Abstract
Functional deregulation of transcription factors has been found in many types of tumors. Transcription factor AML1/RUNX1 is one of the most frequent targets of chromosomal abnormalities in human leukemia and altered function of AML1 is closely associated with malignant transformation of hematopoietic cells. However, the molecular basis and therapeutic targets of AML1-related leukemia are still elusive. Here, we explored immediate target pathways of AML1 by in vitro synchronous inactivation in hematopoietic cells. We found that AML1 inhibits NF-κB signaling through interaction with IκB kinase complex in the cytoplasm. Remarkably, AML1 mutants found in myeloid tumors lack the ability to inhibit NF-κB signaling, and human cases with AML1-related leukemia exhibits distinctly activated NF-κB signaling. Furthermore, inhibition of NF-κB signaling in leukemic cells with mutated AML1 efficiently blocks their growth and development of leukemia. These findings reveal a novel role for AML1 as a cytoplasmic attenuator of NF-κB signaling and indicate that NF-κB signaling is one of the promising therapeutic targets of hematologic malignancies with AML1 abnormality.
Introduction
Functional disruption of tumor-suppressive transcription factors, such as p53, has been widely found in many types of tumors. Although great efforts have been made to reactivate wild-type p53,1 therapeutic interventions to impaired transcription factors still need innovative strategy. To overcome this difficulty, we need to seek the treatable and pathogenetic targets for deregulated transcription factors.
Transcription factor AML1, also known as RUNX1, is one of the most frequent targets of chromosomal abnormalities in human leukemia.2 Functional impairment of AML1 caused by point mutation also is reported in patients with leukemia or myelodysplastic syndrome (MDS).3-6 Patients who have AML1 mutations are reported to be accompanied with poor prognosis.5,6 Genetic disruptions of AML1 also are known to cause familial platelet disorder, with predisposition to acute myelogenous leukemia (AML).7 AML1 is a critical regulator in hematopoiesis and has an essential function in the establishment of definitive hematopoiesis, differentiation of lymphocytes, maturation of megakaryocytes, and regulation of hematopoietic stem cells (HSCs).8-11 Point mutations of AML1 have been found throughout the length of this gene in myeloid tumors such as AML or MDS; most of the mutants loose the potential to activate gene transcription, whereas some mutants show a dominant-negative effect over AML1 function.4,12 The types of mutations are similar between AML and MDS. Significantly, it was recently reported that AML1 mutants cause MDS/AML in a mouse bone marrow transplantation (BMT) model.13 To date, many target genes of AML1 have been reported.14,15 However, the signaling pathways involved in the pathogenesis of AML1-related leukemia are still elusive.
t(8;21) produces the chimeric protein AML1/ETO and is one of the most frequent chromosomal translocations found in AML. AML1/ETO is constituted of the N-terminal AML1-derived part and the C-terminal ETO part that contain a DNA-binding domain and a corepressor-binding domain, respectively. Besides the defect of trans-activation potential, AML1/ETO is a potent repressor of gene transcription and acts as a dominant-negative mutant of AML1.
Nuclear factor-κB is a dimeric complex of transcription factors mainly consisting of p65 (RelA)/p50 (NFKB1) or RelB/p52 (NFKB2). There are 2 major pathways in NF-κB signaling: the canonical pathway that broadly modulates cell proliferation, survival, or inflammation; and the noncanonical pathway that mainly controls lymphogenesis or B-cell maturation.16 In the canonical pathway, p65 and p50 (NFKB1) constitute NF-κB complex and are localized in the cytoplasm with IκB in a steady state. Once inflammation is induced, TNF-α stimulates its receptor that in turn activates the IκB kinase (IKK) complex. Then, IκB is phosphorylated by the activated IKK complex and subsequently degraded through the ubiquitin-proteasome pathway, resulting in nuclear translocation of p65/p50 and transactivation of NF-κB target genes. In the noncanonical pathway, RelB stably binds to p100 (NFKB2) in the cytoplasm. During the maturation of B cells, B cell–activating factor of the TNF family (BAFF) stimulates its receptor and activates IKK complex that phosphorylates and processes p100 to p52, resulting in nuclear translocation.17 Although these 2 pathways usually act in distinct physiologic conditions, their regulations are closely related, as indicated by the fact that transcriptional control of NFKB2 is directly regulated by p65.18 Recently, deregulated activation of NF-κB signaling has been reported to be responsible for many types of tumors, including hematologic malignancies.19-22
In this study, we revealed that AML1 represses NF-κB signaling in the hematopoietic cells. This repression is achieved by inhibition of the kinase activity of IKK through physical interaction between AML1 and IKK. Remarkably, aberrant activation of NF-κB signaling is induced not only by targeted disruption but also by leukemia-related gene alteration of AML1. Furthermore, we demonstrate that blocking the NF-κB signaling efficiently represses proliferation of leukemic cells with mutant AML1, including chimeric proteins, and development of AML1-related leukemia in mice.
Methods
Mice
AML1 cKO mice were described previously.11 Homozygous AML1 floxed mice (AML1 f/f) and Mx-Cre–expressing homozygous AML1 floxed mice [AML1 f/f Mx (+)] were kept at the Animal Center for Biomedical Research (University of Tokyo, Tokyo, Japan), according to the institutional guidelines. NOD/Shi-scid, IL-2γ null (NOG) mice were purchased from the Central Institute for Experimental Animals (Kawasaki, Japan).
Retrovirus production, BM harvest, and serial replating assays
To produce oncoprotein-expressing retrovirus, Plat-E packaging cells23 were transiently transfected with retroviral constructs, as described previously.24,25 To produce green fluorescent protein (GFP)– or Cre recombinase fused to the estrogen receptor (CreER)-GFP–expressing retrovirus, we used MP34 packaging cells transduced with pGCDNsam-enhanced (e)GFP (a gift from H. Nakauchi and M. Onodera, University of Tokyo) or pGCDNsam-eGFP-CreER. The viral supernatant was collected from the transfected Plat-E cells, filtered, and used for BM progenitor infection. Six- to 8-week-old female C57/B6 mice were primed with 3 mg of 5-fluorouracil 5 days before BM harvest (day 1). BM cells flushed from the tibia and femur were isolated by density centrifugation over Histopaque-1083 (Sigma-Aldrich) and were allowed to rest overnight in RPMI-1640 with 10% FCS, 1% penicillin streptomycin (PS), and cytokines (50 ng/mL SCF, 50 ng/mL megakaryocyte growth and development factor, 50 ng/mL fusion protein of IL-6R and IL-6, and 50 ng/mL Flt-3 ligand). The following day (day 2), the BM cells were combined with retroviral supernatant and RetroNectin (Takara). On day 4, the infected BM were harvested and resuspended in cold PBS (final concentration, 1 × 104/μL). Serial replating assays were performed at a density of 104/mL replated in duplicate every 7 days in Methocult 3434 (StemCell Technologies). BMS-345541 (Calbiochem) was reconstituted with DMSO (Sigma-Aldrich) and added to methylcellulose of the third generation colony-forming cells.
