Abstract
In human B-acute lymphoblastic leukemia (B-ALL), RAG1-induced genomic alterations are important for disease progression. However, given that biallelic loss of the RAG1 locus is observed in a subset of cases, RAG1's role in the development of B-ALL remains unclear. We chose a p19Arf−/−Rag1−/− mouse model to confirm the previously published results concerning the contribution of CDKN2A (p19ARF /INK4a) and RAG1 copy number alterations in precursor B cells to the initiation and/or progression to B-acute lymphoblastic leukemia (B-ALL). In this murine model, we identified a new, Rag1-independent leukemia-initiating mechanism originating from a Sca1+CD19+ precursor cell population and showed that Notch1 expression accelerates the cells' self-renewal capacity in vitro. In human RAG1-deficient BM, a similar CD34+CD19+ population expressed p19ARF. These findings suggest that combined loss of p19Arf and Rag1 results in B-cell precursor leukemia in mice and may contribute to the progression of precursor B-ALL in humans.
Introduction
Aberrant V(D)J recombination and persistent, nonspecific RAG1 endonuclease activity are reportedly key events in the induction of genomic alterations, which contribute to the oncogenic transformation of hematopoietic precursor cells.1 In contrast, RAG1 deletions have been observed in precursor B-cell leukemia (but not in precursor-T cell leukemia) at a noninsignificant frequency.2 Because of these partially conflicting results the impact of (1) RAG1 misexpression (leading to leukemogenic DNA cleavage) and (2) biallelic copy number losses of RAG1 genes on the initiation or progression of precursor B-ALL remains unclear. There is indirect evidence to suggest that Rag-induced genomic alterations are not essential for leukemia initiation: p53 and nonhomologous-end-joining (NHEJ) factor (such as Ku80, DNA-Pkc, Artemis) double-knockout mice develop aggressive pro-B-cell leukemia with Rag1-dependent IgH/c-myc translocations.3 However, in some cases, B-ALL still occurs when Rag activity is absent and IgH/c-myc translocation is suppressed.4-7 Similarly, in an Eμ–c-myc transgenic mouse model, loss of Rag1 increased (rather than suppressed) the incidence of B-cell lymphoma, although the mechanism has not been clarified.8 Thus, it is likely that both aberrant Rag1 activity and Rag1 deficiency contribute to ALL, albeit with different underlying mechanisms and target cell populations.
To study the role of Rag1 loss of function in the initiation of precursor B-ALL, we developed a murine p19Arf−/−Rag1−/− double-knockout model. We chose to inactivate these particular genes for 2 reasons. First, loss of the RAG1 locus occurs with significant frequency in precursor B-ALL,2 and Rag1 deficiency contributes to leukemogenesis in mouse models only when combined with secondary oncogene activation or tumor suppressor gene inactivation.8,9 Second, p19ARF loss-of-function mutations are involved in a high percentage of human precursor B-cell leukemia (16 of 47 tested cases in Mullighan et al2 ). One possible explanation for the predisposition to precursor B-ALL in the absence of RAG1 is that the Rag-deficiency–induced arrest in B-cell differentiation leads to alterations in the phenotype and frequency of the various lymphoid precursor populations in the BM.10
In the present study, we demonstrated that the combined lack of Rag1 and p19Arf results in the emergence of a new subset of Sca1+CD19+ B-cell precursor cells in the BM. This cell fraction contains leukemia-initiating cells (LICs) that are specifically characterized by abundant Notch1 expression.
Methods
Mice
The p19Arf−/− mice were generously provided by Dr Charles Sherr (St Jude's Hospital, Memphis, TN) and backcrossed with Rag1−/− mice (Charles Rivers Laboratories) to obtain p19Arf−/−Rag1−/− double-deficient mice. Recipient WT mice were obtained from Charles Rivers Laboratories. Rag2−/−γc−/− animals were generously provided by Dr Jim di Santo (Pasteur Institute, Paris, France). Mice expressed CD45.1 (recipient) or CD45.2 (donor) alloantigens for determination of chimerism in transplantation experiments. Animals were housed in a specific, pathogen-free animal facility at Necker Children's Hospital/René Descartes University (Paris, France). All experiments and procedures were performed in compliance with the French Ministry of Agriculture's Regulations for Animal Experiments (Act 87847, October 19, 1987, as modified in May 2001).
Ex vivo expansion of BM cells and transplantation
Before use as donors, mice were closely monitored for preexisting hematologic abnormalities. BM was obtained from femurs and tibiae of 4- to 6-week-old mice, RBC lysis was performed with 0.75% NH4Cl solution in tris(hydroxymethyl)aminomethane buffer, Sca1+ cells were selected by labeling with an anti–Sca1-PE Ab (BD Pharmingen) and subsequent coupling to an anti-PE magnetic selection kit (Miltenyi Biotec).
The Sca1+CD19+ or Sca1+CD19− population was selected by FACS cell sorting. Sca1+CD19− cells were preactivated for 24 hours in plates coated with fibronectin (RetroNectin CH-296; Takara Biomedicals) and Stemspan medium supplemented with 5% FBS, 100 ng/mL murine SCF (mSCF; Abcys), 100 ng/mL murine FMS-like tyrosine kinase 3-ligand (mFlt3-L; R&D Systems), 100 ng/mL thrombopoietin (TPO; R&D Systems), 50 ng/mL mIL-6 (Abcys), and 10 ng/mL mIL-11 (R&D Systems). Cells were mock-cultured or submitted to 3 transduction cycles with an MFG-GFP vector at a multiplicity of infection (MOI) of 11. Construction of the MFG-GFP vector and production of viral supernatant has been described elsewhere.11 A sample of 5 × 105 “A” or “AR” cells was injected IV into 10 Gy-irradiated WT mice. For secondary transplantations, 5 × 106 leukemic cells from 2 different AR mice were transplanted into 6 nonirradiated Rag2−/−γc−/− animals after RBC lysis.
Flow cytometry
Analysis of immunologic reconstitution in AR transplanted mice was performed by retro-orbital bleeding 15 weeks after transplantation. Mice were euthanized at the first signs of illness and their thymus, spleen, and BM were harvested. Flow cytometric analysis was performed on organs and peripheral blood after RBC lysis (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Analysis was performed on a FACSCalibur machine (BD Pharmingen) using Flow Jo software.
Histology
Spleen tissue samples from (1) recipient mice which developed pro-B-cell leukemia, (2) healthy AR mice, and (3) a WT control mouse were harvested and fixed in PBS/10% formalin, embedded in paraffin, and cut into 2-μm-thick slices. Slides were conventionally stained with hematein-eosin-safran reagent and were interpreted independently by 2 pathologists (N.B. and J.B.).
RT-PCR analysis
cDNA was synthesized using the RNeasy Kit (QIAGEN). Primer sequences, annealing temperatures, and fragment size are indicated in supplemental Methods (supplemental Table 2).
