note that SFRP promoter methylation was associated with mRNA down-regulation in these cell lines. (B) Semiquantitative RT-PCR analysis of SFRP expression in MV4-11 cells after treatment with the DNA demethylating agent 5′-AZA. Amplification of GAPDH cDNA served as internal control. (C) Representative MSP analysis of the 4 SFRP promoters in 5 childhood patients with AML (AML-1 to -5). The results of positive (LP1 and HeLa cell lines) and negative (NPB, normal peripheral blood) controls are also shown.
In our cohort of 59 childhood patients with AML, 36 (61%) of them did not show any SFRP promoter methylation. Among patients with SFRP methylation (n = 23), 15 of them had 1 methylated SFRP gene, 7 patients had 2 methylated genes, and 1 patient had 3 methylated genes (Figure 1C). None of the patients showed concurrent methylation of the 4 SFRP promoters. The methylation frequency of SFRP1, SFRP2, SFRP4, and SFRP5 promoter was 34% (20 of 59), 17% (10 of 59), 0% (0 of 59), and 3% (2 of 59), respectively. Aberrant SFRP1 methylation was found to be significantly associated with concurrent methylation of SFRP2 or SFRP5 promoter (P = .005).
Clinicopathologic significance of SFRP promoter methylation in childhood patients with AML
To investigate the clinicopathologic significance of SFRP methylation in childhood patients with AML, we first compared patients who had ≥ 1 methylated SFRP gene (n = 23) with those patients without any SFRP methylation (n = 36; Table 1). SFRP promoter methylation correlated significantly with an increased age (median age, 12 years vs 3.5 years; P = .0001) and a male sex (P = .034). In addition, SFRP methylation occurred predominantly in patients with AML (FAB M0/M1/M2; P = .0005). Similar to previous findings in adult patients with AML,6 SFRP methylation was absent in all the 7 patients with childhood APL (P = .036; Table 1). Next, we investigated if the associations were contributed by specific SFRP genes. SFRP5 methylation was not analyzed because of the small number of patients with this alteration. Both SFRP1 and SFRP2 methylations were significantly associated with an increased age and the FAB M0/M1/M2 subtypes (Table 1). Moreover, methylation of SFRP1 (P = .035) and SFRP2 (P = .004) promoters was found to correlate with the presence of CEBPA double mutations. All the 3 double CEBPA mutations were a combination of N-terminal and C-terminal mutations. No significant association with other genetic lesions that involved FLT3, KIT, and NPM1 was observed. Interestingly, when analyzed individually, we observed strong association of SFRP1 and SFRP2 methylation with distinct cytogenetic subtypes: SFRP1 with t(8;21) (P = .002) and SFRP2 with a normal karyotype (P = .002; Table 1).
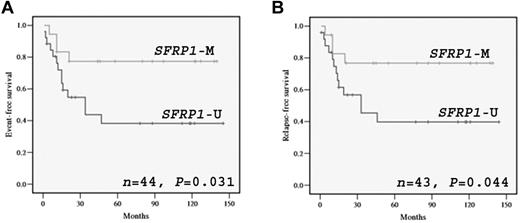
Among the 51 childhood patients with non-APL with survival data, 49 (96%) achieved induction remission. Seventeen patients (33%) relapsed and 11 (22%) died. The median follow-up period was 42 months for all patients and 79 months for the survivors. None of the potential prognostic factors, including age, sex, presentation white blood cell counts, cytogenetics, FAB subtypes, FLT3 mutations, KIT mutations, NPM1 mutations, and CEBPA double mutations, alone was found to have significant effects (P < .05) on EFS, RFS, and OS. In addition, no significant effect of SFRP methylation on survivals in this patient cohort was found. The survival rates did not differ significantly between patients with one or multiple methylated SFRP genes. However, when patients with adverse cytogenetics (n = 7) were excluded, a significant association of SFRP1 methylation with better EFS (P = .031; estimated 5-year EFS rates: 78% vs 38%; Figure 2A) and RFS (P = .044; estimated 5-year RFS rates: 78% vs 40%; Figure 2B) was observed. In this patient subset (n = 44), patients with SFRP1 methylation (3 of 18; 17%) had a significantly lower relapse rate than those without the methylation (13 of 26; 50%; P = .03). Cytogenetics posed no significant effect on EFS and RFS in this subset. In multivariate analysis, SFRP1 methylation remained as an independent prognostic factor for both EFS (P = 0.043; HR, 3.161; 95% confidence interval [CI], 1.037-9.632) and RFS (P = .041; HR, 3.234; 95% CI = 1.046-9.993). No significant effect of SFRP1 methylation on OS was observed in this patient subset.
Kaplan-Meier analysis of EFS and RFS. Kaplan-Meier analysis of EFS (A) and RFS (B) based on SFRP1 promoter methylation in childhood patients with non-APL with nonadverse cytogenetics. M and U represent methylation and unmethylation, respectively. One patient in this cohort failed to achieve CR and was omitted in the RFS analysis.
Kaplan-Meier analysis of EFS and RFS. Kaplan-Meier analysis of EFS (A) and RFS (B) based on SFRP1 promoter methylation in childhood patients with non-APL with nonadverse cytogenetics. M and U represent methylation and unmethylation, respectively. One patient in this cohort failed to achieve CR and was omitted in the RFS analysis.