Microarray analysis
These procedures were performed as described previously.25 Lineage-, Sca-1+, c-Kit+ (LSK) cells derived from AML1 f/f mice were transduced with GFP or CreER-GFP–expressing retroviruses. GFP-positive cells were sorted by an FACSAria cell sorter (BD Biosciences). Total RNA was prepared from sorted GFP+ cells after 24 or 48 hours from adding 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) using the RNeasy micro kit (QIAGEN). Amplification and biotin labeling of fragmented cDNA was carried out using the NuGen Ovation Biotin labeling system (NuGEN Technologies) in duplicate. Labeled probes were hybridized to the Mouse Genome 430 2.0 Array (Affymetrix) and scanned with a GeneChip Scanner 3000 7G (Affymetrix). Expression data were extracted from image files produced on GeneChip Operating software 1.0 (Affymetrix). Normalization and expression value calculation were performed using DNA-Chip Analyzer (www.dchip.org).26 Normalized data were filtered for minimal expression and then tested for gene set enrichment using gene set enrichment analysis (GSEA) Version 2.0 (www.broad.mit.edu/gsea/).27 GSEA enrichment results were filtered for statistical significance using a nominal P value threshold of .05. We also used gene expression data of 285 individuals with AML (www.ncbi.nlm.nih.gov/geo; accession GSE1159 [NCBI GEO])28 and expression data of AML1/ETO transduced human cord blood cells (www.ncbi.nlm.nih.gov/geo; accessions GSE8023, GSE7011 [NCBI GEO]).29,30
Nuclear translocation assay
The HEK293T cells were cultured on glass coverslips and transfected with Lipofectamine (Invitrogen) according to the manufacturer's instructions. Thirty-six hours later, conditioning medium was replaced with starvation medium containing 0.5% FCS medium. Twelve hours later, 100 ng/mL TNF-α (Sigma-Aldrich) was added to the cells for 5 to 20 minutes. The cells were washed 3 times with PBS and fixed in 3.7% formaldehyde in PBS for 20 minutes. After washing with PBS, the cell membrane was permeabilized by treatment with 0.2% Triton in PBS for 10 minutes, and the cell membrane was blocked by treatment with 1% BSA in PBS for 40 minutes. The cells were treated with mouse anti-FLAG monoclonal antibody (dilution, 1:200; Sigma-Aldrich) and rabbit anti-p65 monoclonal antibody (dilution, 1:100; Santa Cruz Biotechnology) for 3 hours. After washing with PBS, the cells were stained with Alexa Fluor 555 goat anti-mouse IgG (dilution, 1:500; Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (dilution, 1:500; Invitrogen), and TO-PRO3 (dilution, 1:1000; Invitrogen) for 1 hour. The cells were washed and treated with ProLong Gold antifade reagent (Invitrogen), and the proteins were visualized using a confocal microscope (63×/1.4 NA oil objective, TCS-LS, Leica). FLAG-tagged AML1 mutants were inserted into pME18S expression vector.24
Quantification of nuclear p65
Nuclear intensity of p65 was quantified with ImageJ Version 1.41o software (National Institutes of Health).31 Mean nuclear intensities of 10 cells from 2 pictures were quantified. Intensities were normalized by background subtraction.
Quantitative real-time PCR
Real-time PCR was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) or LightCycler 480 (Roche Diagnostics) according to the manufacturers' instructions. Results were normalized to GAPDH levels. PCR primers used for quantitative PCR were as follows: Nfkb2f, ACCAAGCTCCATGCTAATGTGAAT; Nfkb2r, GGGTGTGTTTCCAGCAAAGGT; GAPDHf, TGGTGAAGCAGGCATCTGAG; GAPDHr, TGCTGTTGAAGTCGCAGGAG; Cd74f, GCAGTGGCTCTTGTTTGAGA; Cd74r, TTCCTGGCACTTGGTCAGTA; Nfkbiaf, CGAGGAGTACGAGCAAATGG; Nfkbiar, TGATTGCCAAGTGCAGGA; Nfkbief, CGACTCTCTGCTGCTGAATG; Nfkbier, GGTCATCGAAGGGCAAATAA; Plekf, CCGGCCTACCTGCACTACTA; Plekr, CAGCCTCTCAAGTGAACTGC; Tnfaip3f, AAGCTCGTGGCTCTGAAAAC; Tnfaip3r, CCCCACATGTACTGACAAGC; Birc3f, AGAGAGGAGCAGATGGAGCA; Birc3r, TTTGTTCTTCCGGATTAGTGC; Il2rgf, AGGCGAGCTGTACAGAAGCTA; Il2rgr, CTGGGATTCACTCAGATTGCT; Relbf, GCCTTGGGTTCCAGTGAC; and Relbr, TGTATTCGTCGATGATTTCCAA.
Flow cytometric analysis
Cells were sorted with an FACSAria cell sorter, and analysis was performed on an FACS LSRII flow cytometer (BD Biosciences). Intracellular staining of phospho-p65 was done according to the manufacturer's protocol (Cell Signaling Technology). To gate AML1-transduced cells, we used Pcdef3-IRES-GFP or Pcdef3-IRES-GFP-AML1 vector.
Immunoprecipitation and Western blotting
For immunoprecipitation, cell lysates were incubated with the anti-FLAG M2 monoclonal antibody (Sigma-Aldrich), anti-AML1 monoclonal antibody (Cell Signaling Technology), normal rabbit IgG (Santa Cruz Biotechnology) for 1 hour at 4°C. Then, the samples were incubated with protein G-Sepharose (GE Healthcare) for 1 hour at 4°C. The precipitates were washed twice with the TNE buffer32 , twice with high salt–containing wash buffer (50mM Tris-HCl, pH 7.5, 500mM NaCl, 0.1% Nonidet P-40, and 0.05% sodium deoxycholate), and once with low salt–containing buffer (50mM Tris-HCl, pH 7.5, 0.1% Nonidet P-40, and 0.05% sodium deoxycholate), and they were subjected to sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analysis by Western blotting. Antibodies used in immunoblotting were as follows: anti-FLAG (Sigma-Aldrich), anti-hemagglutinin (HA; Roche Diagnostics), anti-AML1 (Cell Signaling Technology), anti-IKKα (Cell Signaling Technology), anti-IKKβ (Cell Signaling Technology), anti-IκBα (Cell Signaling Technology), anti-p65 (Cell Signaling Technology), anti–NF-κB2 p100/p52 (Cell Signaling Technology), and anti–β-actin (Cell Signaling Technology). ECL detection (GE Healthcare) was carried out according to the manufacturer's recommendations. Protein levels were quantified with ImageJ Version 1.41o software.31 Nuclear fraction or cytoplasmic fraction was prepared using a nuclear extract kit (Active Motif) according to the manufacturer's instructions. Splenic B cells were separated by an auto-MACS separation column (Miltenyi Biotec) according to the manufacturer's protocol. These B cells were stimulated with 200 ng/mL recombinant human BAFF (BioSource) for 8 hours. HA-tagged AML1 mutants were inserted into pME18S expression vector.24 FLAG-tagged IKKα and IKKβ were gifts from Dr T. D. Gilmore (Boston University, Boston, MA).
Leukemogenesis assays in vivo
These procedures were performed as described previously.13 In brief, BM mononuclear cells were prestimulated and infected for 60 hours with the retroviruses harboring AML1 S291fsX300-IRES-GFP. Then, 0.2 to 1.2 × 106 infected BM cells (Ly5.1) were injected through the tail vein into C57/B6 (Ly5.2) recipient mice (8 weeks of age) that had been administered a sublethal dose of 5.25 Gy of total body γ-irradiation. In survival assays, 2 × 106 S291fsX300-induced leukemic spleen cells or 1 × 105 MLL/ENL-induced leukemic spleen cells were injected into sublethally irradiated (7.5 Gy) C57/B6 (Ly5.2) mice. Bortezomib (LC Laboratories) was administrated at doses of 1.0 mg/kg, given by intraperitoneal bolus injection twice weekly.
