Abstract
Mutations in the uroporphyrinogen III synthase (UROS) gene cause congenital erythropoietic porphyria (CEP), an autosomal-recessive inborn error of erythroid heme biosynthesis. Clinical features of CEP include dermatologic and hematologic abnormalities of variable severity. The discovery of a new type of erythroid porphyria, X-linked dominant protoporphyria (XLDPP), which results from increased activity of 5-aminolevulinate synthase 2 (ALAS2), the rate-controlling enzyme of erythroid heme synthesis, led us to hypothesize that the CEP phenotype may be modulated by sequence variations in the ALAS2 gene. We genotyped ALAS2 in 4 unrelated CEP patients exhibiting the same C73R/P248Q UROS genotype. The most severe of the CEP patients, a young girl, proved to be heterozygous for a novel ALAS2 mutation: c.1757 A > T in exon 11. This mutation is predicted to affect the highly conserved and penultimate C-terminal amino acid of ALAS2 (Y586). The rate of 5-aminolevulinate release from Y586F was significantly increased over that of wild-type ALAS2. The contribution of the ALAS2 gain-of-function mutation to the CEP phenotype underscores the importance of modifier genes underlying CEP. We propose that ALAS2 gene mutations should be considered not only as causative of X-linked sideroblastic anemia (XLSA) and XLDPP but may also modulate gene function in other erythropoietic disorders.
Introduction
Congenital erythropoietic porphyria (CEP, MIM #263700), also known as Günther disease, is a rare inborn error of metabolism resulting from a partial deficiency in uroporphyrinogen III synthase (UROS; EC 4.2.1.75), the fourth enzyme of the heme biosynthetic pathway.1,2 UROS catalyzes the rapid cyclization of the linear tetrapyrrole hydroxymethylbilane (HMB) into the asymmetric macrocycle uroporphyrinogen III (URO III).
CEP is transmitted as an autosomal-recessive trait as the result of mutations in the UROS gene. According to the Human Gene Mutation Database (Cardiff University) and updates,3-5 43 mutations, spread along the 10 exons of the UROS gene, have been reported, including missense or nonsense mutations, splicing defects, and large deletions/insertions. Mutations in the erythroid-specific promoter region have also been described.3 A common missense mutation in exon 4, p.C73R, is found in approximately 30% of disease alleles found in CEP patients. Nevertheless most of the other mutations have been reported in only one or a very small number of cases.4
Because of enzyme deficiency, affected homozygotes or compound heterozygotes manifest excessive nonenzymatic conversion of HMB to uroporphyrinogen I, some of which is converted to coproporphyrinogen I. These isomer I porphyrinogens cannot be converted to heme and accumulate in erythrocytes, undergo oxidation to uroporphyrin (URO I) and coproporphyrin (COPRO I), and induce hemolysis.5 The released porphyrins are distributed throughout the body, with substantial amounts being deposited in skin and bones. URO I is a highly photo-catalytic molecule. Therefore, exposure of the skin to sunlight results in blistering, vesicle formation, and in some cases leads to severe scarring and deformities, predominantly of the face and hands.1,2 Porphyrinuria is usually present from the first months of life, with typical red-wine urine present in the patient. Other typical features of CEP include splenomegaly and erythrodontia.6
However, the clinical manifestations of CEP vary greatly and range from hydrops fetalis, which occurs as the result of severe hemolytic anemia in utero, to benign forms with only mild cutaneous involvement. Clinical profiles of patients with CEP can be classified into 3 classes, as proposed by Desnick and Astrin1 : (1) Severe, birth-onset forms responsible for fetal ascites, transfusion dependency, and mutilating skin lesions; (2) moderate forms, including latent chronic hemolysis, and limited cutaneous involvement; and (3) mild and late-onset forms mainly restricted to cutaneous symptoms or associated with late-onset thrombocytopenia.
The degree of impairment of UROS activity with the concomitant reduction in URO III levels and increased accumulation of photoreactive URO I and derived porphyrins may correlate with the severity of the disease. Homoallelism for the C73R mutation was found to cause significant decreases in erythrocyte UROS activity and very low residual-specific activity of the purified mutated enzyme produced in Escherichia coli.7,8 This mutation has been found in the most severe cases of CEP, whereas several patients with compound heterozygosity have shown mild-to-moderate phenotypes depending on allele combinations.9 However, striking deviations from the expected phenotype-genotype correlation have been observed among several CEP cases. Indeed, a few patients with the same UROS genotype were reported with a dramatically different phenotype,10 even within the same family,11 leading us to hypothesize that sequence variations involving still-unknown modifier genes or environmental factors could modulate CEP phenotypic expression, at least in some patients.
Recently, we described a new type of porphyria, X-linked dominant erythropoietic protoporphyria (XLDPP, MIM# 300752), characterized by a high proportion of zinc-protoporphyrin in erythrocytes.12 This rare condition is induced by gain-of-function mutations within the C-terminal region of 5-aminolevulinate synthase 2 (ALAS2, EC 2.3.1.37), the erythroid-specific isoform of the first and rate-regulating enzyme of the heme biosynthetic pathway.13 Therefore, we hypothesized that CEP phenotypic variations could be modulated by ALAS2 gene variations, affecting ALAS2 activity in the bone marrow. We tested this hypothesis in 4 Spanish CEP patients harboring the same compound heterozygous UROS genotype but with different levels of clinical severity.
Methods
Patients
We studied 4 unrelated Spanish CEP patients, 2 women and 2 men. One of these patients (patient 1) has been documented in a previous report,10 whereas the other 3 are newly described (patients 2-4; Table 1). Three patients were identified through referral to a specialist porphyria center in Barcelona (Hospital Clínic of Barcelona) and one was studied at the University of Bordeaux. All were of Western European ancestry. They presented variable clinical severity and were classified according to the method of Desnick and Astrin.1 The CEP patients and their relatives were informed of their status, they were asked to enroll in a study that would include genotyping and pedigree studies, and all provided their informed consent. The study was conducted in accord with the World Medical Association Declaration of Helsinki ethical principles for medical research involving human subjects and its subsequent amendments.
Porphyrin analysis
The diagnosis of CEP was determined by classic biochemical analyses of porphyrins in urine and erythrocytes, isomer I/III ratio, enzymatic assays (UROS activity), and UROS gene sequencing according to the European Porphyria Network guidelines and quality control schemes (http://www.porphyria-europe.org).2 Erythrocyte porphyrins were measured as previously described.14 The percentage of zinc-protoporphyrin was calculated from fluorescence emission spectra of ethanol or acetone extracts of erythrocyte hemolysates. Porphyrins in stool were extracted according to Lockwood et al15 and were analyzed by HPLC according to Lim and Peters.16 Porphyrins in urine and the respective proportion of isomer I and III were similarly analyzed by reverse-phase HPLC.17 UROS activity was measured in erythrocytes with the coupled-enzyme assay of Tsai et al.18
DNA analysis
Genomic DNA was extracted from peripheral blood after the salting-out procedure.19 Analysis of UROS and ALAS2 genes (GenBank accession numbers for human ALAS2 mRNA: NM000032.4 and protein NP_000023.2; for human UROS mRNA: NM000375.2 and protein NP 000366.1) was performed by bidirectional sequencing. Gene promoters, exons, and exon-intron junctions were amplified by PCR (Master Mix; Promega Co). PCR products were purified (QIAquick gel extraction kit; QIAGEN), and both strands were sequenced with the use of fluorescent ddNTPs (BigDye) on an ABI Prism 3130XL Genetic Analyzer (Biosystems) and analyzed with ABI PRISM GeneMapper software version 3.0. Analysis of X-chromosome inactivation was performed as previously described20 with the use of the androgen receptor polymorphism as a marker.
Prokaryotic expression vectors
To investigate the effect of the Y586F mutation on ALAS2 activity, the mutated enzyme was expressed in E coli by the use of 2 different expression systems. First, the c.1757A > T mutation was introduced into pMALc2-AE2 (ie, a wild-type ALAS2 [ALAS2 WT] expression plasmid)21 by site-directed mutagenesis by use of the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and the following oligonucleotides: 5′-GGGGCCCCAGTATGTCACCACCTTTGCCTGAGAAGCC-3′ and its complementary antisense. Competent E coli BL21 DE3 cells (Invitrogen, Life Technologies) were transformed with the generated expression plasmid. Second, to obtain larger amounts of purified WT and Y586F ALAS2 variant and to determine their steady-state kinetics parameters, the corresponding coding sequences were PCR-amplified and were subcloned into pGF23 plasmid. Their expression was placed under control of the alkaline phosphatase promoter.22 The 5′-end PCR primer contained the codons for first N-terminal amino acids of the mature human ALAS2 and the sequence for the SalI site primer, whereas the 3′end PCR primer contained the codons for last C-terminal amino acids of the mature human ALAS2 and the sequence for the BamHI site.
The PCR-amplified ALAS2 WT and Y586F-encoding products and pGF23 plasmid were digested with BamHI and SalI (Invitrogen, Life Technologies). The digestion products were separated on an agarose gel and the pGF23 vector, and inserts were purified from the gel with the kit DNA extraction from agarose gel (Nucleospin Extract II; Macherey-Nagel, distributed by Clontech Laboratories Inc). After ligation (T4 DNA ligation vector kit), the ligation reaction was transformed into XL1-Blue (Stratagene) and the new expression plasmids pSD1 and pSD2 for ALAS2 WT and Y586F were isolated, respectively. The entire ALAS2 WT- and Y586F-encoding regions were sequenced to confirm that only the desired sequence had been introduced.
ALAS2 enzymatic activity
The previously characterized pMALc2 vectors expressing ALAS2 WT (normal enzyme), ALAS2 C344X (negative control), and delAT (gain-of-function control) were used as different controls.12 ALAS2 activities of the different recombinant proteins (controls and Y586F) were determined in the bacterial lysates as previously described.23 The reaction assay was run for 20 minutes at 37°C, and the specific activity (SA) was expressed as pmol of ALA generated/mg protein/hour. Activity of Y586F and delAT are reported as a percentage of the WT value as calculated by the following formula: 100 × [SA(mut) − SA(C344X)]/[SA(WT) − SA(C344X)]. Results were the mean of 3 experiments. Western blots with antibody directed against the MBP tag of the recombinant enzyme (E8030S; New England Biolabs/Ozyme)23 were performed in denaturing conditions before and after reaction to confirm that the expression level of recombinant WT and mutant proteins were similar.
Protein purification
WT ALAS2 and the Y586F variant and were purified from E coli BL21(DE3) cells harboring pSD1 and pSD4, respectively, as previously described,22 with slight modifications. First, the harvested bacterial cells were resuspended in 20mM potassium phosphate, pH 8.0, containing 1mM EDTA, 5mM β-mercaptoethanol, 10% glycerol, 1 mg/mL protamine sulfate, and 20μM pyridoxal 5′-phosphate (PLP). Second, the Y586F variant and WT ALAS2 proteins were fractioned with 45% ammonium sulfate precipitation. Third, the 45% ammonium sulfate protein precipitate was resuspended in 20mM potassium phosphate, pH 8.0, containing 1mM EDTA, 5mM β-mercaptoethanol, 10% glycerol, 20μM PLP, and 5% DMSO. Fourth, elution of Y586F variant from the DEAE-Sephacel resin was with 20mM potassium phosphate, pH 8.0, containing 1mM EDTA, 5mM β-mercaptoethanol, 10% glycerol, 20μM PLP, and 50mM KCl, whereas the elution of WT ALAS2 required that the concentration of KCl was increased to 100mM. Protein concentration was determined by use of the bicinchoninic acid assay with BSA as standard,24 and the protein purity was evaluated by SDS gel electrophoresis.
UV-visible absorption spectroscopy
Absorption spectra were recorded by the use of a Shimadzu UV 2100U dual-beam spectrophotometer at 25°C. To remove free PLP (ie, PLP nonspecifically bound to ALAS), before spectra were acquired, the purified enzymes (ALAS2 WT and Y586F) were dialyzed in 20mM HEPES, pH 7.5, containing 10% glycerol. Spectra were recorded for both the free holoenzymes (ALAS2 WT or Y586F at 10μM) and the holoenzymes on glycine binding by use of the dialysis buffer (ie, 20mM HEPES, pH 7.5, containing 10% glycerol) as blanck. The spectral data files were imported as ASCII files and plotted with Sigma Plot (Version 10.0).
Steady-state kinetics
ALAS activity of purified mature WT ALAS2 and Y586F variant was determined by the use of a continuous spectrometric assay at 30°C.25 Enzyme concentration (ALAS2 WT or Y586F) in the reaction assays was 0.5μM. The enzymatic activity data were constructed as matrixes of 5 glycine and 5 succinyl-coenzyme A (CoA) concentrations from which apparent maximal rates were determined by fitting the data to the Michaelis-Menten equation by use of the nonlinear regression analysis software program SigmaPlot (Version 10.0). Secondary plots were used to obtain Km (ie, KmGly, KmSCoA) and kcat values as described in Gong et al.26
Rapid scanning stopped-flow spectroscopy and presteady-state kinetic data analysis
Rapid scanning stopped-flow kinetic measurements were performed with the use of a model RSM-1000 stopped-flow spectrophotometer (OLIS Inc). This instrument has a 2-ms dead time, a 4.0-mm path length, and a temperature-controlled observation chamber. In general, scan spectra covering the wavelength range of 325-545 nm were collected at a rate of 62 scans per second. A fixed 0.6-mm slit and a 16 × 0.2-mm scandisk were used to collect data. A circulating water bath controlled thermostatically at 30°C was used to maintain the temperature of the loading syringes (containing the reactants) and the stopped-flow cell compartment. Reactant concentrations in the 2 loading syringes were 2-fold greater than the final concentrations in the observation chamber. (These final concentrations are reported in the figure legends.) The reaction buffer for the experiments was 20mM HEPES, pH 8.0, containing 10% glycerol. The reaction of either ALAS2 WT or Y586F with glycine and succinyl-CoA was performed under multiple turnover conditions, and the time courses for a species absorbing at 510 nm (ie, a quinonoid intermediate) were analyzed by fitting to an equation for a 2-step process as previously described.27
Statistical analysis
Data were analyzed with Microsoft Excel, SPSS, and Minitab (Version 13) software. The significance of differences between quantitative variables and proportions was assessed with the Mann-Whitney U test and the Student t test.
Results
Patient 1 had already been reported by To-Figueras et al,10 whereas patients 2-4 are now described. In all patients, separation and quantification of porphyrins by HPLC showed a typical CEP pattern for urine and erythrocytes with accumulation of porphyrin series I isomers. However, the concentration of porphyrins both in urine and erythrocytes showed large differences between patient 1 and patients 2-4, although all of them harbored the same C73R/P248Q UROS genotype (Table 1; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). UROS activity in erythrocytes was almost identical for all patients (Table 1).
ALAS2 genotyping in CEP patients
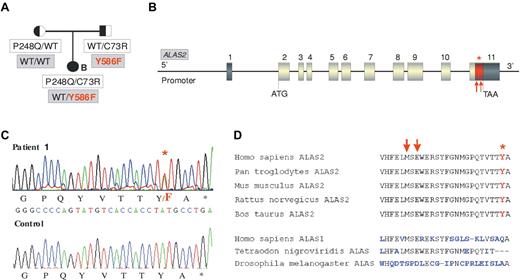
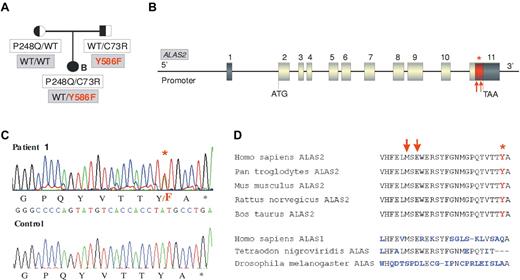
We then sequenced the ALAS2 gene to test the possibility that a mutation at this locus could modulate the CEP phenotype. In 3 CEP patients (patients 2-4), the gene had the WT sequence, whereas one patient (patient 1) presented a novel heterozygous c.1757 A > T mutation in exon 11. This mutation is predicted to affect the penultimate C-terminal amino acid of ALAS2 resulting in a p.Y586F change (Table 1; Figure 1A-C). This tyrosine in the C-terminal domain of ALAS2 is highly conserved (Figure 1D). To exclude a rare polymorphism, 100 unrelated control patients were sequenced but the Y586F mutation was not found (data not shown). The affected girl (patient 1) did not show any skewed X inactivation (data not shown). Pedigree analysis revealed that the father was carrier of the Y586F mutation in addition to being an heterozygous carrier of the UROS C73R mutation, whereas the mother was WT for ALAS2 and an heterozygous P248Q carrier for the UROS gene (Figure 1A). Biochemical analyses of erythrocytes and urine of both parents did not show significant accumulation of porphyrins or deviation from the normal I/III isomer ratios. The father presented a greater erythroid protoporphyrin level than the mother (+29%), but this was still in the normal range (796 nmol/L vs 619 nmol/L; normal range, < 1500 nmol/L).
ALAS2 mutation in the family of patient 1. (A) Pedigree of the CEP family with both ALAS2 and UROS mutations. Cosegregation of UROS mutations (white boxes) and ALAS2 mutations (gray boxes) is shown. (B) Representation of ALAS2 gene: Dark boxes represent untranslated regions and gray boxes coding exons. The red part of the last exon corresponds to the domain in which mutations causing XLDPP have been found (red arrows). The red star corresponds to the Y586F substitution found in the CEP proband (patient 1). (C) C-terminal mutation in ALAS2. Sequence analysis of genomic DNA from CEP patient 1 demonstrating c.1757 A > T (Y586F) mutation in the ALAS2 gene. (D) Comparison of the C-terminal sequence of ALAS enzymes from different species. Red arrows indicate the location of XLDPP mutations and the red star indicates the location of the Y586F substitution.
ALAS2 mutation in the family of patient 1. (A) Pedigree of the CEP family with both ALAS2 and UROS mutations. Cosegregation of UROS mutations (white boxes) and ALAS2 mutations (gray boxes) is shown. (B) Representation of ALAS2 gene: Dark boxes represent untranslated regions and gray boxes coding exons. The red part of the last exon corresponds to the domain in which mutations causing XLDPP have been found (red arrows). The red star corresponds to the Y586F substitution found in the CEP proband (patient 1). (C) C-terminal mutation in ALAS2. Sequence analysis of genomic DNA from CEP patient 1 demonstrating c.1757 A > T (Y586F) mutation in the ALAS2 gene. (D) Comparison of the C-terminal sequence of ALAS enzymes from different species. Red arrows indicate the location of XLDPP mutations and the red star indicates the location of the Y586F substitution.
Clinical characterization of the CEP patient with digenism
Patient 1 was diagnosed with CEP at 10 months of age. Pink urine had been observed since the neonatal period. The phenotype was dominated by skin photosensitivity, including abnormal skin fragility, bullae, erosions, blistering, and scarring. Physical examination revealed erythrodontia, hyperpigmentation on her face and hands, and facial hypertrichosis. The spleen was palpable 3 cm below the left costal margin with no hepatomegaly. Blood counts revealed anemia and markers of hemolysis (Table 1). The red cells were hypochromic and microcytic with anisocytosis, poikilocytosis, polychromasia, and basophilic stippling (Table 1). White blood cell and platelet counts were normal. Compared with the 3 other CEP patients with the same UROS genotype (patients 2-4), this patient demonstrated an earlier onset of the disease associated with a more severe phenotype (Table 1). The patient was reevaluated at a clinical follow-up when she was 6 years of age. Because she had been diagnosed with CEP, the patient had been thoroughly protected from sunlight exposure. However, hyperpigmentation, hypertrichosis, erythrodontia, and small atrophic scars on hands and face secondary to previous bullae were found. Hematologic parameters showed a slight improvement (Hb 12 g/dL; hematocrit 35%, mean corpuscular volume, 70 fL). Total porphyrins in erythrocytes had decreased from 27 050 nmol/L to 13 480 nmol/L. Iron status parameters and markers of hemolysis showed no significant changes compared with the initial examination (data not shown).
Functional studies of the Y586F-mutated ALAS2 protein
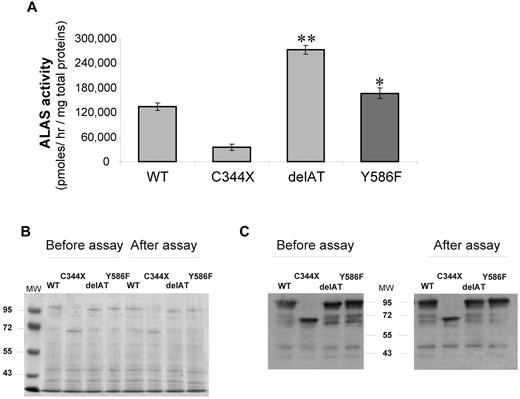
The Y586F ALAS2 mutation was predicted by computer modeling to be possibly damaging when PolyPhen software (data not shown) was used. Moreover the ALAS2 sequences are highly conserved at the carboxyterminus, and the replacement of this amino acid might be deleterious (Figure 1D). To confirm this prediction, we expressed the mutated enzyme in E coli. Activity of the mutant ALAS2 enzyme was significantly increased by 34% (P < .05) in the bacterial lysate with the Y586F variant compared with that with the WT protein (Figure 2A). However, this activity was much lower than that of the bacterial lysate with the delAT mutant used as the ALAS2 “gain-of-function” positive control, suggesting that a milder effect is associated with the Y586F mutation. Coomassie staining (Figure 2B) and immunoblotting (Figure 2C) were performed to confirm that the level of expression of the each recombinant proteins was similar and that the proteins were not degraded during the incubation period.
Prokaryotic expression of WT and mutated ALAS2 proteins. (A) Rates of ALA formation by bacterial lysates expressing either WT ALAS2, negative control with a misense mutation (C344X), a gain of function control (delAT), and Y586F-mutated ALAS2; data are expressed as mean ± SD for 3 experiments. *P < .05; **P < .001 (Student t test). (B) Coomassie blue stain of a 8% acrylamide gel with lysates of E coli cells expressing either WT or mutated ALAS2 cDNAs and run on acrylamide gel. Lysates were prepared either after the sonication step (ie, before assay) or after the assay (20 minutes at 37°C). (C) Western blot analysis of lysates of E coli cells expressing either WT or mutated ALAS2 cDNAs. After protein transfer, detection was performed by incubating the membranes with a primary antibody directed against the MBP tag. A prestained protein ladder was used as molecular weight markers (MW).
Prokaryotic expression of WT and mutated ALAS2 proteins. (A) Rates of ALA formation by bacterial lysates expressing either WT ALAS2, negative control with a misense mutation (C344X), a gain of function control (delAT), and Y586F-mutated ALAS2; data are expressed as mean ± SD for 3 experiments. *P < .05; **P < .001 (Student t test). (B) Coomassie blue stain of a 8% acrylamide gel with lysates of E coli cells expressing either WT or mutated ALAS2 cDNAs and run on acrylamide gel. Lysates were prepared either after the sonication step (ie, before assay) or after the assay (20 minutes at 37°C). (C) Western blot analysis of lysates of E coli cells expressing either WT or mutated ALAS2 cDNAs. After protein transfer, detection was performed by incubating the membranes with a primary antibody directed against the MBP tag. A prestained protein ladder was used as molecular weight markers (MW).
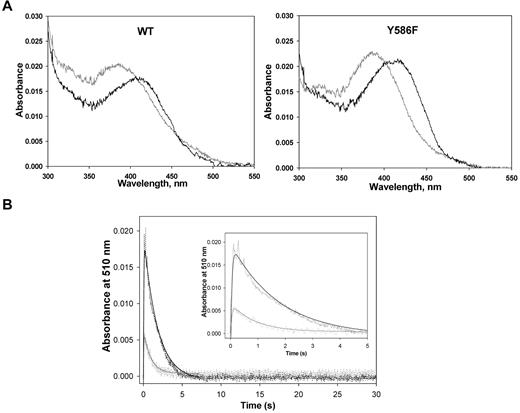
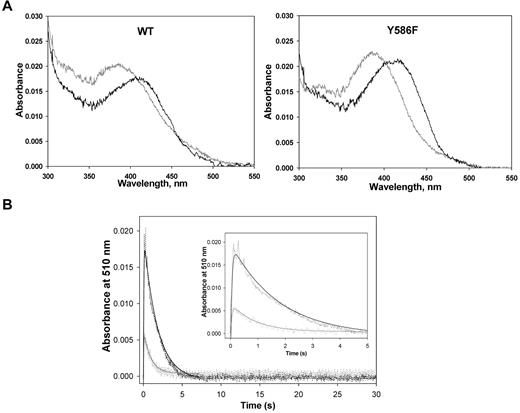
Purification of WT ALAS2 and Y586F from E coli cells harboring expression plasmids pSD1 and pSD4, respectively, yielded enzymes with specific activities of ∼ 0.19 nmol ALA.mg−1s−1 and ∼ 0.44 nmol ALA.mg−1s−1 at 30°C. The purified recombinant WT and Y586F ALAS2 exhibited similar UV-visible absorption spectra (Figure 3A).27 On the addition of glycine, a 420-nm absorption species was apparent, which was assigned to the Schiff base linkage between the pyridoxal 5′-phosphate cofactor and the glycine substrate—the formation of an external aldimine with the addition of the amino acid substrate is typical among pyridoxal 5′-phosphate–dependent enzymes.27,28 Although the UV-visible absorption spectra for the ALAS2 and Y586F holoenzymes were similar, the absorbance ratio between the 410- and 330-nm species (A410/A330) differed between the 2 enzymes. Presumably, the 410-nm absorption species corresponds to the ketoenamine form of the internal aldimine between the cofactor and an active site lysine residue, and the 330-nm absorption species relates to a substituted aldimine, as previously described for murine ALAS2.29 The more prominent absorbance at ∼ 398 nm than at 410 nm in the absorption spectra of the holoenzymes indicates that part of the PLP cofactor, bound to either ALAS2 WT or Y586F, is not covalently bound. Previously, an Absmax ∼ 398 nm was observed for murine ALAS variants in which the active site lysine involved in the Schiff base linkage with the PLP cofactor is mutated to alanine or histidine, as the cofactor is bound to the enzyme, albeit not covalently.30,31 The subtle differences between the absorption spectra of ALAS2 WT and Y586F (eg, different A410/A330 ratios) likely reflect a slightly different nature of their active sites.
Enzymatic studies of the recombinant ALAS2 enzymes. (A) UV-visible absorption spectra (300-550 nm) of ALAS2 (WT) and Y586F2. Both proteins were at 2.0μM in 20mM HEPES, pH 7.5, containing 10% glycerol, and the spectra were recorded at 25°C. In both cases, the spectrum in gray is for the holoenzyme, whereas the black spectrum is for the holoenzyme in the presence of 100mM glycine. (B) Time courses for the reaction of WT ALAS2 (90μM; black) and Y586F (70μM; gray) with a mixture of glycine (100mM) and succinyl CoA (120μM) as monitored by following the changes in absorbance at 510 nm. The inset displays the first 5 seconds of the time courses. The collected data (dashed lines) were fitted to a 2-exponential equation with rate constants k1 and k2 for the formation and decay phases of the quinonoid intermediate (ie, 510-nm absorption species), respectively.27 The rate constants for the WT ALAS2-catalyzed reaction are k1 = 22.1 ± 1.1 seconds−1 and k2 = 0.57 ± 0.02 seconds−1, whereas for the Y586F-catalyzed reaction are k1 = 20.0 ± 3.2 seconds−1 and k2 = 1.11 ± 0.10 seconds−1.
Enzymatic studies of the recombinant ALAS2 enzymes. (A) UV-visible absorption spectra (300-550 nm) of ALAS2 (WT) and Y586F2. Both proteins were at 2.0μM in 20mM HEPES, pH 7.5, containing 10% glycerol, and the spectra were recorded at 25°C. In both cases, the spectrum in gray is for the holoenzyme, whereas the black spectrum is for the holoenzyme in the presence of 100mM glycine. (B) Time courses for the reaction of WT ALAS2 (90μM; black) and Y586F (70μM; gray) with a mixture of glycine (100mM) and succinyl CoA (120μM) as monitored by following the changes in absorbance at 510 nm. The inset displays the first 5 seconds of the time courses. The collected data (dashed lines) were fitted to a 2-exponential equation with rate constants k1 and k2 for the formation and decay phases of the quinonoid intermediate (ie, 510-nm absorption species), respectively.27 The rate constants for the WT ALAS2-catalyzed reaction are k1 = 22.1 ± 1.1 seconds−1 and k2 = 0.57 ± 0.02 seconds−1, whereas for the Y586F-catalyzed reaction are k1 = 20.0 ± 3.2 seconds−1 and k2 = 1.11 ± 0.10 seconds−1.
The turnover number (kcat) of Y586F was enhanced by more than 2-fold in relation to that of the WT enzyme (Table 2), and the catalytic efficiencies for the glycine and succinyl-CoA substrates were 2.5- and 4-fold greater for Y586F than ALAS2 WT, respectively. These findings are consistent with the accumulation of the ALA-derived porphyrin observed in the bacterial cells harboring the expression plasmid for Y586F and in patient 1.
The reaction of either ALAS2 WT (90μM) or Y586F (70μM) with 100mM glycine plus 120μM succinyl-CoA was analyzed by rapid stopped-flow, scanning absorption spectroscopy. The kinetic traces for the species absorbing at 510 nm (quinonoid intermediate) are shown in Figure 3B. The first 5 seconds of either reaction could be described by a 2 exponential process with rates of 22.1 ± 1.1 seconds−1 and 0.57 ± 0.02 seconds−1 for quinonoid intermediate formation and decay in the WT enzyme-catalyzed reaction and rates of 20.0 ± 3.2 seconds−1 and 1.11 ± 0.10 seconds−1 for quinonoid intermediate formation and decay in the reaction catalyzed by Y586F (Figure 3B inset). The similar rate for the formation of the quinonoid intermediate and an approximately 2-fold increase in its rate of decay in the reaction of the mutated enzyme over that of ALAS2 WT is consistent with a more rapid formation of ALA and release from the enzyme.
Furthermore, these results agree with those obtained for hyperactive variants of murine ALAS2, for which the decay of the quinonoid intermediate was shown to be accelerated in the reactions catalyzed by the hyperactive variants compared with that of the WT enzyme.32 It is also important to note that the thermostabilities of ALAS2 WT and Y586F are similar (data not shown), supporting the finding that the Y586F variant is more active than the WT enzyme on a per-molecule basis. Together these results show a gain-of-ALAS2 function associated with the Y586F missense mutation, albeit the gain-of-function appears slightly less significant than that observed with the C-terminal deletions of ALAS2 responsible for XLDPP.
Discussion
Phenotypic heterogeneity is a common finding in CEP, but correlations between genotype and phenotype are difficult to establish, mostly because of the rarity of the disease; up to now, only 43 disease-causing mutations have been published.5 The 4 patients studied here were compound heterozygous for the same C73R/P248Q UROS genotype but presented major differences in their phenotypes. We exploited this situation to explore the role of ALAS2 as a modifier gene, and we identified a gain-of-function mutation that appeared to account for the most severe phenotype present in one of our patients.
The relatively high prevalence of the P248Q UROS mutation was previously observed in Spain,10 and the appearance, reported in this study, of new independent CEP patients with a P248Q allele confirmed that this mutation is highly prevalent among Spanish CEP carriers.10,33,34 The second missense mutation, C73R is commonly found in CEP patients without a reported founder effect.35 Homozygosity for this prevalent mutation is uniformly manifested by a severe phenotype characterized by intense hemolysis, severe photomutilation, and reduced life expectancy.1,34 Because of the very complex genotype-phenotype relationship and the influence of lifestyle on the CEP phenotype, clinical comparison of different CEP cases with a similar genotype is not simple. In this study the follow-up of patient 1, 5 years after her diagnosis, showed that phenotypic expression of the disease was slightly attenuated. This evolution confirmed observations made in other Spanish CEP patients in which the hematologic condition improved with age. Thus, CEP presents significant phenotypic variations during the lifetime because of the influence of numerous endogenous and exogenous factors.
The demonstration by Phillips et al36 of an authentic CEP disease because of a molecular defect in a transacting gene involved in the regulation of the heme biosynthetic pathway (GATA1) provides evidence that CEP is not a monogenic disease and may be caused by a dysregulation of an erythroid specific transcription factor. However, GATA1 sequences were found normal in the 4 CEP patients in this study (data not shown). This genetic heterogeneity also suggests that the CEP phenotype could be modulated by modifier genes.
Recently, a previously unrecognized form of erythropoietic porphyria, XLDPP, was described.12 This porphyria results from increased activity of the ALAS2 enzyme. In the absence of other gene mutations affecting the heme biosynthetic pathway, ALAS2 gain-of-function leads to the production of protoporphyrin IX in excess of iron availability and in quantities sufficient to cause photosensitivity despite the normal activity of ferrochelatase.2,37 The finding of a CEP patient with a missense mutation within the C-terminal region of the ALAS2 gene is intriguing. The XLDPP-causing mutations reported so far are small deletions leading to a truncation of the C-terminal part of the enzyme. To date, the only missense mutations described close to this region lead to loss of function (R559H,38 R560H,39 S568G,40,41 and R572H23 ) and have been found in patients with X-linked sideroblastic anemia (XLSA, MIM #300751).
The novel Y586F mutation corresponds to a substitution of the penultimate C-terminal amino acid of ALAS2, and we demonstrate that it is associated with an increase in the activity of the enzyme. Indeed, the substitution of this C-terminal tyrosine in ALAS2 causes an increase in kcat value and catalytic efficiencies toward both succinyl-CoA and glycine substrates. The rates of product formation and release from the enzyme may be also accelerated. The C-terminal region of ALAS2 is highly conserved between species and has diverged through evolution from ALAS1 (Figure 1D), thus suggesting that the terminal residues of the mature ALAS2 protein may contribute to a yet-unknown specific role in erythropoiesis. Nevertheless, the Y586F missense mutation induces a gain-of-function less potent than the one observed in deletions causing XLDPP. This missense mutation alone would probably produce a mild effect in vivo. However, the C73R mutation may also limit the availability of protoporphyrin IX for ferrochelatase. No carrier of the Y586F ALAS2 mutation without any UROS mutation was available in the family. Therefore, even if it is likely that Y586F alone may induce a slight zinc-protoporphyrin accumulation, this remains to be proven.
A different outcome is to be expected in CEP patients in whom ALAS2 gain-of-function is combined with a UROS defect. The enhanced kinetic properties of the mutated ALAS2 enzyme could cause an increased production of the linear tetrapyrrole HMB, the substrate of UROS. This increased formation of HMB associated with a defective UROS could contribute to amplifying the URO I and COPRO I accumulation in erythrocytes (Figure 4), thus augmenting hemolysis and subsequent release of phototoxic porphyrins into the circulation responsible for the aggravated phenotype. Clinical data support this hypothesis. At first examination, the Y586F carrier presented a moderate phenotype with hemolytic anemia, and CEP was diagnosed in the first months of life. In contrast, the 3 other patients with the same UROS genotype were diagnosed later and presented a milder phenotype at examination with significantly lower levels of porphyrins both in urine and in erythrocytes compared with the Y586F carrier. In conclusion, our study demonstrates that abnormal ALAS2 activity should be considered, not only as responsible for XLSA or XLDPP, but also as a possible aggravating factor for erythropoietic diseases when it is increased in bone marrow cells.
Proposed mechanism for the role of ALAS2 as a modifier gene in CEP. An enhanced kinetic property of the mutated ALAS2 enzyme would cause an increased production of the linear tetrapyrrole HMB, the substrate of UROS. The increased supply of HMB associated with a defective UROS enzyme would lead to the aggravated CEP phenotype. (A) normal individual; (B) CEP patient wild-type for ALAS2; and (C) CEP patient with ALAS2 mutation.
Proposed mechanism for the role of ALAS2 as a modifier gene in CEP. An enhanced kinetic property of the mutated ALAS2 enzyme would cause an increased production of the linear tetrapyrrole HMB, the substrate of UROS. The increased supply of HMB associated with a defective UROS enzyme would lead to the aggravated CEP phenotype. (A) normal individual; (B) CEP patient wild-type for ALAS2; and (C) CEP patient with ALAS2 mutation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the family members for their cooperation; Dr Vasco Pereira Da Silva, Sylvie Simonin, and Anne Marie Robreau Fraolini for expert laboratory assistance; Prof Bernard Grandchamp for informative and helpful discussions related to clinical and physiopathologic details in genetics and hematology; and Jean-Pierre Laigneau for illustrations.
This work was supported by Grant Public Health and Consumer Protection Directory Public Health Executive Agency from the European Commission, Brussels, Belgium, and by grant number ANR-GIS Maladies rares-07-MRAR-008-01 (to S.D., S.L., L.G., J.-C.D., and H.P.) Paris, France; by grant 10GRNT4300073 from the American Heart Association (to G.C.F); and by a grant from the Spanish “Fondo de Investigación Sanitaria” (FIS; to J.T.-F., C. Badenas, and C.H.).
Authorship
Contribution: All authors participated in designing and performing the research, writing the report, and checking the final version of the manuscript; S.D., C. Badenas, C.D., and L.G. performed molecular biology; J.T.-F. and S.L. performed porphyin analysis; J.T.-F., C. Badenas, and J.-C.D. examined the patients; J.C. and G.C.F. performed enzymatic investigations; C.G., H.d.V., C.H., G.C.F., and H.P. supervised the study and discussed the results; and S.D., C. Beaumont, G.C.F., and H.P. designed the figures and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Carole Beaumont, Inserm U773, Universite Paris Diderot, site Bichat, 16 rue Henri Huchard, 75018 Paris, France; e-mail: carole.beaumont@inserm.fr.