Abstract
Expression of a BCR is critical for B-cell development and survival. We have identified 4 patients with agammaglobulinemia and markedly reduced but detectable B cells in the peripheral circulation. These B cells have an unusual phenotype characterized by increased expression of CD19 but no BCR. The cells are positive for CD20, CD22, and CD38, but not for Annexin 5 or activation markers, including CD69, CD83, or CD86. EBV lines derived from these B cells lack functionally rearranged immunoglobulin heavy-chain transcripts, as shown by PCR–rapid amplification of cDNA ends (PCR-RACE). Analysis of BM from 2 of the patients showed a severe reduction in the number of pro-B cells as well as pre-B cells. Functionally rearranged heavy-chain transcripts were identified, indicating that machinery to rearrange immunoglobulin genes was intact. Flow cytometry of B-lineage cells suggested accelerated acquisition of maturation markers in early B-cell precursors and increased phosphorylation of signal transduction molecules. Further, expression of TdT, a molecule that is normally down-regulated by a functional pre-BCR complex, was decreased. We hypothesize that the accelerated maturation, increased expression of CD19, and lack of a BCR were due to the constitutive activation of the BCR signal transduction pathway in these patients.
Introduction
B-cell development is critically dependent on appropriate signaling through the pre-BCRs and BCRs. Mutations that decrease the function of any of the components of these receptors, including μ heavy chain, Igα, Igβ, and the proteins that make up the surrogate light chain result in a severe block in B cell differentiation at the pro-B to pre-B-cell transition.1-5 Defects in proteins required for downstream signaling through these receptors, including Btk and BLNK, also result in a selective B-cell deficiency.6-8 However, mutations in these proteins do not account for all of the patients with isolated defects in B-cell development.
Premature or excessive signaling through the pre-BCR or BCR may also cause a reduction in the number of peripheral B cells. Early studies in transgenic mice showed that expression of the membrane form of a rearranged immunoglobulin heavy chain resulted in markedly reduced numbers of B-lineage cells in the BM and in the periphery.9 This was attributed, at least in part, to rapid transit through early stages of differentiation. Rearranged μ heavy chains encoding high-affinity or activating antibodies for auto-antigens result in clonal deletion of immature B cells.10 Enhanced activation of the BCR pathway also causes a reduction in the number of B cells. Mice bearing a transgene for a constitutively active form of Btk (E41K) have a B-cell deficiency that is more severe than that seen in mice that lack Btk.11 More indirectly, increased expression of CD19, a molecule that enhances signaling through the BCR, also results in a marked decrease in the number of peripheral B cells.12 Genetic alterations that enhance B-cell signaling have been reported in patients with systemic lupus erythematosus,13,14 but not in patients with agammaglobulinemia.
We have found that some patients with isolated defects in B-cell development have a small number of B cells in the peripheral circulation. The phenotype of these cells may provide clues about the nature of the gene defect. A subset of patients, including all reported patients with mutations in the μ heavy chain, have undetectable CD19+ cells in the blood with a level of sensitivity of 0.01%.15 By contrast, most patients with mutations in Btk have between 0.01% and 0.5% CD19+ cells in the blood. These cells express high-density surface IgM, but have decreased intensity of HLA DR and CD21.16 Some of the patients with B-cell defects of unknown etiology have a small number of CD19+ cells with a phenotype similar to that seen in patients with mutations in Btk; others have a few B cells that resemble B cells from healthy controls except that they lack CD27+ B cells. We have analyzed 4 unrelated patients with agammaglobulinemia and < 3% CD19+ cells who have a very distinctive B-cell phenotype characterized by high-intensity CD19 and the absence of a BCR.
Methods
Patients
All of the patients included in this study were enrolled in a St Jude Children's Research Hospital institutional review board–approved protocol to identify and characterize gene defects resulting in immunodeficiency. Written consent was obtained from patients and/or parents in accordance with the Declaration of Helsinki. Clinical and laboratory information on each patient was obtained from the referring physician using a standard survey form. Patient number 1 was reported previously.17
Flow cytometry
Peripheral blood lymphocytes and BM cells were separated by Ficoll Hypaque centrifugation and stained as described previously.2,3 All patient samples were stained and analyzed in parallel with a sample from a healthy donor. In some experiments, both the patient and control samples had been previously frozen. No difference was noted between the fresh and frozen samples. The following reagents were used: PE CD19, APC CD19, APC CD22, PE CD20, Per CP Cy 5.5 CD20, PE CD10, FITC CD5, FITC CD25, FITC CD69, PE CD69, PE CD123, and FITC annexin V (BD Biosciences); FITC goat anti-IgM, PE goat R(ab)2 anti-IgM, PE goat anti-IgD, and FITC goat anti-κ and anti-λ (SouthernBiotech); FITC CD21 (Biosource); FITC CD38, PE CD37, PE CD117, PE CD179a, PE CD127, and FITC CD23 (Beckman Coulter); PE CD24 (Caltag); PE CD79a (Dako); PE CD86, FITC CD83, and APC CD62L (eBioscience); and FITC TdT (SuperTechs). PE-labeled antibodies to phosphorylated PLCγ2, BLNK, NFκB, ERK, and total phosphotyrosine were obtained from BD Biosciences.