Orthotopic cell line model and in vivo bioluminescence imaging
These procedures were performed as described previously.33 In brief, 1 million SKNO-1 cells were injected into sublethally irradiated (2.0 Gy) NOG mice via tail vein. One week after injection, bortezomib was administrated at doses of 1.0 mg/kg, given by intraperitoneal bolus injection twice weekly. Total body bioluminescence was determined by in vivo bioluminescence imaging (IVIS Lumina2; Caliper Life Sciences) and quantitated using Living Image 2.60 software (Caliper Life Sciences). Luciferase positive SKNO-1 cells were kindly provided by Dr Andrew L. Kung (Dana-Farber Cancer Institute, Boston, MA).
In vitro kinase assay
These procedures were performed as described previously.34 In brief, HEK293T cells transiently expressing either AML1 or mock vector were treated with 100 ng/mL TNF-α (Sigma-Aldrich) for the indicated times and then harvested. Cell extracts were precipitated with anti-IKK antibody (Cell Signaling Technology) for 1 hour at 4°C, and protein G-Sepharose was added for 1 hour at 4°C. Immune complexes were washed twice with lysis buffer and then washed twice with kinase buffer (20mM HEPES, pH 7.4, 1mM MnCl2, 5mM MgCl2, 10mM β-glycerol phosphate, 0.1mM sodium orthovanadate, 2mM NaF, and 1mM dithiothreitol). Kinase assays were performed for 30 minutes at 30°C in 20 μL of kinase buffer containing 5 μCi of [γ-32P]ATP (GE healthcare) and 1 μg of GST-IκBα (Millipore) as a substrate. The reaction mixtures were resolved by SDS-PAGE and then detected by autoradiography and quantified with ImageJ Version 1.41o software.31
Statistical analysis
To compare data between groups, unpaired Student t test was used when equal variance were met by F-test. When unequal variances were detected, the Welch t test was used. Differences were considered statistically significant at a P value of less than .05. To analyze the survival curve, log-rank (Mantel-Cox) test was used. These analysis were done using Prism 5 (GraphPad Software).
Results
NF-κB signaling is activated in AML1-deficient cells
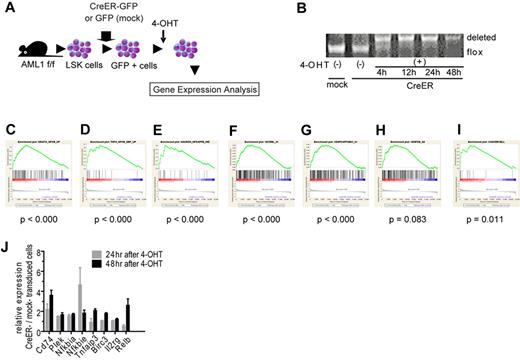
To seek the therapeutic target in AML1-related leukemia, we investigated the deregulated signaling pathway induced by loss of AML1. We analyzed the change of gene expression profiles in immature hematopoietic cells induced by AML1 inactivation. We purified LSK cells as hematopoietic stem and progenitor cells from AML1 conditional knockout (cKO) mice and retrovirally transduced CreER into them.35 Immediately after deleting AML1 by the activation of CreER with 4-OHT, we collected them and analyzed their gene expression profiles with oligonucleotide microarray (Figure 1A, and supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As a control, we used LSK cells infected with an empty retrovirus and treated them with 4-OHT. We confirmed that AML1 was efficiently deleted after the addition of 4-OHT (Figure 1B). To analyze their gene expression profiles, we used GSEA with molecular signature database.27,36 First, we analyzed the expression profiles with all curated gene sets (C2) and found that sets of NF-κB–related genes are significantly enriched in AML1-deficient cells (AML1 flox/flox, CreER+, 24 and 48 hours after adding 4-OHT; Figure 1C-E and supplemental Figure 1B-D).37-39 Next, we analyzed them with all motif gene sets (C3), and again we found that NF-κB–regulated genes are significantly enriched in the AML1-deficient cells (Figure 1F-H and supplemental Figure 1E-G). In addition, the target genes of NF-κB signaling detected in hematopoietic malignant cells were distinctly enriched in AML1-deficient cells (Figure 1I and supplemental Figure 1H).21 We confirmed the elevated expression of NF-κB target genes by real-time PCR (Figure 1J). Because we added 4-OHT both to mock- and CreER-transduced cells, the effects of 4-OHT to our gene expression analysis are unlikely.
NF-κB signaling is activated in AML1-deficient cells. (A) Schematic representation of the gene expression analysis. LSK cells from AML1 cKO mice (AML1 f/f) were transduced CreER-GFP or GFP expressing retroviruses. GFP+ cells were sorted and were subjected to gene expression analysis 24 or 48 hours after adding 4-OHT. (B) PCR genotyping of BM cells from AML1 cKO mice after adding 4-OHT. (C-I) NF-κB–related gene sets enriched in AML1-defecient cells (AML1 flox/flox, CreER+ 24 and 48 hours after adding 4-OHT) by GSEA Version 2.0. (C) A gene set that includes genes up-regulated by NF-κB (HINATA_NFKB_UP). (D) A gene set that includes genes which are up-regulated TNF-α treatment (TNFA_NFKB_DEP_UP). (E) A gene set that includes Ras-inducible, NF-κB–regulated genes (HANSON_NFKAPPB_IND). (F) A gene set that includes genes with promoter regions containing REL motif (V$CREL_01); (G) a gene set which includes Genes with promoter regions containing p65 motif (V$NFKAPPAB65_01). (H) A gene set that includes genes with promoter regions containing NF-κB motif ($NFKB_Q6). (I) Target genes of NF-κB.21 Nominal P value was estimated by GSEA software. (J) mRNA expression for NF-κB target genes in LSK cells transduced with CreER or mock from AML1 f/f mice 24 or 48 hours after 4-OHT addition. Error bars show mean ± SEM.
NF-κB signaling is activated in AML1-deficient cells. (A) Schematic representation of the gene expression analysis. LSK cells from AML1 cKO mice (AML1 f/f) were transduced CreER-GFP or GFP expressing retroviruses. GFP+ cells were sorted and were subjected to gene expression analysis 24 or 48 hours after adding 4-OHT. (B) PCR genotyping of BM cells from AML1 cKO mice after adding 4-OHT. (C-I) NF-κB–related gene sets enriched in AML1-defecient cells (AML1 flox/flox, CreER+ 24 and 48 hours after adding 4-OHT) by GSEA Version 2.0. (C) A gene set that includes genes up-regulated by NF-κB (HINATA_NFKB_UP). (D) A gene set that includes genes which are up-regulated TNF-α treatment (TNFA_NFKB_DEP_UP). (E) A gene set that includes Ras-inducible, NF-κB–regulated genes (HANSON_NFKAPPB_IND). (F) A gene set that includes genes with promoter regions containing REL motif (V$CREL_01); (G) a gene set which includes Genes with promoter regions containing p65 motif (V$NFKAPPAB65_01). (H) A gene set that includes genes with promoter regions containing NF-κB motif ($NFKB_Q6). (I) Target genes of NF-κB.21 Nominal P value was estimated by GSEA software. (J) mRNA expression for NF-κB target genes in LSK cells transduced with CreER or mock from AML1 f/f mice 24 or 48 hours after 4-OHT addition. Error bars show mean ± SEM.