Integration site analysis
Genomic DNA was purified using a phenol-chloroform extraction protocol. Integration site analysis was performed as described elsewhere.12 In brief, to determine vector integration sites in the mouse genome, DNA fragments from host-vector junctions were prepared using ligation-mediated PCRs. Each DNA sample (1-1.5 mg) was digested with 5 different enzymes. The digested samples were ligated to linkers and then amplified in nested PCRs. To sequence all the samples in a single experiment, primers containing unique 4-bp barcodes were used in the second PCR step. The PCR products were gel-purified, pooled, and pyrosequenced (454 Life Sciences).
BrdU incorporation assays
To measure cell proliferation, mice were twice injected IP with 1 mg BrdU (Sigma-Aldrich, B5002-1G). Five hours after injection, mice were euthanized and BM was harvested, stained with Sca1+ PE and CD19 APC Abs and fixed in PBS with 0.01% Tween/1% paraformaldehyde (PFA). After a 48-hour incubation, cells were permeabilized in PBS with 0.5% Tween and stained with an anti-BrdU–FITC Ab (BD Biosciences).
Multiplex PCR
Multiplex RT-PCR analysis was performed according to the method published by Peixoto et al,13 with some minor modifications. We considered Mrp-S21 (coding for a protein within the 28S ribosome subunit) to be a constitutive housekeeping gene. The selected PCR primers are listed in supplemental Table 1. Cells were sorted in 96-well plates containing PBS–diethyl pyrocarbonate (Sigma-Aldrich) and stored at −80°C. After cell lysis, RNA was reverse-transcribed using gene-specific 3′ primers. The first-round PCR was subsequently performed on the same plate by addition of a premixed PCR buffer containing 3′ and 5′ primers for all 8 genes and operation of 15 cycles. The first-round PCR products were aliquoted into new PCR tubes (2% per second-round PCR). The second-round PCR was performed separately for each individual gene using semi-nested primers (48 cycles). The PCR products were resolved on a 1.5% agarose ethidium bromide gel.
In vitro cell-culture assays
BM was enriched for the CD19+ population using a MACS magnetic cell sorting kit (Miltenyi Biotec). Subsequently, the Sca1lowckitlowCD19+ population was sorted by FACS (on a FACSAria machine from BD Biosciences). For coculture experiments, sorted cells were cultured on a murine OP-9 stroma cell line expressing (or not) murine Notch-ligand delta1 (OP9-delta1) in α MEM (Invitrogen) supplemented with Hyclone FBS (Perbio), glutamine (Invitrogen), 2 ng/mL mIL7 (R&D Systems), and 5 ng/mL mFLT3L (R&D Systems). The cells were replated every 7 days. At each replating, round cells were washed in PBS and a portion was taken for FACS analysis to establish the expression of B-cell Ags (B220/CD19) or T-cell Ags (CD4/CD8/TCR).
Expression analysis of human RAG1−/− BM cells
After the provision of informed consent, BM from a RAG1-deficient patient and a healthy control was isolated. Mononuclear cells were isolated by Ficoll and CD34+ selection was performed using a human CD34 MicroBeadKit (Miltenyi Biotec). The CD34+CD19+ and CD34+CD19− populations were isolated by FACS and cDNA was synthesized. The TaqMan primer probe set for the human CDKN2A (p19Arf) locus (assay Hs99999189_m1) was purchased from Applied Biosystems.
Statistical analysis
All statistical analyses were performed with GraphPad Prism software (GraphPad Software Inc). Survival was analyzed according to the Kaplan-Meier method and survival curves were compared in a log-rank test.
Results
A Rag1−/− background and loss of p19Arf act synergistically to promote pro-B-cell leukemia
Double-knockout p19Arf−/−Rag1−/− (AR) mice were born in the anticipated Mendelian proportion. As expected, mature B and T cells were totally absent, whereas normal NK monocytes and polynuclear cell counts were observed.
In p19Arf−/− (A) mice, we observed an overall mortality rate of 50% at 50 weeks after birth. This was mainly because of the development of solid tumors, such as sarcomas and carcinomas. Seventeen percent of the A animals developed hematopoietic tumors with a predominantly T-cell phenotype (own data and Kamijo et al14 ). R mice had an overall survival rate of > 90% at 28 weeks after birth.8 We did not consistently observe solid or hematopoietic tumors in R mice after 52 weeks of follow up in own studies.11 The overall mortality rate for AR mice (65% at 50 weeks) did not differ significantly from that seen in A mice. The AR mice also developed predominantly solid tumors. Hematologic lymphoproliferation occurred in 26% of AR animals but (in contrast to A mice) these tumors all had a pro-B-cell phenotype (CD19+IgM−; Figure 1A, Table 1).
Phenotype of pro-B-cell leukemia in AR mice. AR and A mice were kept in a pathogen-free animal facility and were euthanized at the first signs of illness. (A) Kaplan-Meier analysis of overall survival as a percentage of AR mice (◊, n = 19) and A mice (■, n = 17) without any treatment over a 52-week follow-up period postbirth. A log-rank test was used to compare survival in the 2 cohorts (P < .3887). (B) Macroscopic splenomegaly in diseased AR* mice, compared with spleens of healthy, age-matched A, AR, WT, and R mice, as well as a CD19+IgM− cell population as the percentage of total MNCs in the spleen and BM of diseased AR mice (AR*) and age-matched healthy AR mice. (C) Hematein-eosin staining of spleen sections from an AR* mouse, an age-matched healthy AR mouse, and a WT mouse at 100× and 400× magnification. Lymphoblast infiltration (*) and atypical mitoses (**) are present in the AR* spleen sections. Normal, extramedullar hematopoiesis showing erythroblasts and megakaryocytes is present in the healthy AR and WT mice. (D) FACS analysis of BM and spleen from a diseased AR* mouse and healthy, age-matched AR and WT mice gated on the white blood cell (WBC) populations after RBC lysis. Dot plots indicate the percentage of the immature lymphoblastoid CD19+IgM− (AR*), healthy immature CD19+IgM− (AR), and mature CD19+IgM+ (WT) B-cell compartments in the spleen and the immature (CD19+IgM−) populations in the BM of the indicated mice, respectively. (E) Histograms indicate the surface expression (according to an immunofluorescence analysis) of IL7Rα, CD25, BP1, CD24, and B220 gated on the CD19+IgM− population of an AR pro-B-cell leukemia sample from the spleen.