Single-cell PCR
Peripheral blood lymphocytes from the patient and a control were stained with PE CD19 and FITC CD38 and then isolated on a MoFlo sorter. Gates that were drawn to separate the patient cells that were brightest for CD19 and CD38 from cells that were dimmer but still positive for CD19 and CD 38 were used to isolate single cells from both the patient and healthy controls. Cells were sorted into a 96-well BioRad Hard-Shell plate. Reagents from the iScript Kit (Bio-Rad) were used to synthesize cDNA according to the manufacturer's instructions with the addition of 0.1% Triton X-100. The first round of nested PCR was done directly in the 96-well plate in a 50-μL volume. Depending on the PCR reaction, 2 or 3 μL of first-round PCR product was added to a new 96-well plate with the addition of positive and negative controls, and 20 μL of the second-round PCR products were analyzed on a 1% agarose gel. The primers for heavy-chain and light-chain rearrangements were taken from Brezinschek et al18 and Wang et al,19 respectively.
PCR-RACE
EBV-transformed lymphocytes were harvested and RNA was extracted using the Qiamp RNA blood mini kit (QIAGEN). A total of 10 μg of RNA was used to produce rapid amplification of cDNA ends (RACE) cDNA using the Applied Biosystems First Choice RLM-RACE kit. Nested PCR was performed according to the kit instructions using nonoverlapping primers. RACE products were cloned into Topo TA vector (Invitrogen) and sequenced using vector primers.
Results
CD19 expression on B cells from patients with defects in B-cell development
The intensity of CD19 on circulating B cells from healthy controls was homogeneous, whereas the expression of this marker was variable and lower in patients with mutations in Btk. Decreased expression of CD19 has also been seen on the B cells from a patient with a hypomorphic mutation in Igβ3 and a patient with a mutation in BLNK (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A subset of patients with defects in B-cell development of unknown etiology also have a small number of B cells with decreased expression of CD19. To determine whether the decreased CD19 could be attributed to the stage of maturation of the B cells, we stained cells with antibodies to CD19, CD24, and CD38 to identify transitional B cells, recent emigrants from the BM.20,21
In most patients with X-linked agammaglobulinemia (XLA), the majority of the CD19+ cells express a CD38 high/CD24 high phenotype, the phenotype that characterizes immature transitional B cells. However, as expected, only 5%-10% of the CD19+ cells from controls expressed this phenotype. The CD19 mean fluorescence intensity (MFI) of the transitional cells from patients with XLA was consistently lower than that of controls (supplemental Figure 2).
BCR− B cells
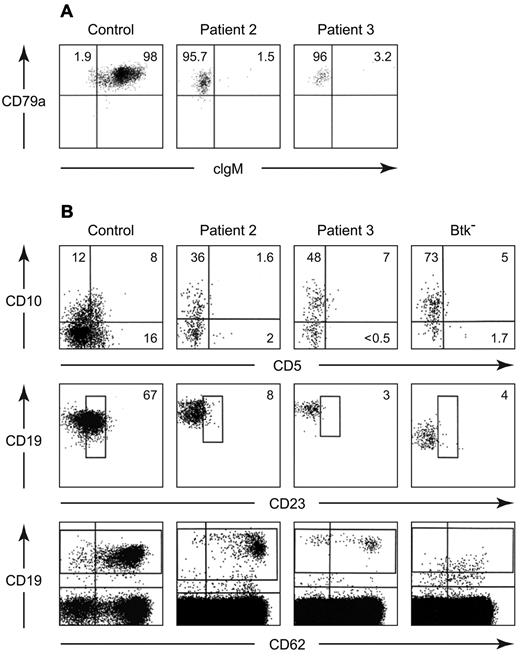
In contrast, increased expression of CD19 was seen on the B cells from 4 patients with defects in B-cell development of unknown etiology (Table 1). Three of the patients were North Americans of European descent and the fourth was Mexican. None had a family history of consanguinity or immunodeficiency. All 4 patients had the early onset of infections, profound hypogammaglobulinemia, and < 3% CD19+ cells in the circulation on multiple occasions (Table 2). More strikingly, these B cells lacked a BCR (Figure 1). Although these CD19 bright cells were positive for other B-cell markers, including CD22, CD20, and cytoplasmic CD79a (Igα), they were negative for surface IgM, IgD, Igκ, and Igλ. They were also negative for cytoplasmic IgM (Figure 2A). Like the majority of B cells from patients with mutations in Btk, the B cells from these patients were predominantly CD38+ and CD21−. The costaining of CD19 and CD38 demonstrated a distinctive pattern in which the cells that were brightest for CD19 were also brightest for CD38 (Figure 1).