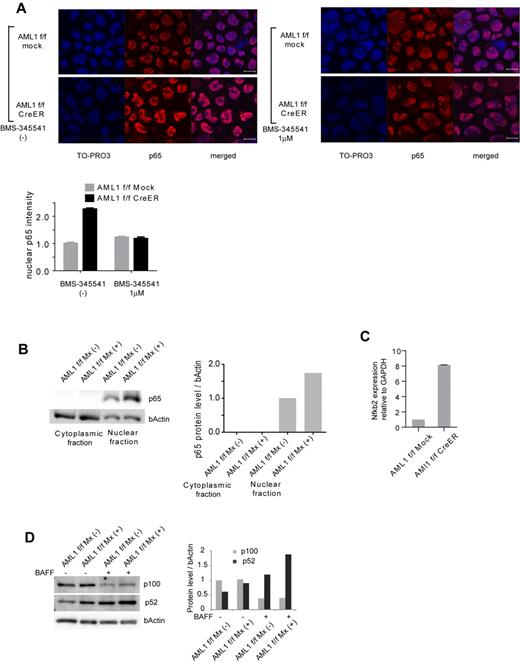
To confirm that NF-κB signaling is activated in AML1-deficient cells, we performed immunofluorescent staining of p65, a major component of NF-κB in the canonical pathway, in AML1-deficient hematopoietic progenitor cells. As shown in Figure 2A, nuclear localized p65 was increased in AML1-deficient cells (AML1 flox/flox, CreER). This increase was attenuated by BMS-345541, an inhibitor of IKK. We also found that nuclear p65 protein was increased in AML1-defficient hematopoietic progenitor cells by Western blotting (Figure 2B). In addition, transcription level of NFKB2, a major component of noncanonical NF-κB pathway, was increased in AML1-deficient cells (Figure 2C), compatible with the transcription of NFKB2 being regulated by p65.18 In the noncanonical NF-κB pathway, cytokines such as BAFF promote processing of p52 from p100 (NFKB2), protein level of which reflects activating status of the pathway. As shown in Figure 2D, both of basal and BAFF-mediated processing of p52 are augmented in AML1-deficient splenic B cells (AML1 flox/flox Mx+) compared with control cells (AML1 flox/flox Mx−) that express intact AML1. These data indicate that both canonical and noncanonical pathways of NF-κB signaling are aberrantly activated in AML1-deficient murine hematopoietic cells.
Canonical and noncanonical pathways of NF-κB signaling are activated in AML1-deficient cells. (A) Immunofluorescent staining of p65 in c-kit + BM cells transduced with CreER or mock from AML1 flox/flox (f/f) mice with or without BMS-345541 (1μM). Scale bar represents 10 μm. Blue indicates TO-PRO3 (nucleus), and red indicates p65. The mean intensity of nuclear localized p65 was quantified with ImageJ Version 1.41o software.31 (B) Fractionated Western blotting of p65 in c-kit + BM cells of AML1-deficient (AML1 f/f Mx+), or control (AML1 f/f Mx−) mice. (C) NFKB2 mRNA expression in BM cells transduced with CreER or mock from AML1 f/f mice 48 hours after 4-OHT addition. Error bars show mean ± SEM (D) Western blotting of NFKB2 (p100 or p52) in B220 + spleen cells from AML1 cKO mice (AML1 f/f Mx+) or control mice (AML1 f/f Mx−) with or without BAFF (200 ng/mL). Protein levels were quantified with ImageJ Version 1.41o software.31
Canonical and noncanonical pathways of NF-κB signaling are activated in AML1-deficient cells. (A) Immunofluorescent staining of p65 in c-kit + BM cells transduced with CreER or mock from AML1 flox/flox (f/f) mice with or without BMS-345541 (1μM). Scale bar represents 10 μm. Blue indicates TO-PRO3 (nucleus), and red indicates p65. The mean intensity of nuclear localized p65 was quantified with ImageJ Version 1.41o software.31 (B) Fractionated Western blotting of p65 in c-kit + BM cells of AML1-deficient (AML1 f/f Mx+), or control (AML1 f/f Mx−) mice. (C) NFKB2 mRNA expression in BM cells transduced with CreER or mock from AML1 f/f mice 48 hours after 4-OHT addition. Error bars show mean ± SEM (D) Western blotting of NFKB2 (p100 or p52) in B220 + spleen cells from AML1 cKO mice (AML1 f/f Mx+) or control mice (AML1 f/f Mx−) with or without BAFF (200 ng/mL). Protein levels were quantified with ImageJ Version 1.41o software.31
AML1 attenuates NF-κB signaling through interaction with IKK complex
Because the NF-κB signaling is activated by the loss of AML1, we investigated whether AML1 represses NF-κB signaling. We performed nuclear translocation assays using HEK293T cells. When HEK293T cells are starved with culture medium containing 0.5% FCS, p65 stays in the cytoplasmic fraction (Figure 3A top left panel). Twenty minutes after adding TNF-α, p65 translocates to the nuclear fraction (Figure 3A top right panel). In AML1-transduced cells, p65 does not translocate to nuclear fraction after adding TNF-α (Figure 3A bottom right panel). These data show that AML1 inhibits nuclear translocation of p65 when expressed in HEK293T cells (Figure 3A). TNF-α–induced phosphorylation of p65 at serine 536, which is important for its activation, also was inhibited by AML1 in those cells (Figure 3B).
AML1 inhibits nuclear translocation and phosphorylation of p65. (A) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 or mock, 20 minutes after the addition of TNF-α. Scale bar represents 10 μm. Green indicates AML1; blue indicates TO-PRO3 (nucleus); and red indicates p65. (B) Intracellular FACS analysis of phospho-p65 (Ser 536) in HEK293T cells transduced with AML1 or mock. GFP-positive fractions were gated. Green line shows phosphorylated p65 protein 5 minutes after TNF-α stimulation. MFI indicates mean fluorescence intensity.
AML1 inhibits nuclear translocation and phosphorylation of p65. (A) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 or mock, 20 minutes after the addition of TNF-α. Scale bar represents 10 μm. Green indicates AML1; blue indicates TO-PRO3 (nucleus); and red indicates p65. (B) Intracellular FACS analysis of phospho-p65 (Ser 536) in HEK293T cells transduced with AML1 or mock. GFP-positive fractions were gated. Green line shows phosphorylated p65 protein 5 minutes after TNF-α stimulation. MFI indicates mean fluorescence intensity.