Phenotype of pro-B-cell leukemia in AR mice. AR and A mice were kept in a pathogen-free animal facility and were euthanized at the first signs of illness. (A) Kaplan-Meier analysis of overall survival as a percentage of AR mice (◊, n = 19) and A mice (■, n = 17) without any treatment over a 52-week follow-up period postbirth. A log-rank test was used to compare survival in the 2 cohorts (P < .3887). (B) Macroscopic splenomegaly in diseased AR* mice, compared with spleens of healthy, age-matched A, AR, WT, and R mice, as well as a CD19+IgM− cell population as the percentage of total MNCs in the spleen and BM of diseased AR mice (AR*) and age-matched healthy AR mice. (C) Hematein-eosin staining of spleen sections from an AR* mouse, an age-matched healthy AR mouse, and a WT mouse at 100× and 400× magnification. Lymphoblast infiltration (*) and atypical mitoses (**) are present in the AR* spleen sections. Normal, extramedullar hematopoiesis showing erythroblasts and megakaryocytes is present in the healthy AR and WT mice. (D) FACS analysis of BM and spleen from a diseased AR* mouse and healthy, age-matched AR and WT mice gated on the white blood cell (WBC) populations after RBC lysis. Dot plots indicate the percentage of the immature lymphoblastoid CD19+IgM− (AR*), healthy immature CD19+IgM− (AR), and mature CD19+IgM+ (WT) B-cell compartments in the spleen and the immature (CD19+IgM−) populations in the BM of the indicated mice, respectively. (E) Histograms indicate the surface expression (according to an immunofluorescence analysis) of IL7Rα, CD25, BP1, CD24, and B220 gated on the CD19+IgM− population of an AR pro-B-cell leukemia sample from the spleen.
AR mice were euthanized as soon as signs of illness were observed. Monomorphic, immature (CD19+IgM−) lymphoblastic proliferation was observed in the BM, liver, spleen, lymph nodes, and blood (Figure 1B). Hematein-eosin staining of spleen sections revealed widespread large cell proliferation, with the loss of red/white pulp differentiation. Clusters of large cells with an irregularly bounded nucleus, a prominent nucleolus and numerous partially atypical mitoses were observed (Figure 1C). The AR CD19+IgM− population displayed a blastoid phenotype, with surface expression of other B-cell lineage markers (such as BP1, IL7Rα, B220, CD24, and CD25; Figure 1D-E). We did not detect any residual VDJ recombination or IgH-c-myc/IgH–n-myc translocations, emphasizing the complete absence of Rag1 activity (supplemental Figure 1A-B).
The occurrence of B-cell precursor leukemia in the AR animals suggested a cooperative mechanism involving Rag1 deficiency and the loss of p19Arf.
The p19Arf−/−Rag1−/− Sca1+CD19+ population initiates leukemia
The occurrence of pro-B-cell leukemia in this mouse model suggested the existence of a new mechanism capable of inducing Rag1-independent pro-B-cell transformation in hematopoietic progenitors.
To further investigate the Rag1-independent transformation process, we designed a model comprising a 5-day, ex vivo expansion of AR Sca1+ hematopoietic precursor cells followed by transplantation into lethally irradiated wild-type (WT) mice (Figure 2A). Fifteen weeks later, mixed blood chimerism was observed, with a normal count of donor-derived NK cells (NK1.1: 200/μL), monocytes and polynuclear cells (CD11b: 1500/μL) but given the Rag1 deficiency only low numbers of recipient-derived mature B and T cells (CD3+: 350/μL; B220+: 100/μL). As expected, animals transplanted with A cells showed full donor chimerism, with normal cell counts in both the lymphoid and myeloid compartments (Figure 2B). With a time to onset of 20-25 weeks posttransplantation, recipients of AR cells developed hematopoietic tumors with 100% lethality by 45 weeks posttransplantation. Eighty percent of mice transplanted with A cells survived more than 52 weeks (P < .0001). The observed cases of leukemia were predominantly of pro-B-cell origin (90% pro-B, 10% pro-T) in the AR transplanted group and were all of T-cell origin in the A transplanted group (Figure 2C, Table 1). Secondary transplantation of the lymphoblastic AR CD19+IgM− pro-B cells again gave rise to monomorphic infiltration of the spleen, liver, and BM in all recipients 2 weeks after transplantation (supplemental Figure 2A). This finding indicated that this lymphoblastic population had a high self-renewal capacity. In view of a possible contribution of ex vivo culture to the development of leukemia, a cohort of mice was transplanted with non–ex vivo–expanded cells. This cohort showed the same leukemia incidence compared with mice transplanted with ex vivo–expanded cells (supplemental Figure 2B).
Transplantation of the AR Sca1+CD19+ population results in leukemia. (A) Experimental design. HSCs (CD45.2) were cultured ex vivo for 5 days in the presence of mSCF, mTPO, mFLT3L, mIL6, and mIL11 (with or without γ retroviral [MFG-GFP] transduction) and then transplanted into 10 Gy-irradiated WT mice (CD45.1). (B) A blood chimerism analysis of recipient animals 15 weeks after transplantation on the basis of CD45.1 (recipient) and CD45.2 (donor) alloantigen expression. The T-cell (CD3+), B-cell (B220+), NK-cell (NK1.1+), and monocyte (CD11b+) subpopulations were analyzed by immunofluorescence. ■ indicates the WBC count/μL (WBC); and □, the CD45.2+ cell count/μL in the blood. Columns represent the mean values and SD for AR mice (n = 11) and A mice (n = 9). (C) Kaplan-Meier survival curves of WT mice transplanted with A-derived Sca1+ mock-transduced (▵, n = 9) and GFP-transduced cells (▴, n = 9) or AR-derived Sca1+ mock-transduced (□, n = 11) and MFG-GFP vector-transduced cells (■, n = 12). A log-rank test was used to compare the groups (P < .0001). (D) Integration site analysis of pro-B-cell leukemia in the AR-GFP cohort. Gel electrophoresis of PCR-amplified genomic vector integration fragments showed a polyclonal integration pattern for the AR-GFP Sca1+ (Sca1+) pool before transplantation (lanes S1 and S2) and a characteristic, mono-oligoclonal integration pattern found in 7 transduced AR pro-B-cell tumors (AR*) (lanes 1, 4, 5, 9, 12, 13, 1x); M represents the DNA ladder. (E) AR Sca1+CD19− GFP-transduced cells were transplanted into lethally irradiated WT recipients. The Kaplan-Meier curve shows a significantly (*) lower incidence of B-cell leukemia in mice transplanted with the AR Sca1+CD19− population (solid line ■, n = 10) than in animals transplanted with the AR Sca1+ population (dashed line ■, n = 12, P < .0003 in a log rank test) at 52 weeks posttransplantation. (F) Kaplan-Meier survival curves of WT mice transplanted with AR-derived Sca1+CD19− GFP-transduced cells (n = 12), AR-derived Sca1+CD19− Rag1-transduced cells (n = 8).