Flow cytometric analysis of peripheral blood lymphocytes from a healthy control, patient 2, and a patient with a mutation in Btk (8-bp deletion in exon 8). Cells were stained with PE-labeled anti-CD19 and separate aliquots were stained with FITC-labeled polyclonal anti-IgM, a combination of anti-κ and anti-lambda, monoclonal anti-CD22, CD38, or CD21. For the control samples, approximately 20 000 events were analyzed; for the patient samples, between 100 000 and 500 000 events were analyzed.
Flow cytometric analysis of peripheral blood lymphocytes from a healthy control, patient 2, and a patient with a mutation in Btk (8-bp deletion in exon 8). Cells were stained with PE-labeled anti-CD19 and separate aliquots were stained with FITC-labeled polyclonal anti-IgM, a combination of anti-κ and anti-lambda, monoclonal anti-CD22, CD38, or CD21. For the control samples, approximately 20 000 events were analyzed; for the patient samples, between 100 000 and 500 000 events were analyzed.
Further characterization of peripheral blood B cells from patients 2 and 3. Cells from a healthy control and the patients were permeabilized and stained with PerCP Cy5.5 anti-CD20, PE anti-CD79a, and FITC anti-IgM (A). The CD20+ cells were analyzed for expression of cytoplasmic CD79a and IgM. All of the cells were positive for CD79a, the percentage of cells positive or negative for cytoplasmic IgM is shown. Maturation and activation markers on peripheral blood B cells from a healthy control, patient 2, patient 3, and a patient with a mutation in Btk (G to C at the 1+ position of the splice donor site for intron 2) were stained with APC anti-CD19, PerCp Cy5.5 anti-CD20, PE anti-CD10, and FITC anti-20 (B). Cells that were positive for CD19 and CD20 were analyzed for expression of CD10 and CD5 (top). The percentages of B cells positive for each marker are shown. The middle and bottom panels show cells stained with PE anti-CD19, FITC anti-CD23, and APC anti-CD62L. The percentages of CD19+ cells that were dimly positive for CD23 are indicated. The pattern of CD62L staining is seen in the bottom panel.
Further characterization of peripheral blood B cells from patients 2 and 3. Cells from a healthy control and the patients were permeabilized and stained with PerCP Cy5.5 anti-CD20, PE anti-CD79a, and FITC anti-IgM (A). The CD20+ cells were analyzed for expression of cytoplasmic CD79a and IgM. All of the cells were positive for CD79a, the percentage of cells positive or negative for cytoplasmic IgM is shown. Maturation and activation markers on peripheral blood B cells from a healthy control, patient 2, patient 3, and a patient with a mutation in Btk (G to C at the 1+ position of the splice donor site for intron 2) were stained with APC anti-CD19, PerCp Cy5.5 anti-CD20, PE anti-CD10, and FITC anti-20 (B). Cells that were positive for CD19 and CD20 were analyzed for expression of CD10 and CD5 (top). The percentages of B cells positive for each marker are shown. The middle and bottom panels show cells stained with PE anti-CD19, FITC anti-CD23, and APC anti-CD62L. The percentages of CD19+ cells that were dimly positive for CD23 are indicated. The pattern of CD62L staining is seen in the bottom panel.
The B-cell phenotype in patients 2 and 3 was analyzed in more detail. The CD19+ cells from these patients were equal to control cells in size and complexity. They were negative for annexin V and did not express the activation markers CD83, CD86, or CD69, or the memory marker CD27. Like B cells from patients with mutations in Btk, the majority of the B cells from patients 2 and 3 were positive for CD10 but negative for CD5 (Figure 2B). Compared with B cells from healthy controls, the B cells from the BCR− patients and the patients with mutations in Btk were dimmer for CD23. Although the percentage of CD19+ cells that expressed low amounts of CD62L were similar in the patients and controls, the cells that were brightest for CD19 in the BCR− patients were dimmest for CD62L (Figure 2B).
EBV-transformed B-cell lines from patients with BCR− B cells
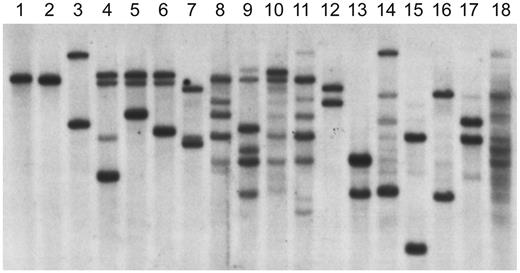
To determine whether the B cells from the patients with increased expression of CD19 were truly BCR−, EBV-transformed B-cell lines were generated from these patients. One cell line each was obtained from patients 1 and 3, and 4 cell lines were obtained from patients 2 and 4. Although the EBV-transformed lines from the BCR− patients were positive for Igα, they were negative for the μ heavy chain and for the κ or λ light chain by Western blot or flow cytometry. The status of the immunoglobulin heavy-chain genes in the B-cell lines was examined by Southern blot using a probe from the J6 region (Figure 3). Because D to J rearrangement on both alleles is one of the earliest events in B-cell development, the germline J6 fragment is replaced by fragments of variable length in normal B-cell lines. Analysis of the line from patient 1 showed only germline J6 fragments. The 4 cell lines from patient 2 showed 2 or 3 rearranged fragments. Three of these 4 cell lines, analyzed at multiple stages, retained a germline DNA fragment. The single cell line from patient 3 demonstrated 2 rearranged fragments. The 4 cell lines from patient 4 were polyclonal and all demonstrated at least some germline fragments. None of the cell lines from patients with mutations in Btk or healthy controls contained any germline fragments.