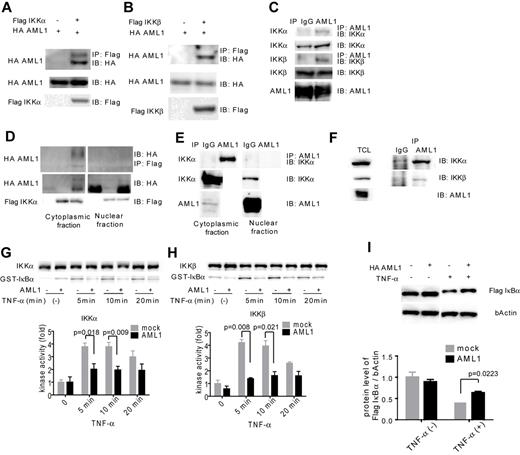
To seek the underlying mechanism for inhibition of NF-κB signaling by AML1, we assessed physical and functional interaction between AML1 and signaling components of NF-κB signaling. IKK is a multimeric complex consisting of IKKα, IKKβ, and IKKγ. The kinase activity of IKK is essential for IκB degradation and nuclear translocation of p65 and is exerted by IKKα and IKKβ. As shown in Figure 4A-B, AML1 was coprecipitated with IKKα and IKKβ when they are expressed in HEK293T cells. These results indicate that AML1 physically interacts with IKKα and IKKβ. Using Jurkat cells, we confirmed that endogenous AML1 forms a complex with IKKα and IKKβ (Figure 4C). Because AML1 is a DNA-binding protein and IKK complex is thought to act mainly in the cytoplasm, we examined where they interact with each other. As shown in Figure 4D, interaction between AML1 and IKK complex was mainly detected in the cytoplasmic fraction in HEK293T cells. Interestingly, protein level of AML1 was increased in the cytoplasmic fraction when IKKα was overexpressed with AML1. We confirmed that endogenous AML1 physically interacts with IKK complex mainly in the cytoplasmic fraction in Jurkat cells (Figure 4E). We also confirmed that AML1 physically interacts with IKK complex in U937 cells, a human myeloid cell line (Figure 4F). Next, we tested whether AML1 affects the kinase activity of IKK complex. Using the in vitro kinase assay, we found that AML1 significantly suppresses the kinase activity of IKKα and IKKβ (Figure 4G-H). In accordance with these findings, AML1 significantly attenuated TNF-α–induced degradation of IκBα in HEK293T cells (Figure 4I).
AML1 physically interacts with IKK complex and inhibits its kinase activity. (A-B) Interaction between AML1 and IKK complex. HEK293T cells were transfected with plasmids encoding for HA-AML1, FLAG-IKKα (A) and FLAG-IKKβ (B), as indicated, and extracts were immunoprecipitated with the antibody against FLAG. Western blots of the input lysate or immunoprecipitates were analyzed using the indicated antibodies. (C) Endogenous interaction between AML1 and the IKK complex in Jurkat cells. Cell extract from the Jurkat cells was immunoprecipitated with the antibody against AML1. Western blots of the input lysate or immunoprecipitates were analyzed using the indicated antibodies. (D) Interaction between AML1 and the IKK complex in cytoplasmic or nuclear fraction in HEK293T cells. (E) Endogenous interaction between AML1 and the IKK complex in cytoplasmic or nuclear fraction in Jurkat cells. (F) Endogenous interaction between AML1 and the IKK complex in U937 cells. Cell extract from the U937 cells was immunoprecipitated with the antibody against AML1. Western blots of the total lysate or immunoprecipitates were analyzed using the indicated antibodies. (G-H) In vitro kinase assays of IKKα (G) or IKKβ (H) in HEK293T cells transduced with AML1 or mock. Kinase activities were detected by autoradiography and quantified with ImageJ Version 1.41o software.31 Error bars show mean ± SEM. (I) Western blotting of IκBα degradation in HEK293T cells transduced with AML1 or mock. Protein levels of IκBα were quantified with ImageJ Version 1.41o software.31 Error bars show mean ± SEM.
AML1 physically interacts with IKK complex and inhibits its kinase activity. (A-B) Interaction between AML1 and IKK complex. HEK293T cells were transfected with plasmids encoding for HA-AML1, FLAG-IKKα (A) and FLAG-IKKβ (B), as indicated, and extracts were immunoprecipitated with the antibody against FLAG. Western blots of the input lysate or immunoprecipitates were analyzed using the indicated antibodies. (C) Endogenous interaction between AML1 and the IKK complex in Jurkat cells. Cell extract from the Jurkat cells was immunoprecipitated with the antibody against AML1. Western blots of the input lysate or immunoprecipitates were analyzed using the indicated antibodies. (D) Interaction between AML1 and the IKK complex in cytoplasmic or nuclear fraction in HEK293T cells. (E) Endogenous interaction between AML1 and the IKK complex in cytoplasmic or nuclear fraction in Jurkat cells. (F) Endogenous interaction between AML1 and the IKK complex in U937 cells. Cell extract from the U937 cells was immunoprecipitated with the antibody against AML1. Western blots of the total lysate or immunoprecipitates were analyzed using the indicated antibodies. (G-H) In vitro kinase assays of IKKα (G) or IKKβ (H) in HEK293T cells transduced with AML1 or mock. Kinase activities were detected by autoradiography and quantified with ImageJ Version 1.41o software.31 Error bars show mean ± SEM. (I) Western blotting of IκBα degradation in HEK293T cells transduced with AML1 or mock. Protein levels of IκBα were quantified with ImageJ Version 1.41o software.31 Error bars show mean ± SEM.
Taken together, AML1 acts as a cytoplasmic attenuator of IKK complex, thus accounting for a mechanistic basis for the inhibition of NF-κB signaling by AML1. Suppression of IKK activity by AML1 results in the inhibition of both nuclear translocation of p65 and activation of NF-κB target genes.
Critical role for NF-κB signaling in the myeloid transformation induced by AML1 mutants
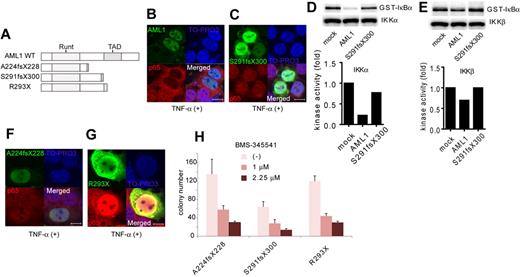
Next, we analyzed the contribution of NF-κB signaling to AML1-related myeloid transformation. We found that 3 types of AML1 mutants, A224fsX228, S291fsX300, and R293X, can transform bone marrow cells in a serial replating assay (Figure 5A and supplemental Figure 2A). A224fsX228 and S291fsX300 were found in human cases with MDS. Similar types of mutants, N209fsX233, R290fsX299, and V292fsX300, were found in cases with de novo AML. R293X was found in both cases with MDS and de novo AML. These mutants belong to the C-terminally truncated type.3,5,40 Among these mutants, it was recently reported that S291fsX300 induce AML in a mouse BMT model, indicating the in vivo transforming activity of this mutant.13 We first assessed the effect of S291fsX300 overexpression on NF-κB signaling. Remarkably, in contrast to the wild type of AML1, S291fsX300 did not inhibit p65 nuclear translocation induced by TNF-α (Figure 5B-C). As is compatible with these findings, S291fsX300 has lost the ability to repress kinase activities of IKKα and IKKβ (Figure 5D-E). In addition, neither A224fsX228 nor R293X inhibited p65 nuclear translocation (Figure 5F-G). In accordance with the inability to inhibit NF-κB signaling, replating capacity of the bone marrow cells introduced with these 3 mutants was highly susceptible to the pharmacologic inhibition of NF-κB signaling by BMS-345541 (Figure 5H). In-frame mutation in the Runt domain such as D171N is another type of AML1 abnormality frequently found in MDS patients (supplemental Figure 2B).12 We found that D171N also did not inhibit p65 nuclear translocation (supplemental Figure 2C). Consistently, the inhibitory effect against IKK activity of D171N was attenuated (supplemental Figure 2D-E). These results suggest that the loss of inhibition of NF-κB signaling is a critical mechanism shared by many types of AML1 mutants in the development of AML.