Transplantation of the AR Sca1+CD19+ population results in leukemia. (A) Experimental design. HSCs (CD45.2) were cultured ex vivo for 5 days in the presence of mSCF, mTPO, mFLT3L, mIL6, and mIL11 (with or without γ retroviral [MFG-GFP] transduction) and then transplanted into 10 Gy-irradiated WT mice (CD45.1). (B) A blood chimerism analysis of recipient animals 15 weeks after transplantation on the basis of CD45.1 (recipient) and CD45.2 (donor) alloantigen expression. The T-cell (CD3+), B-cell (B220+), NK-cell (NK1.1+), and monocyte (CD11b+) subpopulations were analyzed by immunofluorescence. ■ indicates the WBC count/μL (WBC); and □, the CD45.2+ cell count/μL in the blood. Columns represent the mean values and SD for AR mice (n = 11) and A mice (n = 9). (C) Kaplan-Meier survival curves of WT mice transplanted with A-derived Sca1+ mock-transduced (▵, n = 9) and GFP-transduced cells (▴, n = 9) or AR-derived Sca1+ mock-transduced (□, n = 11) and MFG-GFP vector-transduced cells (■, n = 12). A log-rank test was used to compare the groups (P < .0001). (D) Integration site analysis of pro-B-cell leukemia in the AR-GFP cohort. Gel electrophoresis of PCR-amplified genomic vector integration fragments showed a polyclonal integration pattern for the AR-GFP Sca1+ (Sca1+) pool before transplantation (lanes S1 and S2) and a characteristic, mono-oligoclonal integration pattern found in 7 transduced AR pro-B-cell tumors (AR*) (lanes 1, 4, 5, 9, 12, 13, 1x); M represents the DNA ladder. (E) AR Sca1+CD19− GFP-transduced cells were transplanted into lethally irradiated WT recipients. The Kaplan-Meier curve shows a significantly (*) lower incidence of B-cell leukemia in mice transplanted with the AR Sca1+CD19− population (solid line ■, n = 10) than in animals transplanted with the AR Sca1+ population (dashed line ■, n = 12, P < .0003 in a log rank test) at 52 weeks posttransplantation. (F) Kaplan-Meier survival curves of WT mice transplanted with AR-derived Sca1+CD19− GFP-transduced cells (n = 12), AR-derived Sca1+CD19− Rag1-transduced cells (n = 8).
The phenotype of the pro-B-cell leukemia derived from the transplanted AR mice was identical to that seen in nontransplanted AR mice (supplemental Figure 2C-D) and the gene expression profile of the observed leukemia clustered with the gene expression profile of healthy murine hematopoietic populations in Hardy fractions B to C. The absence of a BCR rearrangement was the major criteria to designate the leukemia as pro-B-cell leukemia (supplemental Figure 2E). Because of the lack of a BCR rearrangement, we used γ-retroviral green fluorescent protein (GFP) marking of the stem cells and used integration site tracking as a clonality marker to discriminate a benign polyclonal lymphoproliferation from a monoclonal malignant expansion of lymphoid cells. We transduced AR Sca1+ cells with an MFG-GFP–expressing vector before transplantation and observed mono-oligoclonal dominance in the pro-B-cell leukemia. Integration site mapping revealed 1-3 integration sites (Figure 2D). We observed an integration-pattern characteristic of γ-retroviral vectors (supplemental Figure 3A).12 The leukemia incidence of a cohort of mice transplanted with transduced cells compared with a cohort of mice transplanted with nontransduced cells revealed no difference. Thus, we conclude that the proviral integration does not play a major role in the overall incidence of leukemic events (Figure 2C).
In Rag1-positive human and mouse models of B-ALL, genomic alterations have been shown to be associated with disease progression.1,15 To assess genomic rearrangements in our present model, we performed comparative genomic hybridization (CGH) analysis of 10 AR pro-B-cell leukemias. Chromosome 14 gains were particularly frequent, compared with cases of B-cell leukemia in Rag1-sufficient models. As expected, Rag1-mediated DNA copy number alterations were absent (supplemental Figure 3B).
In light of (1) the leukemic cells' self-renewal capacity, (2) the lack of genomic alterations, and (3) the fact that the AR Sca1+ donor cell population contained 30%-40% of CD19+ cells, we decided to investigate the role of a B cell–committed, LIC population in healthy AR BM. To this end, the AR Sca1+ donor population was depleted of Sca1+CD19+ cells by cell sorting (Sca1+CD19−). This was followed by ex vivo expansion and transplantation into lethally irradiated WT mice. The incidence of pro-B-cell leukemia was significantly lower in the recipient mice (60% at 50 weeks posttransplantation, n = 10) than in a group of animals transplanted with a nondepleted AR Sca1+ cell population (100% at 45 weeks posttransplantation; P < .0003; Figure 2E, Table 1). Transduction of the AR Sca1+CD19− population with a Rag1-γ retroviral vector to complement the Rag1 deficiency led to an increase in leukemia frequency and at the same time to a switch in leukemia phenotype from B- to T-cell leukemia. These results indicate that deficiency in p19Arf and Rag1 synergize in the development of pro-B-cell leukemia in this model (Figure 2F).
Conversely, when we directly transplanted the AR Sca1+CD19+ population (without prior ex vivo expansion) into lethally irradiated WT mice, these cells could be tracked in the BM of recipient mice for > 4 months (supplemental Figure 4A,C). Furthermore, the fact that pro-B-cell leukemia developed 1 year after transplantation into sublethally irradiated WT recipients indicates that the AR Sca1+CD19+ cell population has a competitive advantage over WT cells (supplemental Figure 4B). No donor-derived NK cells (NK1.1), monocytes (CD11b+), and polynuclear cells (Gr1+) were found in the blood of AR Sca1+CD19+ transplanted animals and no donor-derived T-cell precursors were detected in the thymus; these findings clearly show that this population is B-cell lineage-committed and unable to reconstitute the different hematopoietic compartments after transplantation.
p19Arf regulates the cell cycle and apoptosis in the AR Sca1+CD19+ population
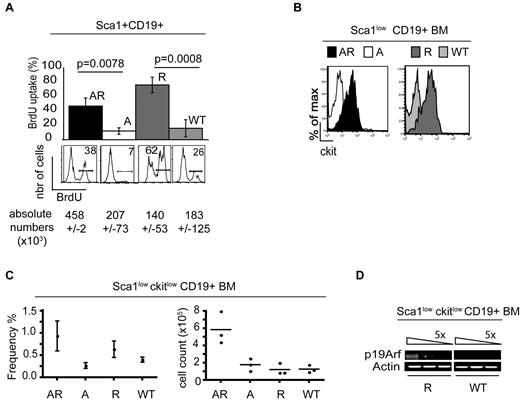
The AR Sca1+CD19+ population displayed a high proliferative capacity, as evidenced by BrdU incorporation assays. In AR BM, 45% of the Sca1+CD19+ cells incorporated BrdU, whereas BrdU incorporation rate for the same cell compartment in WT and A mice was only 10%. Strikingly, an 80% BrdU incorporation rate was seen in the Rag1-deficient Sca1+CD19+ cells (P < .008 in an unpaired t test of AR vs A and P < .0008 for R vs WT). However, loss of p19Arf on a Rag1−/− background led to a 3-fold increase in the absolute Sca1+CD19+BrdU+ cell count—probably as a result of impaired cell-cycle regulation and the lack of apoptosis induction in the AR Sca1+CD19+ population (Figure 3A).
Role of p19Arf in the Rag1-deficient Sca1+CD19+ population. (A) BrdU incorporation is shown both as a percentage of the Sca1+CD19+ population and as a cell count (×103; Sca1+CD19+BrdU+). The FACS histograms show the mean level of BrdU incorporation in percent by the Sca1+CD19+ population (n = 3). (B) The histograms indicate ckit expression gated on the Sca1+CD19+ population in AR, A, R, and WT BM in an immunofluorescence analysis. One of 4 representative experiments is shown here. (C) Frequency (%) and absolute cell numbers (×105) of Sca1lowckitlow CD19+ cells in AR, A, R, and WT BM (P < .037 for AR vs A; n = 4) is indicated. (D) cDNA was synthesized from BM-derived R and WT flow-sorted Sca1lowckitlowCD19+ cells. After 5-fold dilution of the cDNA, RT-PCR assays were used to analyze expression of p19Arf and the control housekeeping gene actin. One of 3 representative experiments is shown here.
Role of p19Arf in the Rag1-deficient Sca1+CD19+ population. (A) BrdU incorporation is shown both as a percentage of the Sca1+CD19+ population and as a cell count (×103; Sca1+CD19+BrdU+). The FACS histograms show the mean level of BrdU incorporation in percent by the Sca1+CD19+ population (n = 3). (B) The histograms indicate ckit expression gated on the Sca1+CD19+ population in AR, A, R, and WT BM in an immunofluorescence analysis. One of 4 representative experiments is shown here. (C) Frequency (%) and absolute cell numbers (×105) of Sca1lowckitlow CD19+ cells in AR, A, R, and WT BM (P < .037 for AR vs A; n = 4) is indicated. (D) cDNA was synthesized from BM-derived R and WT flow-sorted Sca1lowckitlowCD19+ cells. After 5-fold dilution of the cDNA, RT-PCR assays were used to analyze expression of p19Arf and the control housekeeping gene actin. One of 3 representative experiments is shown here.
In line with the high cell-cycle activity observed in the AR and R BM populations, we detected coexpression of ckit in the Sca1+CD19+ population in both AR and R mice (Sca1lowckitlowCD19+) but not in the majority of the corresponding A and WT animals (Figure 3B). The Sca1lowckitlowCD19+ cells accounted for 0.9% and 0.6% of the mononuclear cells in AR and R BM samples, respectively. An absolute 5-fold increase in cell number of the Sca1lowckitlowCD19+ population was only found in the BM of AR mice, compared with A, R, and WT mice (AR vs A; P = .037; Figure 3C). Other B-cell lineage Ags (such as B220, CD24, CD25, and BP1) were expressed at lower levels in the AR and R population than in the corresponding A and WT populations; this suggests that the Sca1lowckitlowCD19+ cell population corresponds to an early lymphoid precursor cell, even though CD19 expression determines B-cell lineage commitment.
Despite the phenotypic and proliferative similarities of the AR and R populations, an absolute increase in cell number and leukemia development was only observed in AR mice. In a RT-PCR analysis, we detected p19Arf mRNA expression in the cell-sorted Rag1-deficient Sca1lowckitlowCD19+ population but not in the WT population, suggesting that p19Arf has a key role in apoptosis and cell-cycle regulation in this specific subpopulation (Figure 3D).
Activation of Notch1 signaling in the AR Sca1lowcKitlowCD19+ population
To further define the developmental stage of the Sca1lowckitlowCD19+ population from AR mice, a comparative, single-cell, multiplex PCR analysis of FACS-sorted Sca1lowckitlowCD19+ cells was performed in AR, A, R, and WT mice. Important transcription factors for B-cell commitment or cytokine receptors were found to be expressed with similar frequencies in the AR Sca1lowckitlowCD19+ population (Il7ra: 85%; E2a: 88%) and the corresponding R population (Il7ra: 72%; E2a: 82%). The expression frequencies were lower in the corresponding A (Il7ra: 52%; E2a: 72%) and WT populations (Il7ra: 40%; E2a: 67%). However, Pu1 was expressed to a greater extent in AR and R cells (50% and 69%, respectively) than in A and WT cells (24% and 20%, respectively). Notch1 was found to be expressed with frequencies of 64%, 49%, 18%, and 17% in the Sca1lowckitlowCD19+ cells from AR, R, A, and WT mice, respectively. These values indicate that AR and R cells harbor the characteristics of earlier progenitors with a broader differentiation capacity. In contrast, other early hematopoietic lineage transcription factors (such as Lmo2 or Gata1) were expressed at similar frequencies in all analyzed cells. The myeloid lineage-specific transcription factor Gmcsfr and the T lymphoid–specific factor Gata3 were expressed in < 5% of all tested cells (Figure 4A).
Transcriptional profiles and the role of Notch–Notch-ligand interaction in the AR and R populations. The AR Sca1lowckitlow CD19+ population was flow-sorted from AR, A, R, and WT BM. (A) Between 30 and 40 single cells (60 for R) from the Sca1lowckitlowCD19+ population from AR (■), A (□), R ( ), and WT (
), and WT ( ) BM were sorted into 96-well plates. Single-cell RT-PCR analysis was performed for the early hematopoietic genes Lmo2 and Gata1, the B-cell lineage genes Pu1, Il7rα, and E2a, the cell-cycle genes Runx1 and c-myB and the gene for the T-cell transcription factor Notch1. The figure illustrates positive gene expression in terms of the percentage of analyzed cells. Mrp-S21 served as an internal control, PBS-loaded wells with no cells served as a negative control and both controls were run for all primer sets. The data shown come from 2 independent cell preparations and RT-PCR experiments. An asterisk (*) indicates a statistically significant difference. (B) RT-PCR of the prototypic Notch1 target gene Hes1, using equal amounts of cDNA in 2 pro-B-cell tumors AR*, in the healthy FACS-sorted Sca1lowckitlowCD19+ AR, A, R, and WT populations and in the thymus of a WT mouse (T). One of 3 experiments (with 2 independent cell preparations) is shown. (C) Sca1lowckitlowCD19+ populations from AR, A, and WT mice were cocultured on an OP-9 stroma cell line expressing murine Notch-ligand delta1 (OPS-9-Delta1) in the presence of mFlT3L and mIL7. Cells were replated every week for > 40 days. Shown is 1 representative experiment of 2. (D) The histograms show the CD19 expression at days 8 (D8) and 33 (D33). One of 3 representative experiments is shown.
) BM were sorted into 96-well plates. Single-cell RT-PCR analysis was performed for the early hematopoietic genes Lmo2 and Gata1, the B-cell lineage genes Pu1, Il7rα, and E2a, the cell-cycle genes Runx1 and c-myB and the gene for the T-cell transcription factor Notch1. The figure illustrates positive gene expression in terms of the percentage of analyzed cells. Mrp-S21 served as an internal control, PBS-loaded wells with no cells served as a negative control and both controls were run for all primer sets. The data shown come from 2 independent cell preparations and RT-PCR experiments. An asterisk (*) indicates a statistically significant difference. (B) RT-PCR of the prototypic Notch1 target gene Hes1, using equal amounts of cDNA in 2 pro-B-cell tumors AR*, in the healthy FACS-sorted Sca1lowckitlowCD19+ AR, A, R, and WT populations and in the thymus of a WT mouse (T). One of 3 experiments (with 2 independent cell preparations) is shown. (C) Sca1lowckitlowCD19+ populations from AR, A, and WT mice were cocultured on an OP-9 stroma cell line expressing murine Notch-ligand delta1 (OPS-9-Delta1) in the presence of mFlT3L and mIL7. Cells were replated every week for > 40 days. Shown is 1 representative experiment of 2. (D) The histograms show the CD19 expression at days 8 (D8) and 33 (D33). One of 3 representative experiments is shown.
Transcriptional profiles and the role of Notch–Notch-ligand interaction in the AR and R populations. The AR Sca1lowckitlow CD19+ population was flow-sorted from AR, A, R, and WT BM. (A) Between 30 and 40 single cells (60 for R) from the Sca1lowckitlowCD19+ population from AR (■), A (□), R ( ), and WT (
), and WT ( ) BM were sorted into 96-well plates. Single-cell RT-PCR analysis was performed for the early hematopoietic genes Lmo2 and Gata1, the B-cell lineage genes Pu1, Il7rα, and E2a, the cell-cycle genes Runx1 and c-myB and the gene for the T-cell transcription factor Notch1. The figure illustrates positive gene expression in terms of the percentage of analyzed cells. Mrp-S21 served as an internal control, PBS-loaded wells with no cells served as a negative control and both controls were run for all primer sets. The data shown come from 2 independent cell preparations and RT-PCR experiments. An asterisk (*) indicates a statistically significant difference. (B) RT-PCR of the prototypic Notch1 target gene Hes1, using equal amounts of cDNA in 2 pro-B-cell tumors AR*, in the healthy FACS-sorted Sca1lowckitlowCD19+ AR, A, R, and WT populations and in the thymus of a WT mouse (T). One of 3 experiments (with 2 independent cell preparations) is shown. (C) Sca1lowckitlowCD19+ populations from AR, A, and WT mice were cocultured on an OP-9 stroma cell line expressing murine Notch-ligand delta1 (OPS-9-Delta1) in the presence of mFlT3L and mIL7. Cells were replated every week for > 40 days. Shown is 1 representative experiment of 2. (D) The histograms show the CD19 expression at days 8 (D8) and 33 (D33). One of 3 representative experiments is shown.
) BM were sorted into 96-well plates. Single-cell RT-PCR analysis was performed for the early hematopoietic genes Lmo2 and Gata1, the B-cell lineage genes Pu1, Il7rα, and E2a, the cell-cycle genes Runx1 and c-myB and the gene for the T-cell transcription factor Notch1. The figure illustrates positive gene expression in terms of the percentage of analyzed cells. Mrp-S21 served as an internal control, PBS-loaded wells with no cells served as a negative control and both controls were run for all primer sets. The data shown come from 2 independent cell preparations and RT-PCR experiments. An asterisk (*) indicates a statistically significant difference. (B) RT-PCR of the prototypic Notch1 target gene Hes1, using equal amounts of cDNA in 2 pro-B-cell tumors AR*, in the healthy FACS-sorted Sca1lowckitlowCD19+ AR, A, R, and WT populations and in the thymus of a WT mouse (T). One of 3 experiments (with 2 independent cell preparations) is shown. (C) Sca1lowckitlowCD19+ populations from AR, A, and WT mice were cocultured on an OP-9 stroma cell line expressing murine Notch-ligand delta1 (OPS-9-Delta1) in the presence of mFlT3L and mIL7. Cells were replated every week for > 40 days. Shown is 1 representative experiment of 2. (D) The histograms show the CD19 expression at days 8 (D8) and 33 (D33). One of 3 representative experiments is shown.
The high frequency of Notch1 detection in the Rag1-deficient Sca1lowckitlowCD19+ population also suggested early hematopoietic characteristics and prompted us to test activation of the Notch1 signaling pathway in this population. An RT-PCR analysis revealed marked Hes1 transcription in the pro-B-cell tumors (AR*) and the AR Sca1lowckitlowCD19+ population but to a lesser extent in the A, R, and WT populations; this clearly indicates the transcription of Notch1 downstream target genes (Figure 4B). We next decided to study the population's Notch1-Notch-ligand–mediated differentiation capacity by performing cocultures on a stroma cell line expressing Notch-ligand δ-1 (OP-9-δ-1). AR cells expanded and expressed CD19+ for more than 50 days when cocultured on an OP-9-delta1–expressing stroma cell line, whereas CD19-expressing cells derived from A and WT populations were not detectable 10-14 days after the start of coculture (Figure 4C-D). To determine whether the observed proliferative effect was really because of activation of Notch1-Notch-ligand signaling pathway, or rather to cytokine supplementation, cells were plated on OP-9 or OP-9-Delta1 cell-lines supplemented with the same cytokines (mFlt3L and mIL7). For the AR population, a higher proliferation index was observed when the cells were plated on OP-9-Delta1, compared with OP-9 (supplemental Figure 5B). Additionally in the presence of a Notch1 signaling pathway inhibitor (γ-secretase inhibitor) the proliferative effect of the AR population on OP-9-Delta1 was diminished, indicating the importance of active Notch1 signaling for the self-renewal capacity of the cells. (supplemental Figure 5A). It is noteworthy that when AR cells were cultured on OP-9-Delta1 stroma, the cells did not generate any DN3 T-cell precursors. Thus, activation of the Notch1 signaling pathway induces proliferation and not differentiation in this population. This particular AR subpopulation formed morphologically B cell–like colonies in semi-solid medium and the colonies could be replated for up to 120 days (compared with 40 and 20 days for A and WT populations, respectively; data not shown).
The fact that (1) Hes1 was highly expressed in the AR population, (2) Notch1-Notch-ligand interaction led to increased proliferation, and (3) coculture on OP-9-Delta1 stroma did not lead to down-regulation of CD19 and apoptosis induction, indicates that Notch1-Notch-ligand interaction mediates a proliferative signal and not a differentiation signal in the AR population. This effect thus contributes to the cells' extended self-renewal capacity.
CD34+CD19+ population with high p19ARF expression in the human RAG1 deficiency
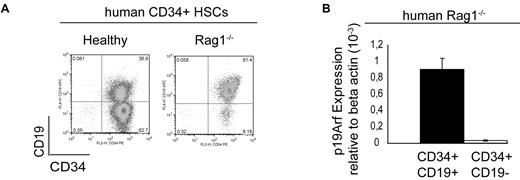
We next sought to identify a corresponding population in the BM of RAG1−/− patients and so harvested mononuclear cells (MNCs) from the BM of RAG1-deficient patients. We observed the accumulation of a CD34+CD19+ population in these patients, compared with MNCs harvested from BM of a healthy individual (Figure 5A). Quantitative RT-PCR using a commercially available p19Arf probe revealed 26-fold higher expression of p19ARF in the RAG1−/− CD34+CD19+ compartment than in the CD34+CD19− compartment; this finding suggests that p19ARF expression also has an important role in this human RAG1−/− CD34+CD19− population (Figure 5B).
p19ARF expression in the human Rag1−/− CD34+CD19+ population. (A) FACS analysis of a Rag1-deficient patient after CD34+ cell sorting of BM-derived MNCs (1 of 6 representative experiments is shown here). (B) TaqMan analysis of p19ARF expression in the cell-sorted human RAG1−/− CD34+CD19+ and CD34+CD19− population indicated 26-fold higher p19ARF expression in the CD34+CD19+ population than in the CD34+CD19− counterpart. One of 2 independent TaqMan analysis of a patient sample is shown here.
p19ARF expression in the human Rag1−/− CD34+CD19+ population. (A) FACS analysis of a Rag1-deficient patient after CD34+ cell sorting of BM-derived MNCs (1 of 6 representative experiments is shown here). (B) TaqMan analysis of p19ARF expression in the cell-sorted human RAG1−/− CD34+CD19+ and CD34+CD19− population indicated 26-fold higher p19ARF expression in the CD34+CD19+ population than in the CD34+CD19− counterpart. One of 2 independent TaqMan analysis of a patient sample is shown here.
Discussion
Persistent Rag1 activity and the resulting genomic alterations have been frequently described and are thought to be part of an important mechanism in the acquisition of genomic alterations which contribute to the development of murine and human B-ALL.1,2 However, there is still debate as to whether the decisive trigger for transformation of hematopoietic precursors into B-ALL is the type and frequency of Rag1-induced genomic alterations or the nature of the target population itself. Thus, the phenotype of BCR-ABL–positive leukemia depends not only on expression of the fusion protein itself but also on the molecular characteristics of the target hematopoietic precursor population.15-17
Here, we described a new Rag1-independent B-ALL initiation mechanism by using a p19ARF−/−RAG1−/− AR double-knockout mouse model. The hematopoietic tumors occurring in AR mice were exclusively cases of pro-B-cell leukemia, which contrasted with the situation in A mice (T-cell leukemia only) and R mice (no tumors).8,11 Transplantation experiments identified a very rare AR Sca1+CD19+ population as the LICs. The population's surface Ag expression pattern as well as the populations transcription profile (as measured in single-cell, multiplex PCR assays) is characteristic of both the B-lineage committed cells (such as Il7ra and E2a) and the early lymphoid progenitor (ELP) to common lymphoid progenitor (CLP) transition (such as Pu1 and Notch1)18,19 This is consistent with earlier findings indicating that lymphoid LICs do not follow the same cellular hierarchy as their physiologic counterparts.20
Activation of the Notch1 signaling pathway inhibits B-cell differentiation and down-regulates CD19 during lymphoid precursor differentiation as we demonstrated for the A and WT populations and as previously described.19,21-23 Expression of Notch1 on the AR Sca1lowckitlowCD19+ population led to activation of the Notch1 signaling pathway in the AR cells, as shown by (1) the expression of the prototypic Notch1 target gene Hes124 and (2) their extended self-renewal capacity when cocultured with OP-9-Delta1 stroma cells but not on OP-9 stromal cells as well as their diminished self-renewal capacity when a Notch1 signaling inhibitor is added to culture, despite the continued expression of CD19 Ag. It is noteworthy that this AR population was B lineage–restricted in transplantation experiments and was thus unable to reconstitute other hematopoietic cell lineages. Nevertheless, the population was able to engraft recipients long-term as shown in transplantation experiments and this is consistent with results obtained from repopulating assays on methocult. Moreover, a new study has recently demonstrated the importance of Notch1 expression and activation of the Notch1-Notch ligand-signaling pathway for the self-renewal capacity and for the long-term engraftment of HSCs. The engraftment is dependent on the expression of specific homing molecules such as VE-cadherin, VEGFR2, and Notch-Notch ligand interaction.25 Taken as a whole, our results support a role of Notch-Notch ligand interaction for long-term engraftment and extended repopulating capacity of a preleukemic AR Sca1lowckitlow CD19+ population. However, it is still of debate whether activation of Notch1 signaling pathway in an outgrown B-ALL has a different effect. Kannan et al recently reported a suppressive effect of Notch1 activation using a cell-line system of B-ALL. This can be indicative of Notch1 providing different effects at the time point of initiation and maintenance of leukemia. Further experiments will be necessary to declare the role of Notch1 at different stages of leukemia development26
The corresponding Sca1lowckitlowCD19+Rag1-deficient B-precursor population expressed p19Arf. Signer et al have described the expression of p19Arf in B-precursor cells at the CLP to pre-B-cell stage in young WT mice.27 Our present results suggest that Rag1−/− early B-cell precursors are at a developmental stage in which precise control of the p19Arf-MDM2-p53 axis is needed to regulate the cell cycle and apoptosis because loss of p19Arf in a Rag1-deficient B-cell precursor subset is enough to drive B-ALL development at a high frequency. Additionally, the shift in hematopoietic cell populations in the BM with the accumulation of a CD19+-precursor population is an explanation for the switch in leukemia phenotype from T in A mice to pro-B-cell leukemia in AR mice. The fact that γ-retroviral correction of Rag1 deficiency in the AR Sca1+CD19− population led to a decrease in the incidence of pro-B-cell leukemia supports the presence of synergy between the loss of p19Arf and loss of Rag1 in this preleukemic subpopulation. It will be interesting to confirm the strength of this pathway in a Rag1−/− mouse model with conditional, B cell–specific loss of p19Arf.
Translation of the Sca1lowckitlowCD19+ population to the human hematopoietic system is difficult because the Sca1+ Ag does not exist in the human stem cell nomenclature. However, CD34+ is expressed on human stem cells and lymphoid precursor cells in a similar way to the Sca1 Ag in the murine HSC compartment. Interestingly, we found that a CD34+CD19+ population accumulated in Rag1-deficient patients. This population was previously described as coexpressing CD36, the pan-B-cell markers CD22 and CyCD79a—none of which have been detected in healthy individuals.10 We demonstrated higher levels of p19ARF expression in this human RAG1−/− CD34+CD19+ cell subset than in the less mature CD34+CD19− population, indicating that p19ARF also plays an important role in the cell-cycle control in the human RAG1−/− CD34+CD19+ population. The functional characteristics of this human precursor have not yet been studied and a predisposition to leukemia (even years after allogeneic stem cell transplantation) in Rag1-deficient patients has not been reported. However, the data from our new murine model parallels the hypothesis put forward by Greaves et al, whereby a CD34+CD38−CD19+ population that is absent from healthy individuals can be detected in patients with TEL/AML1 positive B-cell precursor leukemia.28,29 In addition, genomic alterations involving losses of the Rag1 locus were described in 16% of a cohort of 50 TEL/AML1-positive patients.30 In contrast mutations in the RAG1 locus in T-cell leukemia were absent. These RAG1 mutations are often monoallelic on diagnosis and become biallelic at relapse—indicating a possible selective advantage of RAG1-deficient cells in the progression of B-ALL.2
Further opportunities will involve characterizing NOTCH1 signaling in the human RAG1−/− CD34+CD19+ population by knock-down of p19ARF expression and transplantation into a NOD-SCID xenograft model. This will shed light on the role of NOTCH1 in the process of B-ALL initiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Brian Sorrentino and Albert Zhou for valuable discussions on murine p19Arf models; Dr Isabelle Andre-Schmutz, Dr Chantal Lagresle-Peyrou, Eva-Maria Weick, and Julie Riviere for helpful discussions and advice; Dr Charles Sherr (St Jude's Hospital, Memphis TN) for providing p19Arf−/− mice, and Dr Jim di Santo (Pasteur Institute, Paris, France) for providing Rag2−/−γC−/− mice. We also thank Amelyne David and Julie Piquet for animal work.
J.H. was funded by a fellowship from the Akademie der Naturforscher Leopoldina (BMBF-LPD 9901/8-149) and the “Strategischer Forschungsfond” of the Heinrich-Heine-Unversity Düsseldorf (Düsseldorf, Germany). M.C.-C. was funded by grants from Inserm, the Consert contract 005242, and Agence Nationale de la Recherche (ANR) grant 05-MRAR-004. F.D.B. was supported by National Institutes of Health (NIH) grants AI52845 and AI66290. G.W. was funded by NIH National Institute of Allergy and Infectious Diseases (NIAID) T32 AI07634 (Training Grant in Infectious Diseases) and the University of Pennsylvania School of Medicine Department of Medicine Measey Basic Science Fellowship Award.
National Institutes of Health
Authorship
Contribution: J.H. and E.M. designed the p19Arf−/−Rag1−/−mouse model, performed transplantation experiments, and analyzed the tumors; C.M. performed CGH-analysis and Affymetrix expression analysis of mouse tumors; J.H. and A.B. performed Multiplex-PCR analysis; M.L. and S.R. performed FISH analysis; J.H., G.W., and F.D.B. performed integration site analysis; J.B. and N.B. performed histology; D.T. performed expression analysis; S.K. contributed with scientific advice and discussions; and J.H., A.B., A.F., S.H.-B.-A., and M.C.-C. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marina Cavazzana-Calvo, Inserm U768 and Biotherapy Clinical Investigation Center, Hôpital Necker, rue de Sevres 149, F-75015 Paris, France; e-mail: sec.biotherapie@nck.aphp.fr or m.cavazzana@nck.aphp.fr.
References
Author notes
S.H.-B.-A. and M.C.-C. contributed equally to this work.


![Figure 2. Transplantation of the AR Sca1+CD19+ population results in leukemia. (A) Experimental design. HSCs (CD45.2) were cultured ex vivo for 5 days in the presence of mSCF, mTPO, mFLT3L, mIL6, and mIL11 (with or without γ retroviral [MFG-GFP] transduction) and then transplanted into 10 Gy-irradiated WT mice (CD45.1). (B) A blood chimerism analysis of recipient animals 15 weeks after transplantation on the basis of CD45.1 (recipient) and CD45.2 (donor) alloantigen expression. The T-cell (CD3+), B-cell (B220+), NK-cell (NK1.1+), and monocyte (CD11b+) subpopulations were analyzed by immunofluorescence. ■ indicates the WBC count/μL (WBC); and □, the CD45.2+ cell count/μL in the blood. Columns represent the mean values and SD for AR mice (n = 11) and A mice (n = 9). (C) Kaplan-Meier survival curves of WT mice transplanted with A-derived Sca1+ mock-transduced (▵, n = 9) and GFP-transduced cells (▴, n = 9) or AR-derived Sca1+ mock-transduced (□, n = 11) and MFG-GFP vector-transduced cells (■, n = 12). A log-rank test was used to compare the groups (P < .0001). (D) Integration site analysis of pro-B-cell leukemia in the AR-GFP cohort. Gel electrophoresis of PCR-amplified genomic vector integration fragments showed a polyclonal integration pattern for the AR-GFP Sca1+ (Sca1+) pool before transplantation (lanes S1 and S2) and a characteristic, mono-oligoclonal integration pattern found in 7 transduced AR pro-B-cell tumors (AR*) (lanes 1, 4, 5, 9, 12, 13, 1x); M represents the DNA ladder. (E) AR Sca1+CD19− GFP-transduced cells were transplanted into lethally irradiated WT recipients. The Kaplan-Meier curve shows a significantly (*) lower incidence of B-cell leukemia in mice transplanted with the AR Sca1+CD19− population (solid line ■, n = 10) than in animals transplanted with the AR Sca1+ population (dashed line ■, n = 12, P < .0003 in a log rank test) at 52 weeks posttransplantation. (F) Kaplan-Meier survival curves of WT mice transplanted with AR-derived Sca1+CD19− GFP-transduced cells (n = 12), AR-derived Sca1+CD19− Rag1-transduced cells (n = 8).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/3/10.1182_blood-2010-09-305383/4/m_zh89991174680002.jpeg?Expires=1763596525&Signature=w3B~Z6wzdPEq2gRJTCe2~VOnURqjDbp2wR7n2Km28QJccuN6oXlCiCqa2diwIjNIEboaAoBkcvD3YlDVQyO1~FESEt~FjVM2XHSSg~JzDGQSQIqSF7ukfyQwcnCeno5NNtlJRf8EA84AzD7x2et9g1MDi2YUyBUpxezys8r2TBIgawxTvaNK9JYsc~PxS6JIZT7P06EMSBWbq68UJ8bLCcby2kxu7voGWjJQGtyekI8gwL4Ou1zSples9pAT3YWfS-cC4oVe9uy5lZ2xMOOt~z3wgAlG4~A857lOixCCkmZ7gECcAbA5Eg~lQXzuydx4i7mslUWjyqQ3pwSALjmw9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)