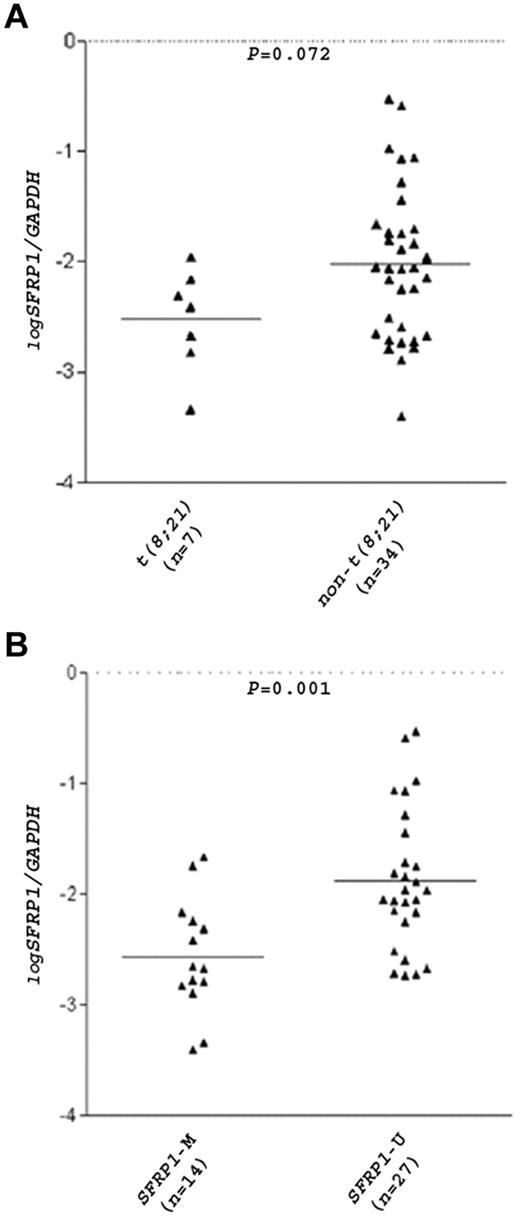
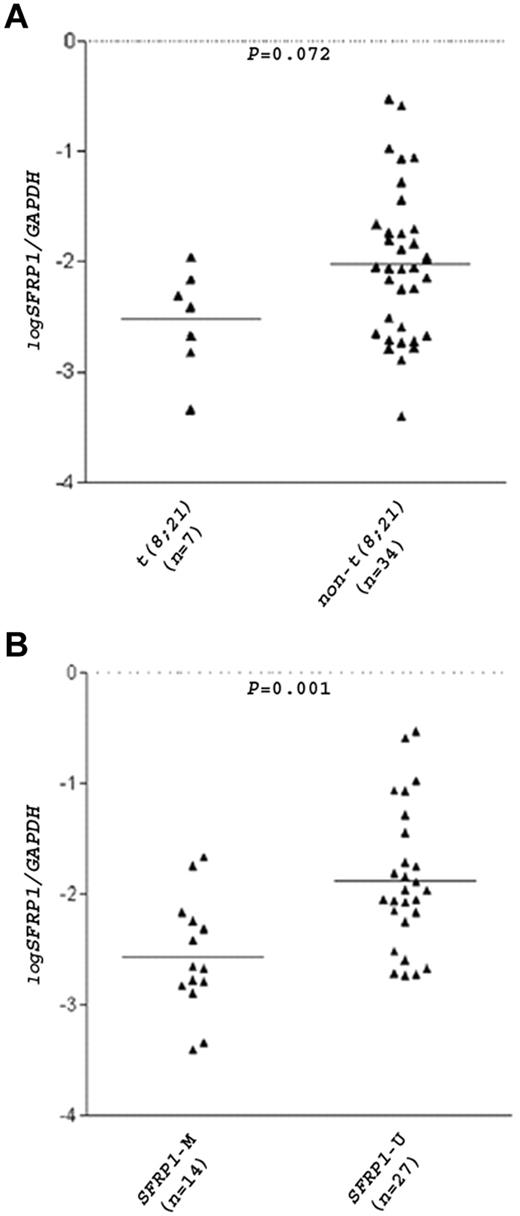
The t(8;21) translocation is closely related to inv(16) in that both chromosome aberrations disrupt the same transcription factor RUNX1, and patients with AML carrying these translocations are collectively termed core-binding factor (CBF)–AML.11 In a total of 30 patients with CBF-AML (16 children and 14 adults), SFRP1 promoter was methylated in 13 of 15 (87%) patients with t(8;21) but only in 6 of 15 (40%) patients with inv(16) (P = .021), indicating preferential SFRP1 methylation in the t(8;21) CBF-AML subgroup. Real-time RT-PCR analysis of 41 patients with AML (27 children and 14 adults) found a substantial reduction of SFRP1 mRNA levels in patients with t(8;21) (mean log SFRP1/GAPDH ratio = −2.53) than in patients with non-t(8;21) (mean log SFRP1/GAPDH ratio = −2.02; P = .072; Figure 3A). Of note, 4 of the 34 patients with non-t(8;21) were of the FAB M2 subtype, and a more substantial difference in SFRP1 levels was observed between these patients with non-t(8;21) M2 (mean expression ratio = −1.49) and the patients with t(8;21) (P = .054). As expected, a significant association between SFRP1 methylation and mRNA down-regulation (P = .001) was found (Figure 3B).
SFRP1 mRNA levels in patients with AML.SFRP1 mRNA levels in 41 diagnostic BM samples from patients with AML (27 children and 14 adults) were measured by real-time RT-PCR and were normalized with GAPDH. The relative quantity of the mRNAs was determined in triplicates with the use of serial dilutions of cDNA from A549 cells as calibration curves. Results are expressed in log10 scale. Patients were divided into 2 groups according to their t(8;21) (A) or SFRP1 methylation (B) status. Each triangle represents one patient, and the number of patients in each group is shown. Horizontal lines indicate the mean SFRP1/GAPDH ratio.
SFRP1 mRNA levels in patients with AML.SFRP1 mRNA levels in 41 diagnostic BM samples from patients with AML (27 children and 14 adults) were measured by real-time RT-PCR and were normalized with GAPDH. The relative quantity of the mRNAs was determined in triplicates with the use of serial dilutions of cDNA from A549 cells as calibration curves. Results are expressed in log10 scale. Patients were divided into 2 groups according to their t(8;21) (A) or SFRP1 methylation (B) status. Each triangle represents one patient, and the number of patients in each group is shown. Horizontal lines indicate the mean SFRP1/GAPDH ratio.
RUNX1-ETO fusion protein bound the SFRP1 promoter and specifically repressed SFRP1 transcription through a consensus RUNX binding site
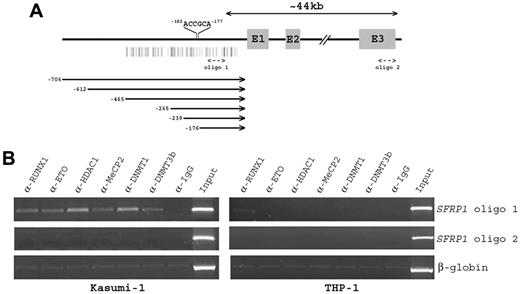
A bioinformatic search found the presence of a consensus RUNX binding site (nt −182/−177) in the 5′-regulatory region of the SFRP1 gene (Figure 4A). We therefore examined the potential occupancy of this RUNX site by the RUNX1-ETO protein in t(8;21)–positive Kasumi-1 and t(8;21)–negative THP-1 cells. ChIP assays showed that DNA sequences flanking the RUNX site (oligo 1) was immunoprecipitated by an anti-RUNX1 N-terminus Ab (Figure 4B). Because Kasumi-1 cells also express RUNX1, we performed ChIP with anti-ETO Ab to distinguish RUNX1-ETO from wild-type RUNX1. The binding of RUNX1-ETO to this chromatin region was confirmed by the detection of ETO immunocomplexes in Kasumi-1 but not THP-1 cells (Figure 4B). The RUNX1-ETO protein has been found to recruit multiple transcriptional corepressors and chromatin remodeling proteins to mediate gene repression.14,15 We found that DNA sequences flanking the RUNX site were also immunoprecipitated by anti-HDAC1, anti-MeCP2, anti-DNMT1, and anti-DNMT3b Abs in SFRP1-silenced Kasumi-1 but not in SFRP1-expressing THP-1 cells (Figure 4B). The specificity of these interactions was indicated by the failure to detect distal SFRP1 sequences lacking RUNX binding sites in the same samples (Figure 4B oligo 2). Together, these findings indicate that the RUNX site on the SFRP1 promoter is a molecular target for the RUNX1-ETO fusion protein.
ChIP analysis of RUNX1-ETO binding to the SFRP1 promoter. (A) Schematic representation of the genomic structure of human SFRP1 gene. The locations of the consensus RUNX binding site (5′-ACCGCA-3′) on the SFRP1 promoter and the 2 oligos used in ChIP analysis are shown. Shaded boxes represent the 3 coding exons (E1-E3), and vertical bars indicate the predicted promoter CpG island. Horizontal lines below the figure indicate the promoter deletions analyzed in Figure 5. The first nucleotide of start codon is assigned as +1. (B) Chromatin from t(8;21)–positive Kasumi-1 and t(8;21)–negative THP-1 cells was immunoprecipitated with the indicated Abs. PCR was performed with oligo 1 primers designed to amplify DNA sequences surrounding the RUNX binding site on the SFRP1 promoter. Oligo 2 primers were designed to amplify a distal region of the SFRP1 gene lacking RUNX site to evaluate the specificity of protein binding. Amplification of β-globin DNA served as a control for nonspecific precipitated sequences.
ChIP analysis of RUNX1-ETO binding to the SFRP1 promoter. (A) Schematic representation of the genomic structure of human SFRP1 gene. The locations of the consensus RUNX binding site (5′-ACCGCA-3′) on the SFRP1 promoter and the 2 oligos used in ChIP analysis are shown. Shaded boxes represent the 3 coding exons (E1-E3), and vertical bars indicate the predicted promoter CpG island. Horizontal lines below the figure indicate the promoter deletions analyzed in Figure 5. The first nucleotide of start codon is assigned as +1. (B) Chromatin from t(8;21)–positive Kasumi-1 and t(8;21)–negative THP-1 cells was immunoprecipitated with the indicated Abs. PCR was performed with oligo 1 primers designed to amplify DNA sequences surrounding the RUNX binding site on the SFRP1 promoter. Oligo 2 primers were designed to amplify a distal region of the SFRP1 gene lacking RUNX site to evaluate the specificity of protein binding. Amplification of β-globin DNA served as a control for nonspecific precipitated sequences.
To investigate the transcriptional control of the SFRP1 gene by the RUNX1-ETO protein, we cloned a 0.7-kb SFRP1 promoter fragment containing the RUNX site into pGL3-Basic and cotransfected the construct with the t(8;21) expression plasmid into U937 myeloid cells. Ectopic expression of RUNX1-ETO but not the functionally related inv(16) fusion protein CBFβ-MYH11 repressed the SFRP1 promoter (Figure 5A). Deletion of the C-terminal region of RUNX1-ETO (RUNX1-ETOΔ469), which is required for oligomerization and corepressor recruitment,31 ablated the repression. We next conducted the reporter assays in U937T-AE and the parental U937T cells. The U937T-AE is a myeloid cell line inducibly expressing RUNX1-ETO in response to tetracycline withdrawal.32 RT-PCR confirmed RUNX1-ETO induction by tetracycline withdrawal in U937T-AE but not U937T cells (Figure 5B). Concordant with results in U937 cells (Figure 5A), RUNX1-ETO induction repressed the SFRP1 promoter by 4.7-fold in U937T-AE cells, whereas only a mild change was observed in the parental U937T cells (Figure 5B). To identify the RUNX1-ETO response region, we performed 5′-deletion mapping on the SFRP1 promoter. Removal of sequences from nt −706 to −239 did not affect the RUNX1-ETO effect as found by the persistence of 2.6-fold repression on −239-Luc (Figure 5C). However, further deletion to nt −176, which involved the consensus RUNX binding site, abolished the RUNX1-ETO repression (Figure 5C). Likewise, mutation of this RUNX site from 5′-ACCGCA-3′ to 5′-ACTCCA-3′ (−239-RUNXmut-Luc) attenuated the RUNX1-ETO effect. These findings indicate the involvement of the consensus RUNX site in mediating the RUNX1-ETO repression. We also investigated whether wild-type RUNX1 could regulate the SFRP1 promoter by cotransfecting −265-Luc with the same amount of pCMV-RUNX1 (encodes AML1B; kindly provided by Prof S. W. Hiebert, Department of Biochemistry, Vanderbilt University School of Medicine) into HeLa cells, which lack RUNX1 expression.25 Overexpression of RUNX1 was found to have a mild effect (1.3-fold repression by RUNX1 vs 2.7-fold by RUNX1-ETO) on the SFRP1 promoter (data not shown). Finally, we examined whether RUNX1-ETO repressed other SFRP promoters, including SFRP2 and SFRP3 promoters. The SFRP2 promoter is similar to SFRP1 promoter in that both are GC rich, whereas the SFRP3 promoter is non-GC rich. Unlike the SFRP1 promoter, no consensus RUNX binding site was found in 1-kb DNA sequences upstream of the SFRP2 and SFRP3 coding regions. Luciferase reporter assays found that RUNX1-ETO did not repress SFRP2 and SFRP3 promoters, indicating SFRP1 as a specific transcriptional target of the RUNX1-ETO protein (Figure 5C).
RUNX1-ETO specifically repressed the SFRP1 gene through a consensus RUNX binding site on the SFRP1 promoter. (A) The SFRP1 promoter-luciferase construct −706-Luc was cotransfected with pCMV-RUNX1-ETO, pCMV-CBFβ-MYH11, or pCMV-RUNX1-ETOΔ469 together with pRL-CMV into U937 cells. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. (B top) Confirmation of RUNX1-ETO induction on tetracycline withdrawal in U937T-AE cells by semiquantitative RT-PCR. Amplification of GAPDH cDNA was done as internal control. No RUNX1-ETO induction was observed in the parental U937T cells. (B bottom) The SFRP1 promoter-luciferase construct −706-Luc was cotransfected with pRL-CMV into U937T and U937T-AE cells. Six hours after transfection, the medium was removed, and cells were cultured for another 48 hours in the presence or absence of tetracycline (Tet) before luciferase measurement. Transfection with the same amount of pGL3-Basic and pRL-CMV was done in parallel. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct with that of pGL3-Basic in each respective group (+Tet or −Tet). The fold of repression is indicated. (C) SFRP1, SFRP2, and SFRP3 promoter-luciferase constructs were cotransfected with pCMV-RUNX1-ETO and pRL-CMV into HeLa cells. Cotransfection with the same amount of empty pCMV was done in parallel. Results are presented as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with pCMV-RUNX1-ETO with that cotransfected with empty pCMV. All numberings are relative to the first nucleotide of the start codon (+1) of each gene. In all experiments, transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. (D) A549 cells were transfected with 10 μg of pCMV or pCMV-RUNX1-ETO with the use of Lipofectamine 2000. RUNX1-ETO expression was validated by RT-PCR (top), and SFRP mRNA levels were determined by real-time RT-PCR and normalized with GAPDH (bottom) 48 hours after transfection. All results are expressed as mean ± SE from at least triplicate assays. *P < .05.
RUNX1-ETO specifically repressed the SFRP1 gene through a consensus RUNX binding site on the SFRP1 promoter. (A) The SFRP1 promoter-luciferase construct −706-Luc was cotransfected with pCMV-RUNX1-ETO, pCMV-CBFβ-MYH11, or pCMV-RUNX1-ETOΔ469 together with pRL-CMV into U937 cells. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. (B top) Confirmation of RUNX1-ETO induction on tetracycline withdrawal in U937T-AE cells by semiquantitative RT-PCR. Amplification of GAPDH cDNA was done as internal control. No RUNX1-ETO induction was observed in the parental U937T cells. (B bottom) The SFRP1 promoter-luciferase construct −706-Luc was cotransfected with pRL-CMV into U937T and U937T-AE cells. Six hours after transfection, the medium was removed, and cells were cultured for another 48 hours in the presence or absence of tetracycline (Tet) before luciferase measurement. Transfection with the same amount of pGL3-Basic and pRL-CMV was done in parallel. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct with that of pGL3-Basic in each respective group (+Tet or −Tet). The fold of repression is indicated. (C) SFRP1, SFRP2, and SFRP3 promoter-luciferase constructs were cotransfected with pCMV-RUNX1-ETO and pRL-CMV into HeLa cells. Cotransfection with the same amount of empty pCMV was done in parallel. Results are presented as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with pCMV-RUNX1-ETO with that cotransfected with empty pCMV. All numberings are relative to the first nucleotide of the start codon (+1) of each gene. In all experiments, transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. (D) A549 cells were transfected with 10 μg of pCMV or pCMV-RUNX1-ETO with the use of Lipofectamine 2000. RUNX1-ETO expression was validated by RT-PCR (top), and SFRP mRNA levels were determined by real-time RT-PCR and normalized with GAPDH (bottom) 48 hours after transfection. All results are expressed as mean ± SE from at least triplicate assays. *P < .05.
We further investigated if RUNX1-ETO is able to inhibit endogenous SFRP1 expression. The A549 lung cancer cell line that endogenously expresses both SFRP1 and SFRP5 mRNAs was selected for analysis. With the use of flow cytometric analysis of green fluorescent protein expression in the transfected cells, we previously demonstrated a transfection efficiency of 53% under our experimental conditions.25 Ectopic expression of RUNX1-ETO reduced SFRP1 mRNA levels by 38% in A549 cells (P < .05; Figure 5D). In contrast, no significant change in SFRP5 mRNA levels was detected.
SFRP1 inhibited canonical Wnt signaling and cellular proliferation of t(8;21)–leukemia cells
The specific transcriptional repression of SFRP1 by RUNX1-ETO led us to investigate the role of this Wnt modulator in t(8;21)–AML. In an attempt to address this issue, we first examined the expression profile of Wnt signaling genes in Kasumi-1 cells and BM from 3 patients with t(8;21)–AML. In all these samples, we found expression of ≥ 8 members of the 13 WNT genes encoding canonical (eg, Wnt1 and Wnt3) and noncanonical (eg, Wnt4 and Wnt11) Wnts and all the 8 frizzled receptors (FZD) examined (Figure 6A). In addition, expression of other core components of the Wnt signaling pathway, including LRP5, LRP6, DVL1, DVL2, APC, AXIN1, CTNNB1, GSK3β, CSNK1A1, CSNK1G1, and LEF1, was consistently detected in all the samples (Figure 6A). The expression profile of these Wnt signaling genes was found to be largely similar in other AML cell lines without t(8;21) (Figure 6A). Next, we asked whether SFRP1 modulated Wnt signaling in t(8;21)–leukemia cells. We used the U937T-AE-Tet cells (U937T-AE cells cultured in the absence of tetracycline) to mimic t(8;21)–leukemia cells for luciferase reporter analysis because we failed to perform transfection on Kasumi-1 cells. The U937T-AE-Tet cells also lack SFRP1 expression as found by RT-PCR (data not shown). Lithium chloride (LiCl) is a specific inhibitor of glycogen synthase kinase-3β,33 which phosphorylates β-catenin, and promotes its degradation. Addition of LiCl potently activated the TCF/LEF reporter in U937T-AE-Tet cells (Figure 6B), indicating the presence of an intact canonical Wnt pathway. Ectopic expression of SFRP1 inhibited basal and LiCl-induced TCF/LEF reporter activity by 32% and 53%, respectively. We also examined whether SFRP1 expression had any effects on noncanonical Wnt signaling pathways. The NFAT- and AP-1–responsive luciferase reporters, which monitor the transcriptional response to activation of the noncanonical NFAT/Ca2+ and JNK/planar cell polarity pathways, were used for analysis. We found no inhibition of transcriptional activity of these 2 reporters in U937T-AE-Tet cells when SFRP1 was ectopically expressed (Figure 6B). In accord with findings from reporter assays, treatment of Kasumi-1 cells with SFRP1-reduced nuclear β-catenin levels (Figure 6C), indicating suppression of the canonical Wnt pathway. In contrast, SFRP1 treatment had no significant effect on the activity of JNK and p38 MAPK (Figure 6C), the latter of which has been implicated in the noncanonical NFAT/Ca2+ pathway.34 Taken together, these findings indicate that SFRP1 selectively inhibits canonical Wnt/β-catenin signaling and its repression by RUNX1-ETO thus promotes this pathway in t(8;21)–leukemia cells. Finally, we investigated the effects of SFRP1 on the growth of these leukemia cells. Treatment of Kasumi-1 cells with recombinant SFRP1 protein inhibited cellular proliferation in a dose-dependent manner (Figure 6D). Likewise, we observed a similar reduction (36%) in proliferation of U937T-AE-Tet cells after 72-hour treatment of 500 ng/mL SFRP1 (data not shown). Real-time RT-PCR analysis of a panel of 13 genes implicated in Wnt signaling and cell growth control found significant down-regulation of MYC, CCND1, and CCND3 mRNA levels after SFRP1 treatment (Figure 6E). The MYC and CCND1 down-regulation was also confirmed at the protein level (Figure 6E). In contrast, treatment with the same concentrations of recombinant SFRP4 protein had no significant effects on proliferation of Kasumi-1 cells, which endogenously express SFRP4. As a control, we conducted similar experiments in another AML cell line THP-1, which expresses both SFRP1 and SFRP4. Treatment with recombinant SFRP1 or SFRP4 protein did not affect THP-1 cell proliferation (Figure 6D).
SFRP1 inhibited canonical Wnt signaling and cell proliferation of t(8;21)–leukemia cells. (A) Expression of Wnt signaling genes in AML cell lines (Kasumi-1, THP-1, NB4, and CMK) and BM samples from 3 patients with t(8;21)–AML was examined by the Human Wnt Signaling Pathway RT2 Profile PCR Array, and PCR products were analyzed by 2% agarose gels. (B) TCF/LEF-, AP-1–, and NFAT-responsive luciferase reporters were cotransfected with SFRP1 expression plasmid into U937T-AE-Tet cells (U937T-AE cells cultured in the absence of tetracycline). These reporters contain constitutively expressing Renilla luciferase construct (40:1) for normalization of transfection efficiency. Cotransfection with the same amount of empty pCMV expression plasmid was done in parallel. LiCl was added to the culture medium 6 hours after transfection, and the cells were incubated for an additional of 24 hours before luciferase measurement. Transfection efficiency was normalized according to the cotransfected Renilla luciferase activity. Results are expressed as mean ± SE from at least triplicate assays and are presented as normalized luciferase activity (firefly luciferase [FLuc]/Renilla luciferase [RLuc]). (C) Analysis of nuclear β-catenin, phospho-JNK, and phospho-p38 MAPK proteins in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 24 hours. Treatment with PBS was done as control. (Top) Nuclear β-catenin levels were examined by Western blot analysis. The same blot was probed with a β-actin Ab, which served as loading control. (Bottom) phospho-JNK (pJNK) and phospho-p38 MAPK (pp38 MAPK) proteins were analyzed by flow cytometry. Representative histograms and the median fluorescence intensity (expressed as mean ± SE) from triplicate assays are shown. (D) WST-1 analysis of cell proliferation of Kasumi-1 and THP-1 cells after 3-day treatment with different concentrations of recombinant SFRP1 and SFRP4 proteins. (E top) Real-time RT-PCR analysis of 13 genes related to Wnt signaling and cell growth control in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 48 hours. Expression was normalized to GAPDH. (Bottom) Confirmation of the MYC and CCND1 down-regulation at protein level in the SFRP1-treated Kasumi-1 cells by Western blot analysis of whole-cell lysates. β-actin was used as a loading control. (D-E) Results are presented as percentage of the PBS-treated control group and expressed as mean ± SE from at least triplicate assays. *P < .05.
SFRP1 inhibited canonical Wnt signaling and cell proliferation of t(8;21)–leukemia cells. (A) Expression of Wnt signaling genes in AML cell lines (Kasumi-1, THP-1, NB4, and CMK) and BM samples from 3 patients with t(8;21)–AML was examined by the Human Wnt Signaling Pathway RT2 Profile PCR Array, and PCR products were analyzed by 2% agarose gels. (B) TCF/LEF-, AP-1–, and NFAT-responsive luciferase reporters were cotransfected with SFRP1 expression plasmid into U937T-AE-Tet cells (U937T-AE cells cultured in the absence of tetracycline). These reporters contain constitutively expressing Renilla luciferase construct (40:1) for normalization of transfection efficiency. Cotransfection with the same amount of empty pCMV expression plasmid was done in parallel. LiCl was added to the culture medium 6 hours after transfection, and the cells were incubated for an additional of 24 hours before luciferase measurement. Transfection efficiency was normalized according to the cotransfected Renilla luciferase activity. Results are expressed as mean ± SE from at least triplicate assays and are presented as normalized luciferase activity (firefly luciferase [FLuc]/Renilla luciferase [RLuc]). (C) Analysis of nuclear β-catenin, phospho-JNK, and phospho-p38 MAPK proteins in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 24 hours. Treatment with PBS was done as control. (Top) Nuclear β-catenin levels were examined by Western blot analysis. The same blot was probed with a β-actin Ab, which served as loading control. (Bottom) phospho-JNK (pJNK) and phospho-p38 MAPK (pp38 MAPK) proteins were analyzed by flow cytometry. Representative histograms and the median fluorescence intensity (expressed as mean ± SE) from triplicate assays are shown. (D) WST-1 analysis of cell proliferation of Kasumi-1 and THP-1 cells after 3-day treatment with different concentrations of recombinant SFRP1 and SFRP4 proteins. (E top) Real-time RT-PCR analysis of 13 genes related to Wnt signaling and cell growth control in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 48 hours. Expression was normalized to GAPDH. (Bottom) Confirmation of the MYC and CCND1 down-regulation at protein level in the SFRP1-treated Kasumi-1 cells by Western blot analysis of whole-cell lysates. β-actin was used as a loading control. (D-E) Results are presented as percentage of the PBS-treated control group and expressed as mean ± SE from at least triplicate assays. *P < .05.
Discussion
Previous studies showed that promoter methylation of SFRPs and other Wnt modulators carried prognostic implications in adult patients with AML.6,7,18 However, the findings obtained so far have been inconsistent. For instance, Jost et al reported that SFRP2 methylation was associated with poorer OS in CBF-AML,6 whereas Griffiths et al showed that SFRP2 and SFRP5 methylations were associated with poorer disease-free survival and OS in cytogenetically normal AML.18 However, Valencia et al reported that methylation of SFRPs and DKKs was associated with a poorer prognosis only in young adult patients with intermediate-risk cytogenetics.7 In this study, we demonstrated for the first time that SFRP methylation also occurred in childhood patients with AML, with incidence comparable to that reported in adult patients with AML.6 In addition, we observed that the prognostic effect of SFRP methylation depended on cytogenetic groups in childhood patients with AML. However, although methylation of Wnt modulators tended to associate with a poor outcome in adult patients with AML, we found that SFRP1 methylation was an independent prognostic factor for prolonged EFS and RFS in childhood patients with non-APL who have nonadverse cytogenetics. Concordantly, SFRP1 methylation was found to correlate with the presence of CEBPA double mutations, which are known to confer a distinctly favorable prognosis.35 Importantly, a similar trend toward better EFS and RFS was also observed when only patients with CBF-AML (n = 16, EFS: P = .055; RFS: P = .055), patients with intermediate cytogenetics (n = 28, EFS: P = .115; RFS: P = .138), or patients with a normal karyotype (n = 15, EFS: P = .084; RFS: P = .073) were analyzed. The discordant results between adult and childhood patients might be in part because of the small number of patients examined and different SFRP genes involved. Larger prospective studies are needed to further analyze the prognostic value of SFRP1 methylation in childhood patients with AML.
Our present study together with those done by other groups strongly suggested that promoter methylation of Wnt modulators was associated with distinct cytogenetic subtypes of AML. Here, we found that SFRP1 methylation was associated with t(8;21) and SFRP2 methylation with a normal karyotype in childhood AML. Concordantly, SFRP2 methylation was significantly associated with CEBPA double mutations, which were also found predominantly in cytogenetically normal patients with childhood AML.36 Similar in adult patients with AML,6 we did not observe SFRP methylation in all 7 childhood patients with APL. Likewise, it was reported that DKK1 promoter was preferentially methylated in CBF-AML but not in patients with APL.37 In contrast, methylation of WIF1 promoter was only found in patients with APL but not in other AML subtypes.38 Together, these observations were consistent with the recent DNA methylation microarray data showing that different biologically distinct AML subtypes were associated with specific methylation signatures.39 In line with this, we observed that among the 12 childhood patients with non-t(8;21) AML with SFRP1 methylation, 8 of them showed concurrent methylation of the SFRP2 promoter. In sharp contrast, none of the 8 SFRP1-methylated patients with t(8;21) showed concurrent methylation of additional SFRP genes, further indicating the specific association of SFRP1 methylation with this AML subtype. The apparent association between methylation of Wnt modulators and specific cytogenetic subtypes might also contribute to the discordant prognostic effects of Wnt modulator methylation in patients with AML because of different patient selection.
Although Wnt signaling is critically involved in normal and malignant hematopoiesis, the role of individual SFRP genes in these processes has remained unclear. It was suggested that these genes might have different roles as indicated by differential expression patterns in hematopoietic cells.40 In particular, SFRP1 expression was recently found to be down-regulated in patients with AML, myelodysplastic syndrome, and acute lymphoblastic leukemia.41 In this study, we found that SFRP1 but not other SFRP genes was a transcriptional repression target of the RUNX1-ETO fusion protein in AML. The repression was mediated through direct binding of RUNX1-ETO to a consensus RUNX site on the SFRP1 promoter. Our ChIP analyses also suggested that RUNX1-ETO recruited DNMTs and HDAC1 to the SFRP1 promoter, implying multiple mechanisms of SFRP1 silencing by the fusion protein. Indeed, previous studies have also reported synergistic up-regulation of SFRP1 expression by HDAC and DNMT inhibitors.42 The recruitment of DNMTs by RUNX1-ETO to the SFRP1 promoter supported our observations of preferential methylation of the SFRP1 promoter in t(8;21)–AML. Nonetheless, the precise mechanism of epigenetic regulation of SFRP1 by RUNX1-ETO remains to be elucidated. Although RUNX1-ETO is generally thought to repress genes that are activated by RUNX1, forced expression of wild-type RUNX1 did not activate but rather mildly repressed the SFRP1 promoter. Our findings further support the model that t(8;21) interferes with normal RUNX1 activity by converting RUNX1 into a constitutive repressor.43
Previous studies showed that SFRP1 can modulate canonical and noncanonical Wnt signaling both positively and negatively.44-47 Here, we demonstrated that SFRP1 selectively inhibited the canonical pathway in t(8;21)–leukemia cells. This observation was also supported by significant down-regulation of canonical Wnt/β-catenin target genes (CCND1 and MYC) but not noncanonical Wnt targets (PCDH8, JUN, ATF2, and SUZ12)34,44,48 in Kasumi-1 cells after SFRP1 treatment. Thus, our present data suggested that SFRP1 repression by RUNX1-ETO enhanced proliferation of t(8;21)–leukemia cells by promoting Wnt/β-catenin signaling. Consistently, earlier studies have also reported an important role of the β-catenin–dependent TCF and LEF signaling in the growth of this specific subtype of leukemia cells.10 However, treatment of Kasumi-1 cells with SFRP1 alone did not affect apoptosis, which could be readily induced by the HDAC inhibitor trichostatin A (our unpublished data, February 2011). Worthy of note is that this cell line also exhibits epigenetic silencing of other Wnt regulators such as SFRP5 and whether concomitant reexpression of these genes is required to induce cell death remains to be addressed. These observations also raise the need of using patient samples for further characterization of SFRP1 actions in t(8;21)–AML.
In conclusion, we demonstrated that SFRP1 was a direct target of the t(8;21) fusion protein in AML. Unlike Wnt activation via plakoglobin induction that is shared by other AML fusion proteins,10 our findings of transcriptional repression of SFRP1 by RUNX1-ETO represent a specific mechanism by which the fusion protein perturbs the canonical Wnt pathway. In addition, our findings suggested that individual Wnt modulators might have distinct roles in different biologic subtypes of AML. Characterizing this complex relation shall help decipher the role of Wnt signaling in AML pathogenesis and identify valuable targets for therapeutic intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof S. W. Hiebert for providing the RUNX1, RUNX1-ETO, and CBFβ-MYH11 expression plasmids and Prof D. E. Zhang for providing the U937T and U937T-AE inducible cell lines. They also thank Ms Angel Pang for her technical assistance during this study.
This work was supported in part by a grant from the Research Council of the Hong Kong SAR, China (project no. CUHK CERG 4415/05M).
Authorship
Contribution: C.K.C. designed and performed research and wrote the manuscript; L.L., S.H.C., and K.N. performed research; S.H.C., N.P.H.C., R.K.L.I., R.S.M.W., M.M.K.S., and C.K.L. collected and analyzed data; and M.H.L.N. designed research and advised on revision of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Margaret H. L. Ng, Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, New Territories, Hong Kong SAR, China; e-mail: margaretng@cuhk.edu.hk.