A critical role of NF-κB signaling in the myeloid transformation induced by AML1 mutants. (A) Schematic presentation of structures of the AML1 mutants. Runt indicates Runt domain, and TAD indicates transactivating domain. (B-C) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 mutants as indicated, 20 minutes after the addition of TNF-α. AML1 (B) and S291fsX300 (C). Scale bar represents 10 μm. (D-E) In vitro kinase assays of IKKα (D) or IKKβ (E) in HEK293T cells transduced with AML1 or S291fsX300, 20 minutes after the addition of TNF-α. Kinase activities were detected by autoradiography and quantified with ImageJ Version 1.41o software.31 (F-G) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 mutants as indicated, 20 minutes after the addition of TNF-α. A224fsX228 (F) and R293X (G). Scale bar represents 10 μm. (H) Colony counts from the serial replating assays of AML1-mutants-transformed cells with BMS-345541. Error bars show mean ± SEM.
A critical role of NF-κB signaling in the myeloid transformation induced by AML1 mutants. (A) Schematic presentation of structures of the AML1 mutants. Runt indicates Runt domain, and TAD indicates transactivating domain. (B-C) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 mutants as indicated, 20 minutes after the addition of TNF-α. AML1 (B) and S291fsX300 (C). Scale bar represents 10 μm. (D-E) In vitro kinase assays of IKKα (D) or IKKβ (E) in HEK293T cells transduced with AML1 or S291fsX300, 20 minutes after the addition of TNF-α. Kinase activities were detected by autoradiography and quantified with ImageJ Version 1.41o software.31 (F-G) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 mutants as indicated, 20 minutes after the addition of TNF-α. A224fsX228 (F) and R293X (G). Scale bar represents 10 μm. (H) Colony counts from the serial replating assays of AML1-mutants-transformed cells with BMS-345541. Error bars show mean ± SEM.
To seek the NF-κB inhibitory domain of AML1, we analyzed a series of deletion mutants of AML1 in nuclear translocation assays of p65 (supplemental Figure 3A). As shown in supplemental Figure 3B, AML1 Δ444, which lacks the domain interacting with a corepressor TLE, inhibited p65 nuclear translocation, whereas AML1 Δ335, AML1a, a truncated isoform of AML1, or AML1 ΔRunt did not (supplemental Figure 3B). These data indicate that C-terminal region with intact Runt domain of AML1 is required for inhibition of p65 nuclear translocation. We also assessed the physical interaction between each mutant of AML1 and IKK and found that all of the employed mutants retain the ability to interact with IKKα (supplemental Figure 3C). In addition, we found that D171N and S291fsX300 can interact with IKKα (supplemental Figure 2F). These results indicate that besides the physical interaction, there exists some additional mechanism that requires the integrity of AML1 for efficient inhibition of NF-κB by AML1.
AML1/ETO-positive leukemia is dependent on NF-κB signaling
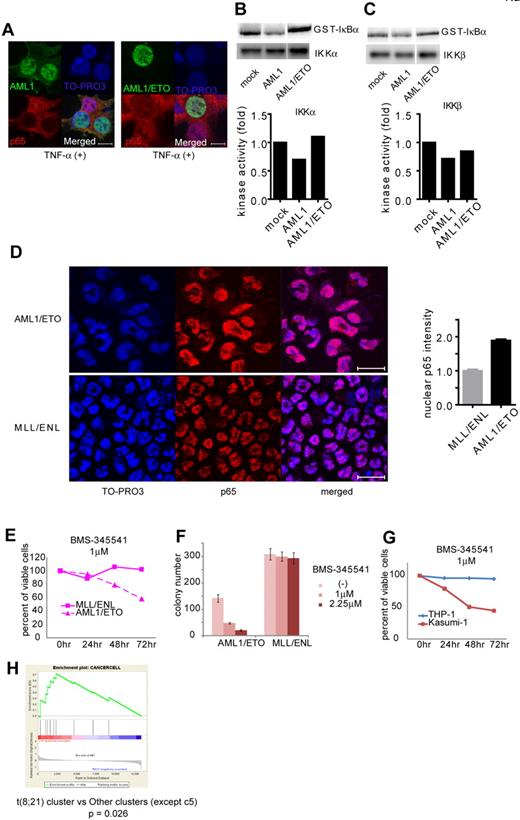
Formation of chimeric genes because of chromosomal translocation is a major cause of AML1 dysfunction that leads to human leukemia. Among them, AML1/ETO, generated in t(8;21) leukemia, is one of the most frequent chimeric genes found in human leukemia. We examined how AML1/ETO affects NF-κB signaling. In contrast to AML1, AML1/ETO could not block nuclear translocation of p65 (Figure 6A). As is compatible with this observation, AML1/ETO has lost the ability to inhibit the kinase activity of IKKα and IKKβ, although it can physically interacted with IKKα (Figure 6B-C and supplemental 4A). In addition, mouse bone marrow cells transformed by AML1/ETO showed enhanced nuclear localization of p65 compared with those immortalized by MLL/ENL (Figure 6D). In agreement with these findings, the growth of AML1/ETO-transformed cells was more susceptible to the NF-κB inhibitor BMS-345541, compared with MLL/ENL-transformed cells (Figure 6E-F). These results indicate a critical role of NF-κB signaling in hematopoietic cell transformation by AML1/ETO.