Heavy-chain gene rearrangements in EBV-transformed cell lines analyzed by Southern blot. DNA from EBV lines was digested with EcoRI and HindIII and probed with a J6 fragment. The DNA in lane 1 is from a T-cell line; the line from patient 1 is shown in lane 2. Lanes 3-6 contain DNA from lines from patient 2. The DNA in lane 7 is from the EBV line from patient 3. The DNA from the 4 polyclonal lines from patient 4 is shown in lanes 8-11. The DNA in lanes 12-14 is from lines from patients with XLA. The DNA in lanes 15-18 is from lines from healthy controls.
Heavy-chain gene rearrangements in EBV-transformed cell lines analyzed by Southern blot. DNA from EBV lines was digested with EcoRI and HindIII and probed with a J6 fragment. The DNA in lane 1 is from a T-cell line; the line from patient 1 is shown in lane 2. Lanes 3-6 contain DNA from lines from patient 2. The DNA in lane 7 is from the EBV line from patient 3. The DNA from the 4 polyclonal lines from patient 4 is shown in lanes 8-11. The DNA in lanes 12-14 is from lines from patients with XLA. The DNA in lanes 15-18 is from lines from healthy controls.
RACE was used to identify heavy- and light-chain gene rearrangements in these EBV lines. An antisense primer hybridizing to the constant region of the μ heavy chain allowed amplification of products from all of the cell lines; however, no functional VDJ rearrangements were identified in any of the lines. Four different sterile μ transcripts were isolated from the line from patient 1; 2 contained sequence from JH1 or the JH1 pseudogene and the remaining 2 contained a small amount of sequence proximal to the μ enhancer as well as the μ constant region. Nine different sequences from the 4 cell lines from patient 2 were identified; 2 full-length VDJ rearrangements were derived from one of the lines (Figure 3 lane 3), one of which was out-of-frame and the other used a pseudo VH gene. Of the 7 sequences isolated from the 3 other cell lines, 5 encoded unique transcripts for D proteins and 2 encoded sterile transcripts that contained J5 or J6 sequence but no D sequence. The transcripts for a D protein (D4-17/J4) and a sterile J5 μ transcript were seen in the EBV line from patient 3. No full-length VDJ rearrangements were seen in the 4 cell lines from patient 4. Two of the cell lines contained transcripts for a D protein as well as sterile transcripts; the remaining 2 lines contained only sterile transcripts.
A total of 6 unique κ transcripts could be found in the lines from patient 2. Four of these were out-of-frame and the remaining 2 appeared to be normal in-frame κ sequences. No rearranged κ transcripts could be found in the EBV lines from patients 1, 3, and 4. Only sterile transcripts for λ were seen in the lines from patients 1, 3, and 4; 2 of the 4 cell lines from patient 2 contained rearranged λ sequences, one was out-of-frame, the other used a pseudogene, the remaining 2 lines contained only germline sequences. All of the cell lines from the controls contained functional rearranged heavy- and light-chain genes.
Single-cell PCR to examine VDJ and VJ rearrangements
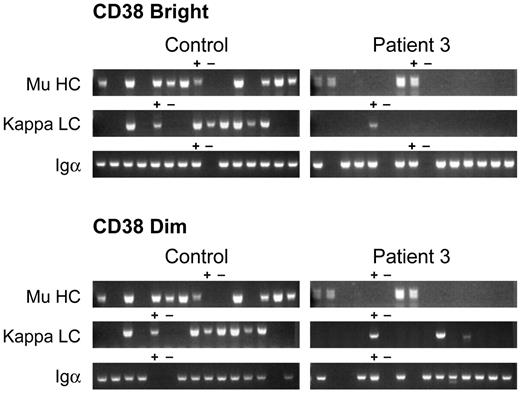
B cells that are transformed by EBV may represent a subset of the peripheral blood B cells. Therefore, single-cell PCR was used to examine VDJ rearrangements in peripheral blood lymphocytes from patient 3 and healthy controls. Two cell populations of CD19+ B cells were analyzed: the cells that were brightest for CD19 and CD38 and the cells that were positive but dimmer for both markers. Cells were sorted into 80 wells of a 96-well plate. The remaining wells were used for positive and negative controls. Nested PCR using sense primers for all 6 VH gene families and antisense primers from the μ constant region was used to analyze heavy-chain gene rearrangements in each of the 2 cell populations. In the cells that were brightest for CD19 and CD38, a rearranged μ heavy chain was amplified from a mean of 59% of wells from 4 healthy controls, 56% of wells from a patient with a mutation in Btk, and 14% of wells from patient 3 (Figure 4). Sequence analysis of 21 clones from a control and 8 clones from patient 3 demonstrated that all of the clones encoded normal-appearing, functional VDJ rearrangements. A rearranged κ transcript was isolated from 58% of control wells and 4% of wells from patient 3. Nine of 11 κ sequences from the control were in-frame, the remaining 2 were out-of-frame. Only one of 3 κ sequences from patient 3 was in-frame. To confirm that the cells analyzed were B cells, nested PCR using primers for Igα (CD79a) was used to document the presence of B cell specific transcripts. A mean of 83% of wells from 3 controls were positive for Igα and 84% of wells from patient 3 were positive.
Immunoglobulin heavy- and light-chain gene rearrangements examined by single-cell PCR. CD19 bright/CD38 bright (A) and CD19 dimmer/CD38 dimmer (B) B cells from a healthy control and patient 3 were analyzed by single-cell RT-PCR. Each lane contains the PCR product from a single cell. Lanes containing positive (+) and negative (−) controls are marked. Igα (CD79a) was amplified from wells to document that the wells from patient 3 contained B cells.
Immunoglobulin heavy- and light-chain gene rearrangements examined by single-cell PCR. CD19 bright/CD38 bright (A) and CD19 dimmer/CD38 dimmer (B) B cells from a healthy control and patient 3 were analyzed by single-cell RT-PCR. Each lane contains the PCR product from a single cell. Lanes containing positive (+) and negative (−) controls are marked. Igα (CD79a) was amplified from wells to document that the wells from patient 3 contained B cells.
In the plates containing cells from the second population of cells, cells that were positive for CD19 and CD38 but slightly dimmer, a mean of 52% of wells from 2 controls were positive for a VDJ rearrangement but only 3% of wells (4/160) from patient 3 were positive. The PCR products from 21 wells from one of the controls and all 4 wells from patient 3 were sequenced. Nineteen unique sequences were isolated from the 21 control wells; 18 of these were in-frame, one was out-of-frame. Four unique sequences were isolated from the patient wells; one of these was in-frame, the remaining 3 wells were out-of-frame. Fifty-eight percent of the wells from the control and 9% of wells from patient 3 were positive for κ transcripts. All 11 sequences from the control were in-frame, whereas only 2 of 5 κ sequences from patient 3 CD38 dim cells were in-frame; 96% of the wells from the control and 76% of wells from patient 3 were positive for Igα.
Early B-cell development
To examine earlier stages of B-cell development, BM aspirates were obtained from patients 2 and 3 and compared with those from healthy donors and patients with mutations in Btk, μ heavy chain, Igα, λ5, or BLNK. Although both samples demonstrated normal cellularity, the percentages of CD19+ cells were markedly decreased in the samples from patients 2 and 3, even compared with samples from other patients with defects in B-cell development (Table 3). In addition, the phenotype of the CD19+ cells from the 2 BCR− patients was very different from that seen in healthy controls or other patients with agammaglobulinemia, but very similar to each other. The cells were brightly stained for CD19 and could not be easily characterized as to stage of differentiation (Figure 5). In the healthy individual, pro-B cells are identified by the coexpression of CD19, CD34, and TdT.22-25 Pre-B cells no longer express either CD34 or TdT, but are positive for the cytoplasmic μ heavy chain. Most of the CD19+ cells from the patients were positive for TdT but negative for CD34 and cytoplasmic μ heavy chain. A very small number of cytoplasmic μ-positive cells, < 1% of the CD19+ cells, could be seen in the sample from patient 2. None were seen in the BM from patient 3. No cells bearing surface immunoglobulin were seen in either patient.
Expression of differentiation markers on BM-derived B-lineage cells from a healthy child, patient 2, and a patient with a mutation in Btk (W588X). Cells were stained with APC CD19, PE CD37, and APC-Cy7a CD20, then permeabilized and stained with FITC TdT. The top panel shows all cells that were positive for either CD19 or TdT with gates drawn to identify TdT+/CD19−, TdT+/CD19+, and TdT−/CD19+ populations. The expression of CD20 and CD37 on each of these populations is shown in the bottom panel.
Expression of differentiation markers on BM-derived B-lineage cells from a healthy child, patient 2, and a patient with a mutation in Btk (W588X). Cells were stained with APC CD19, PE CD37, and APC-Cy7a CD20, then permeabilized and stained with FITC TdT. The top panel shows all cells that were positive for either CD19 or TdT with gates drawn to identify TdT+/CD19−, TdT+/CD19+, and TdT−/CD19+ populations. The expression of CD20 and CD37 on each of these populations is shown in the bottom panel.
The expression of additional differentiation markers was evaluated on 3 subpopulations of BM cells: the least mature TdT+, CD19− cells, which correspond to early pro-B cells; the TdT+, CD19+ cells, which represent more mature pro-B cells; and the TdT−, CD19+ cells which include more mature B-lineage cells. The least mature cells, the TdT+, CD19− cells, from patients 2 and 3 were similar to the TdT+, CD19− cells seen in healthy controls and the patients with mutations in Btk or μ heavy chain. All of these cells were positive for surface CD34 and cytoplasmic VpreB (CD179a). Between 34% and 64% of the cells were positive for receptors for IL-3 (CD123) and IL-7 (CD127) and approximately half were positive for Igα (CD79a). Although the number of B-lineage cells available for analysis was low, there was a suggestion of accelerated maturation in both the BCR− patients and in the patients with defects in μ heavy chain or Btk. The percentage of the TdT+, CD19− cells positive for c-kit (CD117) was 11% in patient 2, 17% in patient 3, 14% in the patient with a mutation in Btk, and 15% in a patient with a defect in μ heavy chain. In contrast, 27% of the control early pro-B cells were positive for c-kit. A higher percentage of these cells was positive for CD20 in patients 2 and 3 (29% and 32%, respectively) than in a patient with XLA (14%), a patient with a mutation in μ heavy chain (< 1%), or a healthy control (< 1%; Figure 5).
The accelerated maturation was more notable in the TdT+, CD19+ cells from patients 2 and 3. In the healthy control, > 95% of the TdT+, CD19+ cells were positive for CD34, whereas only 22% and 31% of the TdT+, CD19+ cells from patients 2 and 3, respectively, were dimly positive for CD34. The total number B-lineage cells expressing CD34 was markedly decreased in both patients (supplemental Figure 3). All of the TdT+, CD19+ cells from the controls and the patients were positive for cytoplasmic Igα and VpreB and approximately half the cells expressed the IL-3 receptor. Expression of CD37, a tetraspan adhesion molecule that is normally expressed on TdT−, CD19+ pre-B cells and more mature B cells but not TdT+, CD19+ pro-B cells, was seen on > 80% of the TdT+, CD19+ cells from patients 2 and 3, but < 5% of the cells from the healthy or disease controls. CD20 was present on the surface of 8%-36% of the healthy and disease control cells, but 56% and 86% of the TdT+, CD19+ pro-B cells from patients 2 and 3, respectively (Figure 5).
In the healthy individual the TdT−, CD19+ subpopulation in the BM includes pre-B cells, immature B cells and mature B cells with approximately half of the cells falling into the pre-B cell stage of differentiation. VpreB was identified in 60% of the TdT−, CD19+ cells from healthy controls, 76%-95% of the cells from patients with mutations in Btk or μ heavy chain but only 11% and 30% of the cells from patients 2 and 3, respectively. Similarly, the IL-7 receptor was seen on 31% of the cells from the healthy control and the μ heavy chain deficient patient, 14% of the cells from the Btk deficient patient and < 1% of the cells from patients 2 and 3. In contrast, < 30% of the cells from the patient and healthy controls were positive for CD37, > 95% of the cells from patients 2 and 3 expressed this marker (Figure 5).
PCR primers that amplify specific VH family members were used to analyze μ heavy-chain rearrangements in cDNA from both the control and the patient BM samples. Of the 75 cloned heavy-chain sequences analyzed from the control, 71 unique sequences were identified and all encoded functional VDJ rearrangements. Thirty-four unique sequences were isolated from 37 clones derived from the BM of a patient with a complete deletion of the μ constant region on 1 allele and a 2-bp deletion in exon 2 on the other allele. Five of these sequences encoded out-of-frame transcripts, 6 contained VH pseudo genes, and 23 encoded functional VDJ rearrangements. There were only 10 different sequences in the 74 clones from patient 2 and 8 different sequences in the 72 clones from patient 3. All of the sequences from both patient 2 and patient 3 were in-frame and coded functional VDJ heavy-chain rearrangements. The length of the CDR3 regions was not significantly different in the patient samples compared with the controls.
Eighteen different κ rearrangements were sequenced from 21 clones from the healthy control, all of which were in-frame. Twenty-seven unique sequences were amplified from the BM of the patient with μ heavy-chain mutations: 12 of these were in-frame and the other 15 were out-of-frame. Only 3 rearranged κ sequences, 2 of which were out-of-frame, were isolated from the BM of patient 2. Two rearranged κ sequences, both of which were out-of-frame, were detected in the BM aspirate from patient 3. Six rearranged λ sequences were derived from the BM of patient 2; 3 of these were in-frame and the other 3 were out-of-frame. No rearranged λ transcripts were isolated from the BM of patient 3.
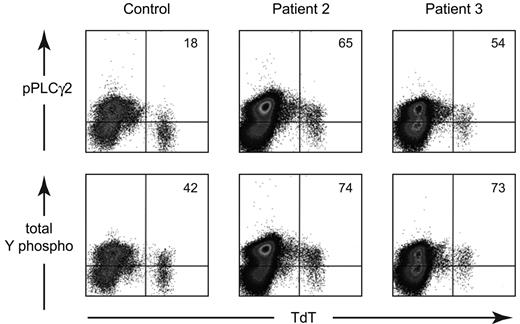
Based on the hypothesis that the increased expression of CD19 and the lack of a BCR were due to constitutive activation through the BCR pathway, we examined the phosphorylation status of signal transduction molecules in the least mature B-lineage cells, the TdT+ cells, in patients 2 and 3. As shown in Figure 6, the MFI for phosphorylated PLCγ2 (pY759) was elevated in the patients, 13.2 for patient 2 and 10.9 for patient 3, compared with the control (7.07). Total tyrosine phosphorylation was also higher in patient 2 (15.3) and patient 3 (14.6) compared with the control (7.11). More detailed analysis of the TdT+ cells from patient 2 showed that these cells were also more brightly stained for phosphorylated BLNK (pY84), NFkb (pS259), and ERK (pT202/pY204). Further, the MFI for TdT was decreased in both patient 2 (224) and patient 3 (181) compared with the control (359) TdT expression is normally down-regulated by expression of a rearranged μ heavy chain and other components of the BCR.26,27
Analysis of the phosphorylation status of early B-lineage cells from a healthy control, patient 2, and patient 3. BM cells were permeabilized and stained with FITC-labeled anti-TdT and PE-labeled phospho-PLCγ2 or total tyrosine phosphorylation. The data are shown as density plots. Quadrants were drawn based on the positive and negative TdT− cells from the control. The percentage of TdT+ cells that was positive for the phospho-antibody is shown in the upper right corner of each plot.
Analysis of the phosphorylation status of early B-lineage cells from a healthy control, patient 2, and patient 3. BM cells were permeabilized and stained with FITC-labeled anti-TdT and PE-labeled phospho-PLCγ2 or total tyrosine phosphorylation. The data are shown as density plots. Quadrants were drawn based on the positive and negative TdT− cells from the control. The percentage of TdT+ cells that was positive for the phospho-antibody is shown in the upper right corner of each plot.
Activating mutations were sought in the genes for μ heavy chain, Igα, Igβ, Btk, BLNK, PLCγ2, Lyn, Blk, syk, all 3 isoforms of AKT, LAT2, SH3BP5 (encodes a Btk binding protein), and CD19. No mutations were detected. Polymorphisms in the promoter and 3′UT regions of CD19 have been reported to affect the amount of CD19 on peripheral B cells.28,29 One of the BCR− patients and several healthy controls were heterozygous for the haplotype associated with increased expression of CD19.
Discussion
We have identified 4 patients with agammaglobulinemia and a small number of B cells with a distinctive phenotype characterized by increased expression of CD19 and the absence of a BCR. These cells are positive for B-cell–specific markers, including CD20, CD22, and cytoplasmic CD79a, but they do not bear surface immunoglobulin. Flow cytometry suggested that these peripheral blood B cells could be divided into 2 populations: one that was intensely stained for CD19 and a second that had more typical expression of CD19. A small number of functionally rearranged μ heavy chains could be identified in rare cells from the first population, whereas 3 of 4 VDJ rearrangements isolated from the second population were out-of-frame. Some of the B cells from the second population were positive for the EBV receptor CD21; it is likely that the EBV-transformed cell lines were derived from these cells. There were no functional heavy-chain VDJ rearrangements in the EBV lines.
Although the amount of cell-surface CD38 and CD21 on the CD19 brightest cells suggested that these cells were less mature than the second population, it is not clear that the second population was derived from the first. It is possible that the CD19 brightest cells do not survive and only cells that have muted signaling through the BCR become part of the second population.
Warnatz et al described a population of CD19 bright/CD21 negative cells that are present in healthy controls but are found in increased numbers in patients with common variable immunodeficiency or system lupus erythematosus.30 These cells, which were described as being partially activated, differ from the BCR− cells described in the present study in that they were larger than naive cells, positive for CD86, negative for CD38, and, most importantly, contained a normal BCR.
Analysis of BM from 2 of the patients showed markedly reduced numbers of CD19+ cells with an asynchrony of expression of B-cell markers. The least mature B-lineage cells, the TdT+ cells, showed enhanced tyrosine phosphorylation and reduced expression of TdT, an enzyme that is normally down-regulated by expression of an intact pre-BCR.26,27 Both of these findings are most compatible with constitutive activation through the pre-BCR or BCR pathway. The capacity to undergo appropriate immunoglobulin gene rearrangements in the BM was intact in these 2 patients. However, it appeared that expression of a pre-BCR or BCR was detrimental to the survival of the B-lineage cells in these patients.
The presence of a BCR on the cell surface is thought to be essential for B-cell survival. Using conditional deletion of the VH region of a μ heavy chain, Rajewsky et al showed that loss of the BCR resulted in the apoptosis of peripheral B cells over a 3- to 6-day period.31 In more recent studies, the same group has shown that a mildly constitutively active PI3K pathway but not an active NF-κB, MEK1, or Rac1 pathway, can rescue BCR− B cells.32 The rescued BCR− B cells, like the B cells from our patients, did not express activation markers but showed decreased expression of CD62L. Older data suggest that there may be other ways for BCR− B cells to survive in the periphery.
In the 1990s, several strains of transgenic mice were produced that bypass the requirement for a BCR. Alt et al showed that mice expressing an Ha-ras transgene on a RAG− or JH− background developed BCR− B cells in the spleen and lymph nodes in numbers that were similar to those of wild-type mice.33 Desiderio et al produced mice transgenic for a constitutively active Blk, a B-cell–specific src family member, on a μMT background.34,35 These mice, which were highly prone to B-cell lymphomas, also had BCR− B cells in the spleen. Finally, Longnecker et al produced mice transgenic for the EBV-derived latent membrane protein 2A (LMP2A).36,37 LMP2A is a transmembrane protein with an N-terminal cytoplasmic ITAM motif that allows it to mimic pre-BCR and BCR signaling.38 On both a wild-type and a RAG− background, the LMP2A mice accumulated BCR− B cells in the periphery. The studies on the LMP2A transgenic mice are particularly relevant to the results reported in the present study. Several strains of LMP2A-transgenic mice were produced, expressing various amounts of LMP2A.37 The mice that expressed the most LMP2A had the lowest number of splenic B cells and the highest proportion of BCR− B cells. These BCR− B cells expressed higher-density surface CD19.
In the present study, the increased intensity of CD19 expression on the surface of the B-lineage cells in the BCR− patients supports the hypothesis that there is constitutive and increased signaling through the BCR pathway. Patients with gene defects that are known to impair signaling through the BCR have B cells with decreased expression of CD19. Decreased expression of CD19 was not seen on B cells from the peripheral blood of Btk-knockout mice (unpublished personal observation), perhaps reflecting the milder effect of Btk mutations in murine B-cell development. In contrast, increased expression of CD19 was noted in transgenic mice with constitutive activation of the BCR pathway mediated by LMP2.37
CD19, along with CD21, CD81, and CD225, forms a transmembrane complex that amplifies signals delivered through the BCR.39-42 The cytoplasmic tail of CD19 contains a series of tyrosines that act as docking sites for VAV, lyn, and PI3K.43 The factors that control the amount of CD19 expressed on the B-cell surface are not well understood. Patients or mice that lack the tetraspanin CD81 demonstrate decreased or absent expression of CD19,44,45 indicating that CD81 either plays a role in escorting CD19 to the cell surface or stabilizes CD19 once it is on the cell surface.46 Studies on cell lines suggest that internalization of CD19 is inversely correlated with the amount of CD21 on the cell surface.47
In cultures of human BM cells, the amount of CD19 on the cell surface is enhanced by stimulation with IL-7,48 indicating that the expression of CD19 is at least partly controlled by the activation status of the cells. It is not clear whether this control occurs at the transcriptional level or the protein level. B cells from individuals who are heterozygous for truncating mutations in CD19 demonstrate reduced expression of CD19.49 Studies in mice that are transgenic for high copy numbers of human CD19 have shown that the amount of CD19 transcribed has a direct effect on the amount of CD19 on the cell surface.50 Mice that are homozygous for the transgenes have almost twice the amount of CD19 on B220+ B cells as heterozygous mice. Overexpression of CD19 is associated with decreased numbers of circulating B cells. Mice with the highest copy number of human CD19 had < 20% of the normal number of B cells in the blood.50
To allow both positive and negative selection, successful B-cell development permits only a narrow window of signaling intensity activated by the BCR. Decreased signal intensity, as is seen in patients with mutations in Btk or BLNK, results in a marked decrease in B-cell numbers. Animal models suggest that increased intensity of signaling through the BCR also results in decreased numbers of B cells. Our data are consistent with the hypothesis that some patients have agammaglobulinemia and absent B cells as a consequence of constitutive activation through the BCR pathway. A better understanding of the control of this pathway may provide ways to manipulate the immune system in patients with autoimmune disease or may aid in vaccine development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for their willingness to participate in these studies; Dana Farmer and Anita Quintana for helpful discussions; Vanessa Howard for editorial assistance; and Betsy Williford and Janis Adams for help in preparation of the manuscript.
These studies were supported in part by grants from the National Institutes of Health (AI25129), the National Cancer Institute (P30 CA21765), the American Lebanese Syrian Associated Charities, and the Federal Express Chair of Excellence.
National Institutes of Health
Authorship
Contribution: A.K.D. designed the experiments, performed the research, analyzed the data, and assisted in manuscript preparation; A.B. performed the research and assisted in manuscript preparation; E.C.-S. designed, performed, and helped to analyze the data related to flow cytometric analysis of the BM; G.T. and F.T.S. provided the clinical data; and M.E.C. designed the experiments, analyzed the data, provided the clinical data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary Ellen Conley, MD, University of Tennessee College of Medicine, St Jude Children's Research Hospital, 226 Danny Thomas Pl, Memphis, TN 38105; e-mail: maryellen.conley@stjude.org.