![Figure 6. SFRP1 inhibited canonical Wnt signaling and cell proliferation of t(8;21)–leukemia cells. (A) Expression of Wnt signaling genes in AML cell lines (Kasumi-1, THP-1, NB4, and CMK) and BM samples from 3 patients with t(8;21)–AML was examined by the Human Wnt Signaling Pathway RT2 Profile PCR Array, and PCR products were analyzed by 2% agarose gels. (B) TCF/LEF-, AP-1–, and NFAT-responsive luciferase reporters were cotransfected with SFRP1 expression plasmid into U937T-AE-Tet cells (U937T-AE cells cultured in the absence of tetracycline). These reporters contain constitutively expressing Renilla luciferase construct (40:1) for normalization of transfection efficiency. Cotransfection with the same amount of empty pCMV expression plasmid was done in parallel. LiCl was added to the culture medium 6 hours after transfection, and the cells were incubated for an additional of 24 hours before luciferase measurement. Transfection efficiency was normalized according to the cotransfected Renilla luciferase activity. Results are expressed as mean ± SE from at least triplicate assays and are presented as normalized luciferase activity (firefly luciferase [FLuc]/Renilla luciferase [RLuc]). (C) Analysis of nuclear β-catenin, phospho-JNK, and phospho-p38 MAPK proteins in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 24 hours. Treatment with PBS was done as control. (Top) Nuclear β-catenin levels were examined by Western blot analysis. The same blot was probed with a β-actin Ab, which served as loading control. (Bottom) phospho-JNK (pJNK) and phospho-p38 MAPK (pp38 MAPK) proteins were analyzed by flow cytometry. Representative histograms and the median fluorescence intensity (expressed as mean ± SE) from triplicate assays are shown. (D) WST-1 analysis of cell proliferation of Kasumi-1 and THP-1 cells after 3-day treatment with different concentrations of recombinant SFRP1 and SFRP4 proteins. (E top) Real-time RT-PCR analysis of 13 genes related to Wnt signaling and cell growth control in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 48 hours. Expression was normalized to GAPDH. (Bottom) Confirmation of the MYC and CCND1 down-regulation at protein level in the SFRP1-treated Kasumi-1 cells by Western blot analysis of whole-cell lysates. β-actin was used as a loading control. (D-E) Results are presented as percentage of the PBS-treated control group and expressed as mean ± SE from at least triplicate assays. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/25/10.1182_blood-2011-05-354712/4/m_zh89991183140006.jpeg?Expires=1765205232&Signature=OuXTIz75h7rM2xbNFR5DDcKGefMYOG~B23a~8LKM9gnMdohj8VrxAgSbNpkLtdxD4ij-JgXACH7BTGhdF18NqVfqzFEpoNiw~Gte1dV9u-w3gtH33sfjja063Ul7WjOhBJVINlgTsRbk7Txlp6Wz7aGYsYHXCciZA-VEhCh4yVaxn2kzNDGmHf7P~xVRFEoHfBbRH32tVw1obs69EJRGYtKZeGdOkrNuk79ri8Fc5txnKMgZsdg7LwU-wr5tD2S-ydJOioXky09F~qKWmQh757D8FkLIybwAomh~ZieC9uhnmY8ecK9BIWbi0ySVSbfogMlIVdLDnT842IEzN4xnHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 6. SFRP1 inhibited canonical Wnt signaling and cell proliferation of t(8;21)–leukemia cells. (A) Expression of Wnt signaling genes in AML cell lines (Kasumi-1, THP-1, NB4, and CMK) and BM samples from 3 patients with t(8;21)–AML was examined by the Human Wnt Signaling Pathway RT2 Profile PCR Array, and PCR products were analyzed by 2% agarose gels. (B) TCF/LEF-, AP-1–, and NFAT-responsive luciferase reporters were cotransfected with SFRP1 expression plasmid into U937T-AE-Tet cells (U937T-AE cells cultured in the absence of tetracycline). These reporters contain constitutively expressing Renilla luciferase construct (40:1) for normalization of transfection efficiency. Cotransfection with the same amount of empty pCMV expression plasmid was done in parallel. LiCl was added to the culture medium 6 hours after transfection, and the cells were incubated for an additional of 24 hours before luciferase measurement. Transfection efficiency was normalized according to the cotransfected Renilla luciferase activity. Results are expressed as mean ± SE from at least triplicate assays and are presented as normalized luciferase activity (firefly luciferase [FLuc]/Renilla luciferase [RLuc]). (C) Analysis of nuclear β-catenin, phospho-JNK, and phospho-p38 MAPK proteins in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 24 hours. Treatment with PBS was done as control. (Top) Nuclear β-catenin levels were examined by Western blot analysis. The same blot was probed with a β-actin Ab, which served as loading control. (Bottom) phospho-JNK (pJNK) and phospho-p38 MAPK (pp38 MAPK) proteins were analyzed by flow cytometry. Representative histograms and the median fluorescence intensity (expressed as mean ± SE) from triplicate assays are shown. (D) WST-1 analysis of cell proliferation of Kasumi-1 and THP-1 cells after 3-day treatment with different concentrations of recombinant SFRP1 and SFRP4 proteins. (E top) Real-time RT-PCR analysis of 13 genes related to Wnt signaling and cell growth control in Kasumi-1 cells treated with 500 ng/mL recombinant SFRP1 protein for 48 hours. Expression was normalized to GAPDH. (Bottom) Confirmation of the MYC and CCND1 down-regulation at protein level in the SFRP1-treated Kasumi-1 cells by Western blot analysis of whole-cell lysates. β-actin was used as a loading control. (D-E) Results are presented as percentage of the PBS-treated control group and expressed as mean ± SE from at least triplicate assays. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/25/10.1182_blood-2011-05-354712/4/m_zh89991183140006.jpeg?Expires=1765221798&Signature=kSx--yLfw5-lbtR~UMf8PE0~pJafjmKsYou38HHfipkL26~HpCLqr-3Jz53OLf12fPZS1qO69xTksmdZM-q826DUDj1B6Snq4ibWYg-QlFuiKE4bBwQ6wlWamRb6odyNHKMkOIAKR9LR06HdaNxaEAGNX8MFR~0fiAkmh3tSeTyFciM9oGH3s1cWuoF29IWuyOv7Pr1gMxRU9b4m79uy39xkH~PLbYENV6jeviREAaL-pDFfl7MTb160RE-4i6byx4UH~OGdT9s2Y88MRtJRFq8dFweaF8aGh7n7A~tvBM9IJkRPpaSEA7LTKXnuAcjoXOsxeHFFyLuv82MSKEDQlA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)