AML1/ETO-induced leukemic cells depend on NF-κB signaling. (A) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 (left) or AML1/ETO (right), 20 minutes after the addition of TNF-α. Scale bar represents 10 μm. (B-C) In vitro kinase assays of IKKα (B) or IKKβ (C) in HEK293T cells transduced with AML1 or AML1/ETO, 20 minutes after the addition of TNF-α. (D) Immunofluorescent staining of p65 in the BM cells transformed with AML1/ETO (top) or MLL/ENL (bottom). Scale bar represents 10 μm. Blue indicates TO-PRO3 (nucleus), and red indicates p65. The mean intensity of nuclear localized p65 was quantified with ImageJ Version 1.41o software.31 (E) Comparison of sensitivities between AML1/ETO-transformed cells with MLL/ENL-transformed cells in liquid culture. (F) Colony counts from the serial replating assay of AML1/ETO- or MLL/ENL-transformed cells with BMS-345541. Error bars show mean ± SEM (G) Comparison of sensitivities between Kasumi-1 cells (AML1/ETO) with THP-1 cells (MLL/AF9) in liquid culture. (H) Enrichment of NF-κB target genes in t(8;21) leukemia cluster (c13) compared with other clusters (except c5).28
AML1/ETO-induced leukemic cells depend on NF-κB signaling. (A) Nuclear translocation assays of p65 in HEK293T cells transduced with AML1 (left) or AML1/ETO (right), 20 minutes after the addition of TNF-α. Scale bar represents 10 μm. (B-C) In vitro kinase assays of IKKα (B) or IKKβ (C) in HEK293T cells transduced with AML1 or AML1/ETO, 20 minutes after the addition of TNF-α. (D) Immunofluorescent staining of p65 in the BM cells transformed with AML1/ETO (top) or MLL/ENL (bottom). Scale bar represents 10 μm. Blue indicates TO-PRO3 (nucleus), and red indicates p65. The mean intensity of nuclear localized p65 was quantified with ImageJ Version 1.41o software.31 (E) Comparison of sensitivities between AML1/ETO-transformed cells with MLL/ENL-transformed cells in liquid culture. (F) Colony counts from the serial replating assay of AML1/ETO- or MLL/ENL-transformed cells with BMS-345541. Error bars show mean ± SEM (G) Comparison of sensitivities between Kasumi-1 cells (AML1/ETO) with THP-1 cells (MLL/AF9) in liquid culture. (H) Enrichment of NF-κB target genes in t(8;21) leukemia cluster (c13) compared with other clusters (except c5).28
To evaluate activation of NF-κB signaling by AML1/ETO in human hematopoietic cells, we first used Kasumi-1 cells, a cell line derived from AML1/ETO-positive leukemia, and we examined their susceptibility to NF-κB inhibition. In Kasumi-1 cells, small amounts of AML1 and AML1/ETO were detected in the cytoplasmic fraction (supplemental Figure 4B). As shown in Figure 6G, proliferation of Kasumi-1 cells was more sensitive to BMS-345541 than that of THP-1 cells, a human leukemia cell line expressing MLL/AF9. Next, we analyzed in silico the previously reported gene expression data of human leukemias by Valk et al.28 As shown in supplemental Figure 4C, NF-κB signaling was strongly activated in cluster 5, which was defined by Valk et al according to gene expression profiles.28 Remarkably, NF-κB signaling was activated in cluster 13, which contains patients with t(8;21), compared with any other clusters except cluster 5 (Figure 6H and supplemental Figure 4D). These results again indicate that NF-κB signaling is activated in AML1/ETO-positive leukemia and suggest that deregulated NF-κB signaling plays a significant role in AML1/ETO-induced transformation of hematopoietic cells.
Bortezomib ameliorates AML1-related leukemia in vivo
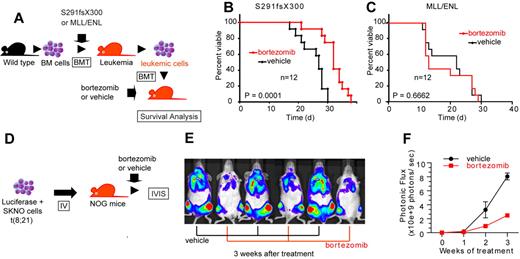
To assess the consequence of NF-κB inhibition on AML1-related leukemia in vivo, we used a mouse BMT model of AML1 S291fsX300.13 We isolated spleen cells from leukemic mice that express AML1 S291fsX300 and transplanted them into sublethally irradiated (7.5 Gy) C57/B6 mice. These mice were treated with twice weekly injections of vehicle or bortezomib, a proteasome inhibitor that broadly inhibits NF-κB signaling (Figure 7A). As shown in Figure 7B, bortezomib significantly prolonged survival of the recipient mice compared with the vehicle-treated mice. Given that NF-κB signaling is not activated in MLL leukemia (Figure 6D-G), we used another leukemia mouse model generated by MLL/ENL as a control that is independent of NF-κB signaling. In contrast to the case with AML1 S291fsX300, bortezomib could not prolong survival of MLL/ENL leukemia mice (Figure 7C).
Bortezomib inhibits the proliferation of leukemic cells with AML1 S291fsX300 or AML1/ETO in vivo. (A) Schematic representation of the following experiments. Spleen cells isolated from S291fsX300-expressing leukemia mice were transplanted into sublethally irradiated (7.5 Gy) recipient mice. These mice were treated with twice weekly injections of vehicle or bortezomib. (B) Survival curves of mice transplanted with S291fsX300-induced leukemic cells treated with vehicle or bortezomib. Each group contains 12 mice. (C) Survival curves of mice transplanted with MLL/ENL-induced leukemic cells treated with vehicle or bortezomib. Each group contains 12 mice. The overall survival of mice in BM transplantation assays was analyzed by log-rank (Mantel-Cox) test. (D) Schematic representation of the following experiments. Luciferase positive SKNO-1 cells were injected into sublethally irradiated (2.0 Gy) NOG mice via tail vein, and these mice were treated with twice weekly injections of vehicle or bortezomib. (E-F) Tumor burden was quantified using in vivo bioluminescence imaging. Each group contains 3 mice.
Bortezomib inhibits the proliferation of leukemic cells with AML1 S291fsX300 or AML1/ETO in vivo. (A) Schematic representation of the following experiments. Spleen cells isolated from S291fsX300-expressing leukemia mice were transplanted into sublethally irradiated (7.5 Gy) recipient mice. These mice were treated with twice weekly injections of vehicle or bortezomib. (B) Survival curves of mice transplanted with S291fsX300-induced leukemic cells treated with vehicle or bortezomib. Each group contains 12 mice. (C) Survival curves of mice transplanted with MLL/ENL-induced leukemic cells treated with vehicle or bortezomib. Each group contains 12 mice. The overall survival of mice in BM transplantation assays was analyzed by log-rank (Mantel-Cox) test. (D) Schematic representation of the following experiments. Luciferase positive SKNO-1 cells were injected into sublethally irradiated (2.0 Gy) NOG mice via tail vein, and these mice were treated with twice weekly injections of vehicle or bortezomib. (E-F) Tumor burden was quantified using in vivo bioluminescence imaging. Each group contains 3 mice.
Next, we examined whether bortezomib can suppress AML1/ETO-induced leukemia in vivo. Because retroviral transduction of AML1/ETO alone cannot induce leukemia in mice,41 we used an orthotopic cell line model of luciferase-positive SKNO-1 cells, a human AML cell line that expresses AML1/ETO (Figure 7D).33 We transplanted SKNO-1 cells into sublethally irradiated NOG mice via tail vein and treated these mice with vehicle or bortezomib. Tumor burden was quantified using in vivo bioluminescence imaging. Mice were intraperitoneally injected with 150 mg/kg luciferin and imaged them with an IVIS imaging system 10 minutes after injection. Total body bioluminescence was determined by in vivo bioluminescence imaging (IVIS Lumina2; Caliper Life Sciences) and quantitated using Living Image 2.60 software. In accordance with our results in vitro and in silico (Figure 6E-H), bortezomib significantly inhibited the growth of SKNO-1 cells in vivo (Figure 7E-F). Although bortezomib may have effects besides the NF-κB signaling inhibition, these results indicate that NF-κB signaling plays a critical role in the pathogenesis of myeloid tumors with deregulated AML1 function in vivo.
Discussion
In this study, we found that targeted disruption, as well as leukemia-related gene alteration, of AML1 results in the aberrant activation of NF-κB signaling. To detect immediate targets of AML1, we analyzed the gene expression profiles of LSK cells just after the synchronous inactivation of AML1 using the CreER system, and we captured the immediate alteration of target gene expression that is sometimes hidden by cell population shift or secondary changes in cellular signaling. Although changes in the expression of individual target genes were relatively subtle, we successfully identified NF-κB signaling to be a target subject to immediate regulation of AML1. The in vitro synchronous inactivation system may provide us with a useful tool that can find unidentified target signaling of transcription factors. Furthermore, by determining expression of several genes related to NF-κB signaling, we can select patients who are candidates for NF-κB–targeted therapy. This strategy should be effective especially when genetic mutations are unknown. Previously, Valk et al made unsupervised cluster analyses and identified 16 clusters of patients with AML on the basis of gene expression profiles.28 We reanalyzed their gene expression data in silico and found that NF-κB signaling is highly activated in a cluster previously identified as “cluster 5.” Valk et al found that cluster 5 was associated with poorer prognoses but that specific genetic changes have not been identified in this cluster.28 In addition to t(8;21)–positive leukemia, the clinical activity of NF-κB inhibitors in AML cases belonging to this cluster warrants further investigation.
Although AML1 mutations that lead to functional impairment have been frequently discovered in AML and MDS, it remains very difficult to develop a molecularly targeted therapy against deregulated AML1 function, as is often the case with leukemia-related transcription factors. We discovered that NF-κB signaling is distinctly activated as a consequence of AML1 mutation found in human leukemia. In the current study, bortezomib, a clinically available drug that can inhibit NF-κB signaling, has shown a significant activity against AML1-related leukemia. A wide variety of small molecules that inhibit NF-κB signaling are now being developed and can become attractive candidates for targeted therapy of AML1-related leukemia in the future.
Although the amount of AML1 in the cytoplasm is lower than that in the nucleus, we show that the physical interaction between AML1 and IKK complex mainly occurs in the cytoplasm. In addition, the finding that AML1 inhibits the nuclear translocation of p65 that is tightly regulated by the cytoplasmic IKK complex supports the cytoplasmic function of AML1. Although nuclear exporting signal was not reported in AML1, IKK may export AML1 to the cytoplasmic fraction. We identified both AML1 and AML1/ETO protein in the cytoplasmic fraction of Kasumi-1 cells in supplemental Figure 4B. The amounts of AML1 and AML1/ETO protein in the cytoplasm are fairly small. One possibility is that these proteins translocate between the cytoplasm and the nucleus in a context dependent manner. Another possibility is that AML1/ETO sequesters some components necessary for AML1 to inhibit IKK complex. A function of transcription factors that is exerted in the cytoplasm is well documented in p53.42 Besides acting in the nucleus as a transcription factor to regulate expression of genes involved in apoptosis and cell cycle regulation, p53 triggers apoptosis and inhibits autophagy in the cytoplasm through a variety of processes, including induction of mitochondrial outer membrane permeabilization, direct regulation BAX activity, inhibition of AMP-dependent kinase, and activation of mammalian target of rapamycin. Revealing cytoplasmic functions of transcription factors will provide a new perspective for the therapy against malignancy with deregulated transcription factors.
Our data, however, do not deny the contribution of the transcriptional function of AML1 in inhibiting NF-κB signaling. It may be possible that AML1 attenuates NF-κB signaling transcriptionally and via the inducible IKK complex like Notch1.20
AML1 mutants including AML1/ETO tested in our study fail to inhibit kinase activity of IKKs, although they can physically interact with IKK complex. Therefore, it is likely that additional mechanism exists for wild-type AML1 to repress IKK activity after binding to IKK complex, which should be inactivated in AML1 mutants. One possible mechanism is that AML1 disturbs the formation of IKK complex. Another possibility is that AML1 blocks the interaction between IKK complex with upstream kinase or downstream substrates, such as IκBα or p65. Importantly, critical for the repression of IKK is C-terminal region, a region that may directly inhibit the IKK complex or to which some cofactor may bind. D171N cannot repress IKK activity, although it possesses the intact C-terminal region, and a conformational change caused by the point mutation may impair the inhibitory effect against IKK. Because AML1 mutants in this study can bind to IKK complex but do not attenuate IKK activity, it is assumed that they exert a dominant-negative effect against wild-type AML1.
In normal hematopoiesis, AML1 suppresses NF-κB signaling and thus may contribute to inhibition of excessive proliferation of hematopoietic cells. Aberrant activation of NF-κB signaling may cause expansion of HSCs in AML1 cKO mice. Although it is reported that neither p65 nor a constitutively active form of IKK increases the number of human cord blood cells when transduced in vitro, intact AML1 may inhibit the NF-κB signaling in these cells.43 It also is reported that NF-κB signaling is more activated in leukemic stem cells than normal HSCs.44 In leukemic cells, the NF-κB inhibitory mechanism including AML1 may be disrupted. Once genetic mutation of AML1 occurs in hematopoietic cells, aberrant activation of NF-κB signaling exerts antiapoptotic and proliferation-promoting effects via activation of BCL-XL or JUNB. Proliferative advantage conferred by activated NF-κB signaling may contribute to the clonal evolution of AML1-mutated cells that leads to leukemic transformation. Conversely, it is known that activation of NF-κB signaling causes myeloproliferative disease via the stroma-mediated signaling.45 Thus, germ line mutation of AML1 that occurs in familial platelet disorder patients may contribute to leukemogenesis in those patients not only via a hematopoietic cell-autonomous function but also via niche-derived signaling.7
Both of AML1 and NF-κB play important roles in lymphocyte differentiation.11,46 Well documented is the pathogenetic significance of NF-κB signaling in autoimmune diseases, especially rheumatoid arthritis (RA).47 Disease-associated single-nucleotide polymorphisms in autoimmune diseases are found in AML1 binding sites of several gene promoters: SLC9A3R1 and NAT9 in psoriasis; SLC22A4 in RA; and PDCD1 in systemic lupus erythematosus.48-50 A single-nucleotide polymorphism in AML1 per se also is associated with RA.49 Along with this study, AML1 may contribute to the pathogenesis of autoimmune diseases through the cytoplasmic function to modulate NF-κB activity as well as the nuclear function as a transcription factor.
In summary, our experiments identified direct regulation of NF-κB signaling by AML1. AML1 negatively regulates NF-κB signaling by inhibiting kinase activity of IKK. Furthermore, mutated forms of AML1 found in MDS or AML fail to inhibit NF-κB signaling. NF-κB signaling can be a promising molecular target for the treatment of AML1-related hematologic malignancies.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs D. E. Zhang (University of California–San Diego), T. D. Gilmore (Boston University), S. W. Hiebert (Vanderbilt University), H. Nakauchi (Institute of Medical Science [IMS], University of Tokyo), T. Kitamura (IMS, University of Tokyo), H. Harada (University of Hiroshima), S. Ogawa (University of Tokyo), T. Nosaka (University of Mie), P. Chambon (Institut de Génétique et Biologie Moléculaire et Cellulaire), and Andrew L. Kung (Dana-Farber Cancer Institute); Kyowa Hakko Kirin Co Ltd; and Wakunaga Pharmaceutical Co Ltd for providing essential materials and instruments; and M. Kobayashi, S. Naito, Y. Sawamoto, and Y. Shimamura for expert technical assistance.
This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology (Japan).
Authorship
Contribution: M.N. conceived of and designed the research, performed experiments and analyses, and wrote the paper; M.S., N.W.-O., S.A., A.Y., A.S., N.N., K. Kataoka, and T.S. performed experiments; M.I., K. Kumano, Y.N., and Y.I. wrote the paper; and M.K. supervised the whole project and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mineo Kurokawa, Department of Hematology and Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: kurokawa-tky@umin.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal