Abstract
The Plasmodium falciparum adhesin PfRh4 binds to complement receptor type-1 (CR1) on human erythrocytes and mediates a glycophorin-independent invasion pathway. CR1 is a complement regulator and immune-adherence receptor on erythrocytes required for shuttling of C3b/C4b-opsonized particles to liver and spleen for phagocytosis. Using recombinant CR1 constructs, we mapped the recognition site for PfRh4 to complement control protein modules 1 to 3 (CCP1-3) at the membrane-distal amino terminus of CR1. This region of CR1 binds to C4b and C3b and accelerates decay of both classic pathway and alternative pathway C3 and C5 convertases. CCP1-3 competed for PfRh4 binding to erythroid CR1 and inhibited the PfRh4-CR1 invasion pathways across a wide range of P falciparum strains. PfRh4 did not bind significantly to other CR1 constructs, including CCP15-17, which is 85% identical to CCP1-3. PfRh4 binding to CR1 did not affect its C3b/C4b binding capability, and we show evidence for a ternary complex between CCP1-3, C4b, and PfRh4. PfRh4 binding specifically inhibited CR1's convertase decay-accelerating activity, whereas there was no effect on factor H-mediated decay-accelerating activity. These results increase our understanding of the functional implications of CR1 engagement with PfRh4 and highlight the interplay between complement regulation and infection.
Introduction
The complement system is a first line of defense against invasion by infectious agents. On pathogen entry into the host and detection of a pathogen-associated or danger-associated molecular pattern, the complement cascade is activated in seconds and results in the production of anaphylatoxins, deposition of opsonic C3 and C4 fragments, and assembly of the potentially cytolytic membrane attack complex. By ensuring that the complement system acts in a directed manner, the regulators of complement activation (RCA) protein family protect self-tissue from complement-mediated attack.1 Interestingly, RCA family members also contribute to cell attachment or invasion strategies of disparate pathogens, including multiple viruses and bacteria.2-6 Recently, complement receptor type-1 (CR1), an erythroid membrane-bound RCA protein, was shown to be a receptor used by the malaria parasite Plasmodium falciparum for invasion of human erythrocytes.7,8
Invasion of human erythrocytes by malaria parasites depends on specific interactions between parasite adhesins and host receptors. In P falciparum, 2 gene families encode important parasite proteins that engage with erythrocyte receptors: the erythrocyte binding-like antigens (PfEBAs; these include EBA-140/BAEBL, EBA-175, EBA-181/JESEBL, and EBL-1); and reticulocyte binding-like homolog proteins (RBPs or PfRhs; these include PfRh1, PfRh2a, PfRh2b, PfRh4 and PfRh5).9-12 During invasion these adhesins localize to the apical tip of the merozoite and interact with specific host receptors to initiate parasite entry. Invasion pathways are defined by examining P falciparum entry into erythrocytes deficient or mutant in host receptors, or treated with enzymes that modify the properties or presence of erythrocyte surface proteins. Commonly used enzymes are neuraminidase (nm) to remove sialic acid residues and trypsin or chymotrypsin to cleave peptide backbones. The PfEBA family of proteins predominantly bind glycophorins on the erythrocyte surface; glycophorin A for EBA-175, glycophorin B for EBL-1, and glycophorin C for EBA-140.13-15 All 3 interactions are sensitive to nm treatment of erythrocytes and thus categorized as sialic acid–dependent invasion pathways. Recent investigations demonstrate that the parasite adhesin PfRh4 binds specifically to CR1 to mediate an invasion pathway that is nm-insensitive.8 The CR1-PfRh4 pathway has therefore emerged as the first major sialic acid–independent alternative to glycophorin-mediated invasion.7,8
CR1 and other members of the RCA family consist almost entirely of independently folding modules called short consensus repeats, sushi domains, or complement control protein modules (CCP).16 The ectodomain of the most common CR1 allelic variant is composed of 30 CCPs, each consisting of approximately 60 to 70 amino acid residues (Figure 1A, reviewed in Krych-Goldberg and Atkinson17 ). The first 28 CCPs are further organized into 4 long homologous repeats (LHR) of 7 CCPs each. The functional domains of CR1 reside predominantly within the 3 most amino-terminal CCPs of LHRs A, B, and C.18,19 Binding and transport of C3b/C4b-opsonized immune complexes (immune adherence) is one of the main functions of erythroid CR1.20 Site 1, which is present within CCP1-3, binds to C4b and weakly to C3b. Site 2, located on both CCP8-10 and CCP15-17, binds both C3b and C4b efficiently.18,21 All 3 sites are highly similar, and CCP8-10 and CCP15-17 are nearly identical. In addition to C3b/C4b-binding, CR1 has multiple complement regulatory activities. Decay-accelerating activity (DAA) for both classic pathway and alternative pathway C3 and C5 convertases resides predominantly within Site 1.22,23 Site 2 is mainly responsible for factor I–mediated cleavage of C3b and C4b.24,25 These activities result in down-regulation of the complement cascade, allowing CR1 to prevent adverse effects of activated complement on erythrocytes and other host cells.
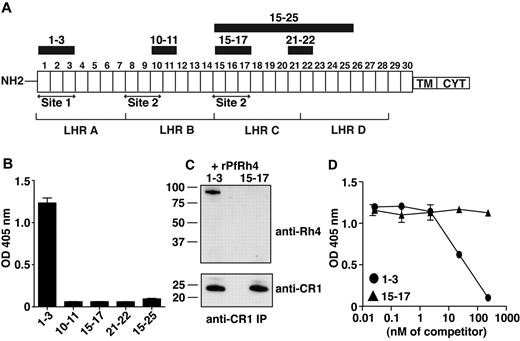
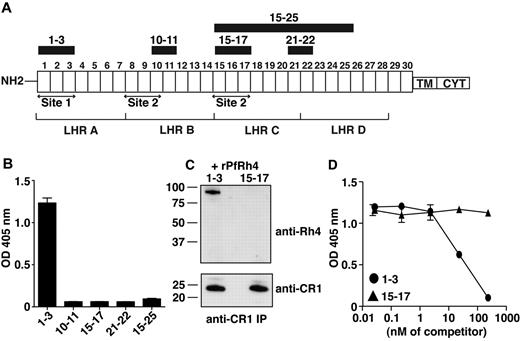
PfRh4 interacts with the N-terminal 3 CCPs of CR1. (A) Schematic of CR1 polypeptide. Each box labeled 1 to 30 represents a complement control protein module (CCP) of 60-70 amino acid residues. The first 28 CCPs are organized based on homology into 4 long homologous repeats (LHRs) A-D, each consisting of 7 CCPs. The functional sites of CR1 are labeled site 1 and site 2. The recombinant CR1 fragments used in this study are indicated as black bars with relevant CCPs labeled above. NH2 indicates amino terminus; TM, transmembrane domain; and CYT, cytoplasmic tail. (B) Recombinant PfRh4 binds to CCP1-3. An ELISA was used to measure the interaction between CR1 fragments and rPfRh4. Microtiter wells were coated with CCP1-3, CCP10-11, CCP15-17, CCP21-22, or CCP15-25 at 0.5 μg/well. Recombinant PfRh4 was added at 0.5 μg/well. Bound PfRh4 was detected with an anti-PfRh4 monoclonal antibody 10C9. (C) Recombinant PfRh4 forms a complex with CCP1-3 but not CCP15-17. Immunoprecipitation experiments were performed in which combinations of recombinant proteins CCP1-3/rPfRh4 or CCP15-17/rPfRh4 were incubated with an anti-CR1 monoclonal antibody 1B4. For Western blot analyses of immunoprecipitated eluates, soluble CR1 fragments were detected with anti-CR1 monoclonal antibody 1B4 and rPfRh4 was detected with 10C9 monoclonal antibody. (D) Binding of sCR1 to rPfRh4 was inhibited by CCP1-3. Microtiter plates were coated with saturating concentrations of rPfRh4 (5 μg/well). CCP1-3 or CCP15-17 at concentrations of 0nM, 0.02nM, 0.23nM, 2.3nM, 23nM, or 234nM was incubated with sCR1 (23nM) before addition to wells. Interaction between sCR1 and PfRh4 was detected using anti-CR1 antibody HB8592, which detected sCR1 and not smaller CR1 fragments. ELISA experiments in panels B and D were repeated with similar results and their y-axis represent A405 nm with error bars showing the range of duplicate readings.
PfRh4 interacts with the N-terminal 3 CCPs of CR1. (A) Schematic of CR1 polypeptide. Each box labeled 1 to 30 represents a complement control protein module (CCP) of 60-70 amino acid residues. The first 28 CCPs are organized based on homology into 4 long homologous repeats (LHRs) A-D, each consisting of 7 CCPs. The functional sites of CR1 are labeled site 1 and site 2. The recombinant CR1 fragments used in this study are indicated as black bars with relevant CCPs labeled above. NH2 indicates amino terminus; TM, transmembrane domain; and CYT, cytoplasmic tail. (B) Recombinant PfRh4 binds to CCP1-3. An ELISA was used to measure the interaction between CR1 fragments and rPfRh4. Microtiter wells were coated with CCP1-3, CCP10-11, CCP15-17, CCP21-22, or CCP15-25 at 0.5 μg/well. Recombinant PfRh4 was added at 0.5 μg/well. Bound PfRh4 was detected with an anti-PfRh4 monoclonal antibody 10C9. (C) Recombinant PfRh4 forms a complex with CCP1-3 but not CCP15-17. Immunoprecipitation experiments were performed in which combinations of recombinant proteins CCP1-3/rPfRh4 or CCP15-17/rPfRh4 were incubated with an anti-CR1 monoclonal antibody 1B4. For Western blot analyses of immunoprecipitated eluates, soluble CR1 fragments were detected with anti-CR1 monoclonal antibody 1B4 and rPfRh4 was detected with 10C9 monoclonal antibody. (D) Binding of sCR1 to rPfRh4 was inhibited by CCP1-3. Microtiter plates were coated with saturating concentrations of rPfRh4 (5 μg/well). CCP1-3 or CCP15-17 at concentrations of 0nM, 0.02nM, 0.23nM, 2.3nM, 23nM, or 234nM was incubated with sCR1 (23nM) before addition to wells. Interaction between sCR1 and PfRh4 was detected using anti-CR1 antibody HB8592, which detected sCR1 and not smaller CR1 fragments. ELISA experiments in panels B and D were repeated with similar results and their y-axis represent A405 nm with error bars showing the range of duplicate readings.
Previous studies show that P falciparum merozoites are engulfed by neutrophils.26,27 Opsonized merozoites treated with normal serum were ingested at a higher rate than those treated with heat-inactivated (or complement-inactivated) serum.26,27 Because merozoites are likely to be decorated with activated complement fragments that are also ligands for erythroid CR1, it is of interest to discover the effect of the binding of PfRh4 to CR1 on these other host–parasite interactions. To this end, we used a variety of soluble recombinant CR1 fragments to map the region within full-length receptors involved in parasite invasion, and to determine the functional implications for CR1 of PfRh4 binding.
Methods
Recombinant proteins
Cloning, production, and purification of recombinant PfRh4 (rPfRh4; previously known as Rh4.9) and a hexaHis-tagged control protein were described previously (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).8,28 The full-length recombinant CR1 ectodomain (sCR1) was a gift from Dr Henry C. Marsh (Celldex Therapeutics, Needham, MA). The DNA encoding the various CCP fragments was cloned into expression vector pPICZαB (Invitrogen), and resultant plasmids were transformed into Pichia pastoris strain KM71H cells (Invitrogen). Purification steps were described previously.29 Nuclear magnetic resonance spectroscopy, where possible subject to size restraints, was used to confirm folding. 1H, 15N heteronuclear single quantum coherence spectra (data not shown) obtained for CCP1-3, CCP15-17, and CCP20-21 were fully consistent with compactly folded protein domains. Dynamic light scattering and analytical ultacentrifugation data (not shown) were consistent with lack of self-association or aggregation of the recombinant proteins.
ELISA
ELISA was performed as described.8 Recombinant PfRh4 binding was detected using anti-PfRh4 monoclonal antibody 10C9. For competitive binding experiments, microtiter wells were coated with rPfRh4, and sCR1 was added at a final concentration of 23nM together with 0nM, 0.02nM, 0.23nM, 2.3nM, 23nM, or 234nM of either CCP1-3 or CCP15-17. Binding of sCR1 was detected using an anti-CR1 monoclonal antibody, HB8592 (ATTC), which recognizes sCR1 but not CCP1-3 or CCP15-17.30
In vitro immunoprecipitation
For immunoprecipitation, recombinant proteins (2 μg/mL) were incubated together in T-NET buffer (1% Triton vol/vol X-100, 150mM NaCl, 10mM EDTA, 50mM Tris, pH 7.4). Immunoprecipitation was performed using anti-CR1 monoclonal antibody 1B4 coupled to Protein G/A Sepharose beads (Crosslink IP Kit; Pierce). Beads were washed and proteins eluted. Eluted proteins were resuspended in sample buffer and analyzed by SDS-PAGE (Invitrogen). For Western blots, rPfRh4 and CR1 constructs were detected using the anti-PfRh4 mouse monoclonal antibody 10C9 and anti-CR1 monoclonal antibody 1B4 (a gift from Prof Ronald Taylor, University of Virginia).8,30
Standard erythrocyte-binding and inhibition assay
Erythrocyte-binding assays were performed as described.31 Competitive binding assays were performed by incubating the relevant recombinant CR1 fragments, C3b or C4b, with culture supernatants before proceeding with standard erythrocyte-binding assays. Final concentrations of CCP1-3, CCP15-17, C3b, and C4b in these assays were calculated as μg of soluble protein per 250 μL of culture supernatants.
Parasite culture
Growth inhibition assay
Surface plasmon resonance
Binding between rPfRh4 and recombinant CR1 fragments was analyzed as described.8 To analyze the influence of rPfRh4 on ability of sCR1 to bind C3b and C4b, 1860 RUs of C3b and 1800 RUs of C4b were immobilized on 2 different flow cells of a CM5 chip. C3b, C4b, factor B and factor D were from Complement Technology. We prepared 2 sCR1 concentration series (0.01μM, 0.1μM, 1μM, 2μM, and 4μM), one of which (un-complexed) was free of rPfRh4, and the other (complexed) of which contained a 2.3-fold molar excess of rPfRh4.
Decay accelerating activity assayed by SPR
A real-time surface plasmon resonance (SPR)–based assay to study convertase decay-accelerating activity was performed.29,36,37 A CM5-sensorchip was used on which 1860 RUs of C3b had been immobilized via standard amine coupling. A mixture of factor B and factor D flowed over the C3b-coated surface for 120 seconds at a rate of 10 μL/min to form C3 convertase. After observing spontaneous decay for 210 seconds, sCR1, a complex of sCR1–rPfRh4, CCP1-3, a complex of CCP1-3–rPfRh4, or FH CCP1-4 (FH1-4) was injected in duplicate for 60 seconds. For comparison, samples flowed over the C3b-coated surface when no convertase was present. Between injections, the surface was regenerated using FH1-4, followed by injection of 1.0M NaCl.
Hemolysis assays
The classic pathway C3 convertase (C4bC2a) was assembled on the surface of antibody-sensitized sheep erythrocytes (EA; Complement Technology) by the sequential addition of purified human components C1, C4 and C2, as previously described.23 These cells (EAC142) were then added to purified CCP1-3 or purified CCP1-3 that had been preincubated with rPfRh4. Samples were next incubated to allow the convertase to decay. The cells were then lysed with guinea pig serum and hemolysis monitored at 414nM. A convertase was not formed in the no-C2 control.
For the alternative pathway convertase, EAC142 were prepared and C3 was added; cation-mediated depletion with EDTA to dissociate C1 and C2a was then carried out.23 The EAC43b cells thus generated were combined with alternative pathway components factor B, factor D, and properdin to form the alternative pathway C3 convertase, C3bBbP. These cells were incubated with CCP1-3 or with CCP1-3, which had been preincubated with rPfRh4. After an incubation to allow the convertase to decay, cells were lysed and hemolysis monitored at 414nM. A convertase was not formed in the absence of factor B.
Results
Recombinant PfRh4 binds to the N-terminal 3 modules of CR1
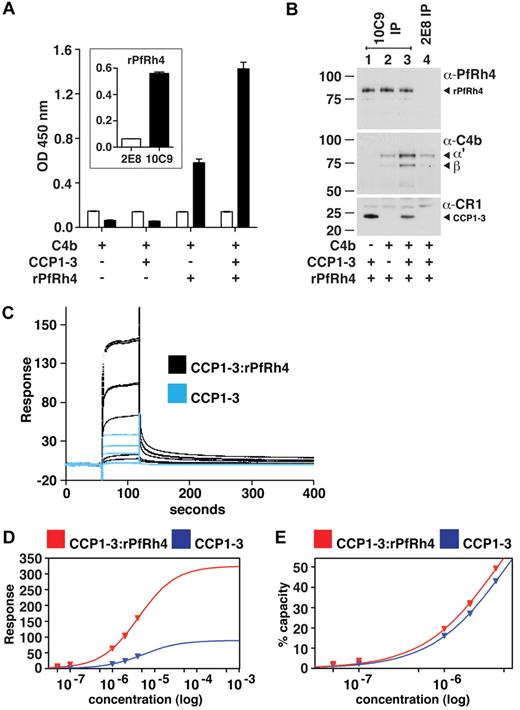
To examine whether PfRh4 binds to CR1's immune regulatory sites, we used ELISA to test the ability of recombinant proteins CCP1-3 (site 1) and CCP15-17 (site 2) to interact with the erythrocyte-binding domain of PfRh4 (Figure 1A-B).31 Recombinant PfRh4 (rPfRh4) bound to CCP1-3, but did not bind detectably to CCP15-17. In addition, as tested by ELISA, rPfRh4 did not bind to CCP10-11 (almost identical to CCP17-18), CCP21-22, or CCP15-25 (Figure 1B). To confirm the specificity of the CCP1-3–rPfRh4 interaction, we performed coimmunoprecipitation experiments in which combinations of either CCP1-3 and rPfRh4 or CCP15-17 and rPfRh4 were incubated with the anti-CR1 monoclonal antibody 1B4 (Figure 1C). Both CCP1-3 and the highly similar CCP15-17 were immunoprecipitated by the anti-CR1 antibody, but rPfRh4 only coprecipitated with CCP1-3.
To assess whether CCP1-3 inhibits the previously reported sCR1:rPfRh4 interaction, considered critical to sialic acid–independent invasion, we performed competition ELISAs (Figure 1D).8 A 23nM sCR1 sample was incubated with rPfRh4, along with a concentration series of CCP1-3 or of CCP15-17. Addition of the 3-module CR1 fragments at < 2.3nM had no detectable effect on the interaction. However, at 23nM (in other words, equimolar with sCR1), CCP1-3 competitively inhibited sCR1 binding to rPfRh4, whereas 23nM CCP15-17 did not. Addition of CCP1-3 in tenfold excess outcompeted sCR1 binding to rPfRh4, but the same excess of CCP15-17 had little detectable effect (Figure 1D). These experiments show that CCP1-3 is the dominant site within CR1 to which PfRh4 binds.
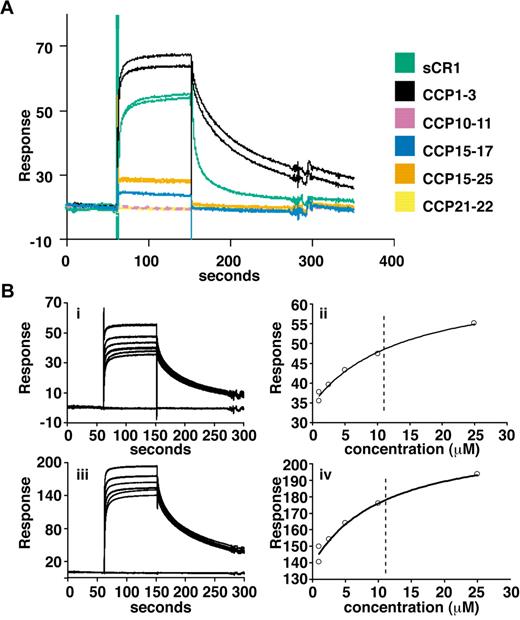
To obtain more quantitative data, surface plasmon resonance (SPR) was used to compare binding of sCR1 and our panel of smaller CR1 derivatives to rPfRh4 amine coupled to a CM5 chip (Figure 2A). CCP1-3 and sCR1 both bound to the immobilized rPfRh4 strongly compared with other fragments tested, including those from LHRs B-D (CCP10-11, CCP15-17, CCP21-22, and CCP15-25; Figure 2A, supplemental Figure 1) that bound weakly. Additional experiments using a concentration series of CCP1-3 and different loadings of rPfRh4 on the chip yielded KD values of 10-11μM (Figure 2B). The SPR data are thus in agreement with the results obtained using coimmunoprecipitation and ELISA, demonstrating that CCP1-3 located at the membrane-distal N-terminus of CR1's ectodomain was the primary region for binding to P falciparum adhesin PfRh4.
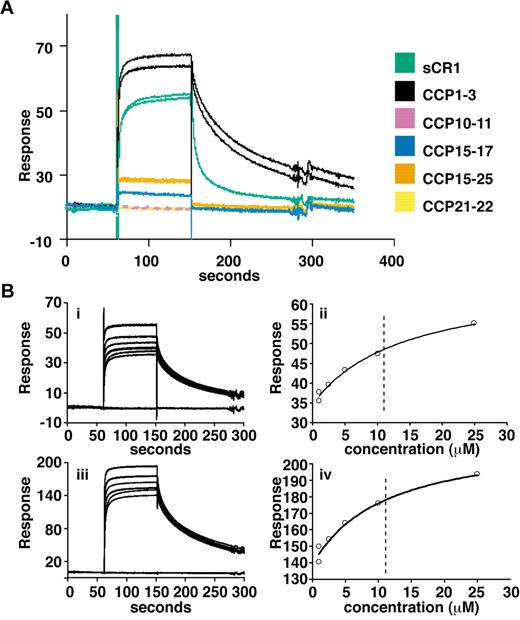
Delineation of PfRh4 binding sites on CR1 and KD measurements by SPR. (A) Recombinant PfRh4 binds to sCR1 and CCP1-3 as shown by SPR. Duplicate injections of sCR1 and smaller CR1 fragments, all at a concentration of 5μM, were performed over a CM5 chip that was coupled with rPfRh4. A small fraction of inactive rPfRh4 present on the chip surface triggers nonspecific background binding for the first analyte samples to be assayed (CCP1-3 and sCR1), as manifested in imperfect reproducibility of the sensorgrams. After saturation of this nonspecific binding capacity, duplicate injections are of acceptable reproducibility (all other constructs). (B) Use of SPR to measure the dissociation constant (KD) of CCP1-3 for PfRh4. The left-hand panels show duplicate sensorgrams for a range of increasing CCP1-3 concentrations (1.0μM, 2.5μM, 5.0μM, 10.0μM, and 25.0μM, bottom to top) flowing over a CM5-chip surface with a loading of 410 RUs (i) and 1480 RUs (iii) of rPfRh4. The right-hand panels show plots of the RUs versus CCP1-3 on 2 different flow cells, coupled with 410 RUs (ii) and 1480 RUs (iv) of rPfRh4. The dashed vertical line indicates the KD fitted to both plots simultaneously. In all panels, blank-subtracted sensorgrams are shown.
Delineation of PfRh4 binding sites on CR1 and KD measurements by SPR. (A) Recombinant PfRh4 binds to sCR1 and CCP1-3 as shown by SPR. Duplicate injections of sCR1 and smaller CR1 fragments, all at a concentration of 5μM, were performed over a CM5 chip that was coupled with rPfRh4. A small fraction of inactive rPfRh4 present on the chip surface triggers nonspecific background binding for the first analyte samples to be assayed (CCP1-3 and sCR1), as manifested in imperfect reproducibility of the sensorgrams. After saturation of this nonspecific binding capacity, duplicate injections are of acceptable reproducibility (all other constructs). (B) Use of SPR to measure the dissociation constant (KD) of CCP1-3 for PfRh4. The left-hand panels show duplicate sensorgrams for a range of increasing CCP1-3 concentrations (1.0μM, 2.5μM, 5.0μM, 10.0μM, and 25.0μM, bottom to top) flowing over a CM5-chip surface with a loading of 410 RUs (i) and 1480 RUs (iii) of rPfRh4. The right-hand panels show plots of the RUs versus CCP1-3 on 2 different flow cells, coupled with 410 RUs (ii) and 1480 RUs (iv) of rPfRh4. The dashed vertical line indicates the KD fitted to both plots simultaneously. In all panels, blank-subtracted sensorgrams are shown.
CCP1-3 inhibits binding of PfRh4 to erythrocytes and PfRh4-CR1 invasion pathway of P falciparum
We showed previously that sCR1 inhibits PfRh4 binding to erythroid CR1.8 To determine whether CCP1-3 had a similar effect, culture supernatants enriched with parasite ligands were preincubated with either CCP1-3 or CCP15-17 before addition of erythrocytes for binding assays. CCP1-3 blocked native PfRh4 binding to erythroid CR1, with approximately 90% inhibition obtained at 8 μg/mL CCP1-3 (Figure 3A). In contrast, the same concentration of CCP15-17 did not affect binding of native PfRh4 to erythrocytes. These results are consistent with selectivity of PfRh4 for binding to site 1 (CCP1-3) over site 2 (CCP15-17) of CR1. In control experiments, we showed that binding of EBA-175, which binds to glycophorin A, to erythrocytes was unperturbed by addition of CR1-derived protein fragments, providing further evidence for specificity of the CR1-PfRh4 interaction (Figure 3A).
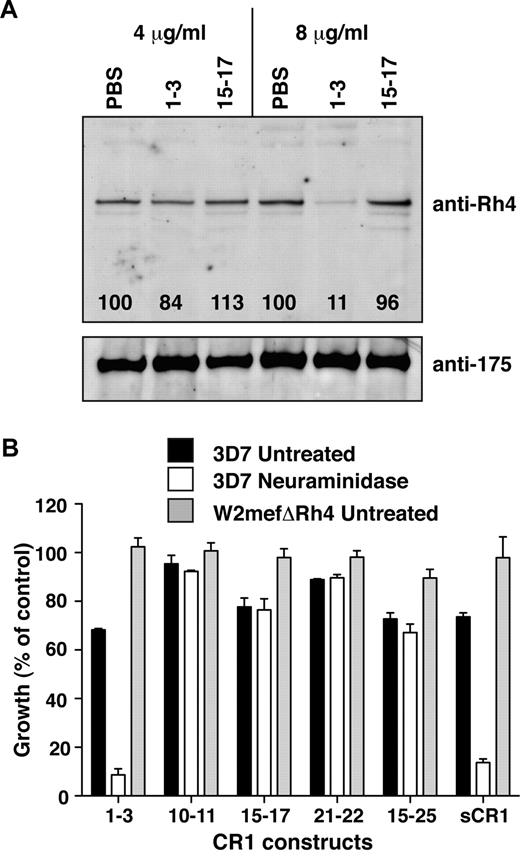
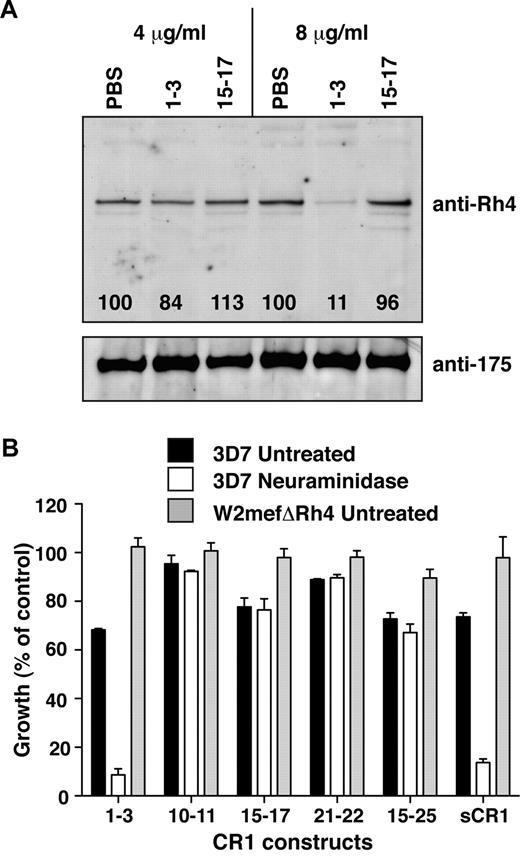
CCP1-3 inhibits native PfRh4 erythrocyte binding and the CR1-PfRh4 invasion pathway. (A) Native PfRh4 binding to erythroid CR1 was inhibited by CCP1-3. Competitive binding assays were performed by incubating either CCP1-3 or CCP15-17 with invasion supernatants at the stated final concentrations of 4 μg/mL or 8 μg/mL. The black numbers in the top panel represent the percentage of PfRh4 binding relative to PBS for each concentration as determined by densitometry. Immunodetection of parasite proteins with anti-PfRh4 or anti-EBA-175 antibodies after erythrocyte binding is shown. (B) The PfRh4 invasion pathway was inhibited in the presence of CCP1-3. Parasite strains W2mefΔRh4 (gray bars, untreated erythrocytes) and 3D7 (black bars for untreated, white bars for nm-treated erythrocytes) were tested in growth assays in the presence of final concentrations of 0.5 mg/mL CCP1-3, CCP10-11, CCP15-17, CCP21-22, CCP15-25, or sCR1. Growth (percentage of control) on the y-axis refers to the percentage of parasitemia in the presence of CR1 constructs relative to the percentage of parasitemia with the addition of PBS (arbitrarily set to be 100%).
CCP1-3 inhibits native PfRh4 erythrocyte binding and the CR1-PfRh4 invasion pathway. (A) Native PfRh4 binding to erythroid CR1 was inhibited by CCP1-3. Competitive binding assays were performed by incubating either CCP1-3 or CCP15-17 with invasion supernatants at the stated final concentrations of 4 μg/mL or 8 μg/mL. The black numbers in the top panel represent the percentage of PfRh4 binding relative to PBS for each concentration as determined by densitometry. Immunodetection of parasite proteins with anti-PfRh4 or anti-EBA-175 antibodies after erythrocyte binding is shown. (B) The PfRh4 invasion pathway was inhibited in the presence of CCP1-3. Parasite strains W2mefΔRh4 (gray bars, untreated erythrocytes) and 3D7 (black bars for untreated, white bars for nm-treated erythrocytes) were tested in growth assays in the presence of final concentrations of 0.5 mg/mL CCP1-3, CCP10-11, CCP15-17, CCP21-22, CCP15-25, or sCR1. Growth (percentage of control) on the y-axis refers to the percentage of parasitemia in the presence of CR1 constructs relative to the percentage of parasitemia with the addition of PBS (arbitrarily set to be 100%).
To investigate further the potential of CCP1-3 to interfere with PfRh4-CR1 invasion pathway, parasite growth was monitored in the presence of our panel of recombinant CR1-derived protein. Invasion of both untreated and nm-treated erythrocytes by 3D7 strain was inhibited by addition of sCR1 (Figure 3B). Invasion of untreated erythrocytes by the parasite strain W2mefΔRh4, which lacks a functional CR1-PfRh4 pathway, was unaffected by the presence of sCR1 (Figure 3B).8 These results confirmed previous studies and acted as a benchmark for experiments with the smaller CR1 fragments.7,8
CCP1-3 was as effective in inhibiting invasion by 3D7 of untreated erythrocytes as sCR1 (Figure 3B). CCP1-3, like sCR1, did not affect W2mefΔRh4 growth. The level of inhibition observed in the case of 3D7 was remarkable when it is considered that multiple alternative invasion pathways remain functional. To study the contribution of PfRh4-CR1 pathway in the absence of various PfEBA-glycophorin pathways, we examined CCP1-3 inhibition of parasite growth in nm-treated erythrocytes. Invasion by 3D7 parasites into nm-treated erythrocytes was inhibited by approximately 90% after the addition of CCP1-3 (Figure 3B). Although CCP10-11, CCP15-17, and CCP21-22 inhibited invasion of untreated erythrocytes to some extent, we did not observe an enhanced inhibitory effect on invasion of nm-treated erythrocytes. These results suggested that the small effects observed on parasite growth of these other non–site 1 fragments were not specific to PfRh4-CR1–mediated invasion pathway (Figure 3B). Taken together, these results show that the CCP1-3–PfRh4 interaction is critical to the PfRh4-mediated invasion pathway of P falciparum into human erythrocytes.
PfRh4-CR1 is the major sialic acid–independent invasion pathway in P falciparum
P falciparum parasite strains have different protein expression levels and carry polymorphism in the genes encoding PfRhs and PfEBAs. These differences determine the varying propensity of strains to invade using alternative invasion pathways, such as glycophorin A-EBA175, rather than CR1-PfRh4.32,33,38,39 To evaluate the importance of CR1-PfRh4 invasion pathway in a range of P falciparum strains, we used CCP1-3 and took advantage of its ability to act as a potent inhibitor of this ligand–receptor interaction.
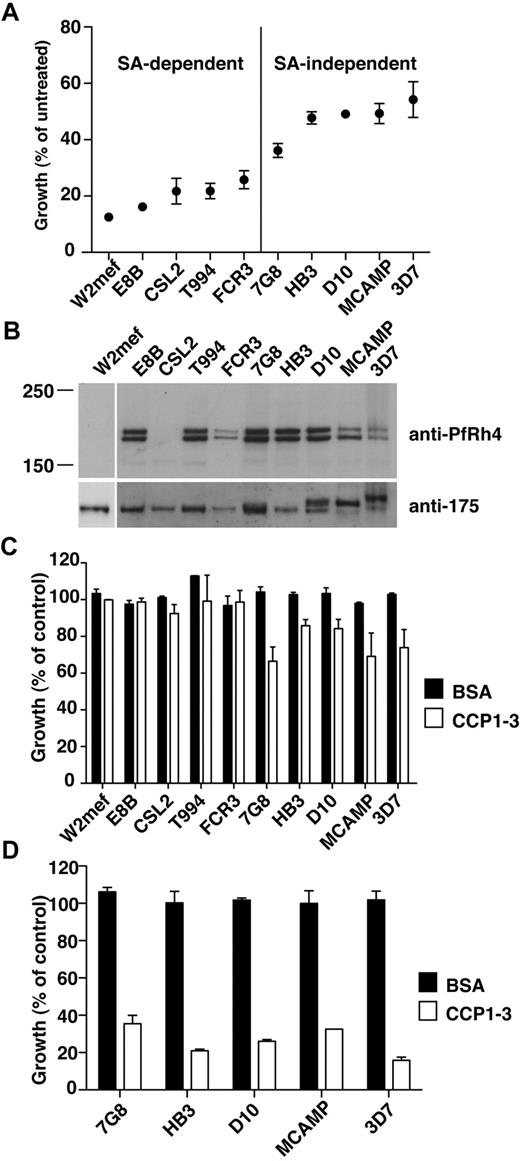
We first determined the ability of various parasite strains to invade nm-treated erythrocytes with respect to the level of production of this adhesin protein. The P falciparum strains W2mef, E8B, CSL2, T994, and FCR3 showed low efficiency of invasion into nm-treated erythrocytes and may therefore be considered sialic acid–dependent (Figure 4A). In contrast, 7G8, HB3, D10, MCAMP, and 3D7 invaded nm-treated erythrocytes with a higher efficiency and were classified as sialic acid–independent (Figure 4A). Western analyses of PfRh4 protein production showed that all sialic acid–independent strains express PfRh4 (Figure 4B). In the case of sialic acid–dependent strains, neither W2mef nor CSL2 showed PfRh4 expression, in agreement with previous work.33 Note that other sialic acid–dependent strains examined in this study (E8B, FCR3, and T994) also express PfRh4, despite showing less invasion into nm-treated erythrocytes (Figure 4A-B).
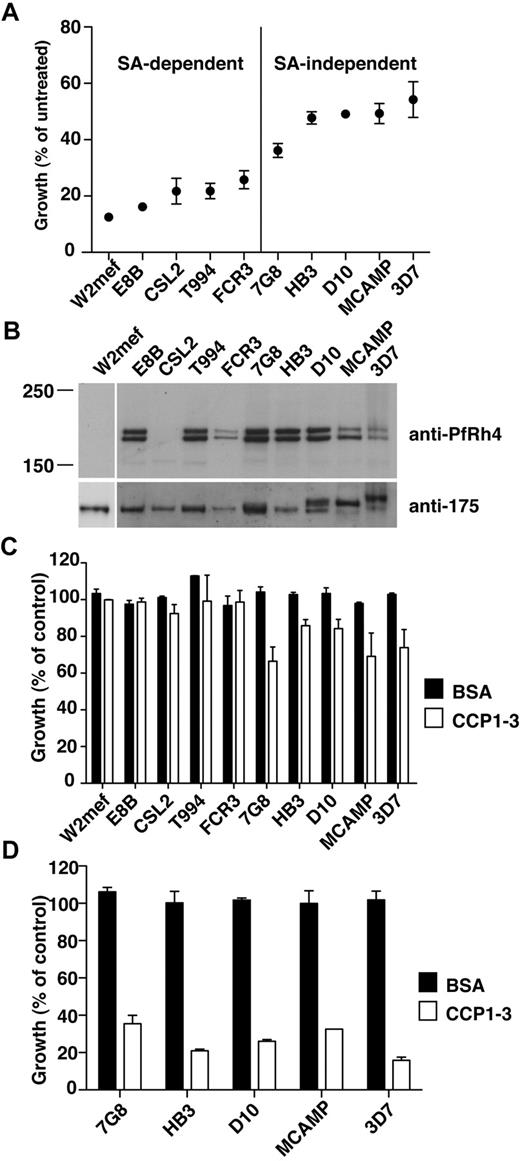
PfRh4-CR1 pathway constitutes the majority of sialic acid-independent invasion events. (A) Sialic acid–dependent and –independent parasite strains. Parasite strains were assayed for their ability to invade nm-treated erythrocytes. The y-axis represents percentage of growth of parasites into nm-treated erythrocytes relative to growth of parasites into untreated erythrocytes. (B) Expression of PfRh4 in P falciparum strains. Western blot of saponin-lysed schizont pellets probed with anti-PfRh4 monoclonal antibody 2E8 (top panel) and anti–EBA-175 rabbit polyclonal antibody (bottom panel). (C) Growth of sialic acid–independent strains in untreated erythrocytes was slightly inhibited in the presence of CCP1-3. Parasite strains were tested in growth assays into untreated erythrocytes in the presence of 0.5 mg/mL final concentration of CCP1-3 (white bars) or BSA (black bars). Growth (percentage of control) on the y-axis refers to the percentage parasitemia in the presence of CCP1-3 or BSA relative to the percentage parasitemia with the addition of PBS, which is arbitrarily set at 100%. (D) Growth of sialic acid-independent strains in nm-treated erythrocytes was inhibited in the presence of CCP1-3. Parasite strains were tested in growth assays using nm-treated erythrocytes in the presence of 0.5 mg/mL CCP1-3 (white bars) or BSA (black bars).
PfRh4-CR1 pathway constitutes the majority of sialic acid-independent invasion events. (A) Sialic acid–dependent and –independent parasite strains. Parasite strains were assayed for their ability to invade nm-treated erythrocytes. The y-axis represents percentage of growth of parasites into nm-treated erythrocytes relative to growth of parasites into untreated erythrocytes. (B) Expression of PfRh4 in P falciparum strains. Western blot of saponin-lysed schizont pellets probed with anti-PfRh4 monoclonal antibody 2E8 (top panel) and anti–EBA-175 rabbit polyclonal antibody (bottom panel). (C) Growth of sialic acid–independent strains in untreated erythrocytes was slightly inhibited in the presence of CCP1-3. Parasite strains were tested in growth assays into untreated erythrocytes in the presence of 0.5 mg/mL final concentration of CCP1-3 (white bars) or BSA (black bars). Growth (percentage of control) on the y-axis refers to the percentage parasitemia in the presence of CCP1-3 or BSA relative to the percentage parasitemia with the addition of PBS, which is arbitrarily set at 100%. (D) Growth of sialic acid-independent strains in nm-treated erythrocytes was inhibited in the presence of CCP1-3. Parasite strains were tested in growth assays using nm-treated erythrocytes in the presence of 0.5 mg/mL CCP1-3 (white bars) or BSA (black bars).
Having established their sialic acid dependencies and PfRh4 expression levels, we examined whether the addition of CCP1-3 affected parasite invasion in these strains. In the case of sialic acid–dependent strains, addition of CCP1-3 did not adversely affect parasite invasion of untreated erythrocytes, suggesting that other pathways are functionally dominant (Figure 4C). For sialic acid–independent strains, we observed some inhibition by CCP1-3 of invasion into untreated erythrocytes (Figure 4C). Invasion of nm-treated erythrocytes by these strains was inhibited 65%-85% by CCP1-3 (Figure 4D). In all assays, the addition of BSA had no effect on parasite growth (Figure 4C-D). This suggests that the PfRh4-CR1 adhesin-receptor interaction was important for sialic acid–independent invasion of erythrocytes for a wide range of P falciparum strains.
C3b/C4b-CR1 interaction is unperturbed by binding of PfRh4
To examine whether PfRh4 binding affects C3b/C4b interaction with CR1, SPR studies were performed in which C3b and C4b were immobilized on the sensor-chip surface by standard amine coupling (Figure 5A-B). Benchmarking experiments confirmed that sCR1 binds significantly more tightly to C3b than C4b and provided KD values for these interactions of 0.2 ± 0.04μM (Figure 5Aii) and 1.2 ± 0.09μM (Figure 5Bii), respectively, which are somewhat stronger than the KD of 2.9 ± 0.2μM for the sCR1-to-rPfRh4 complex.8 When a concentration series of sCR1 with 2.3-fold molar excess of PfRh4 flowed over the chip and KD values were recalculated, they did not change substantially (Figure 5Aiv, Biv).
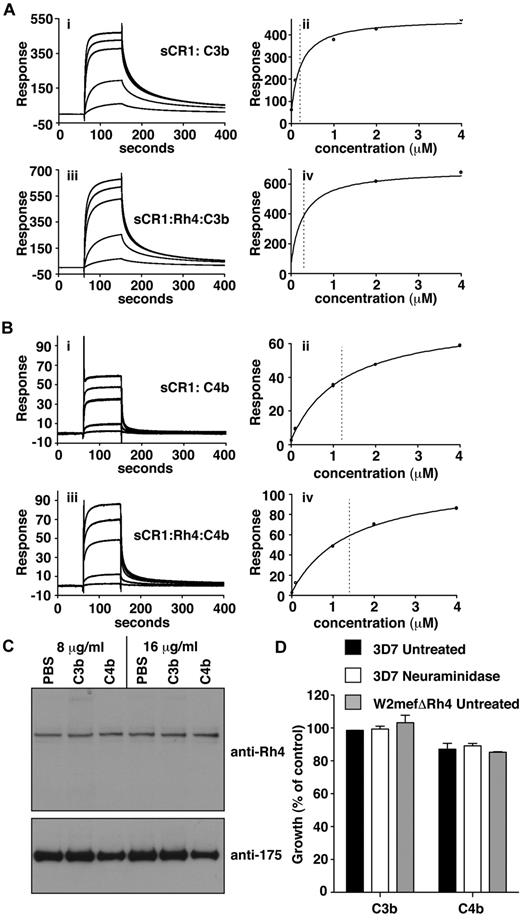
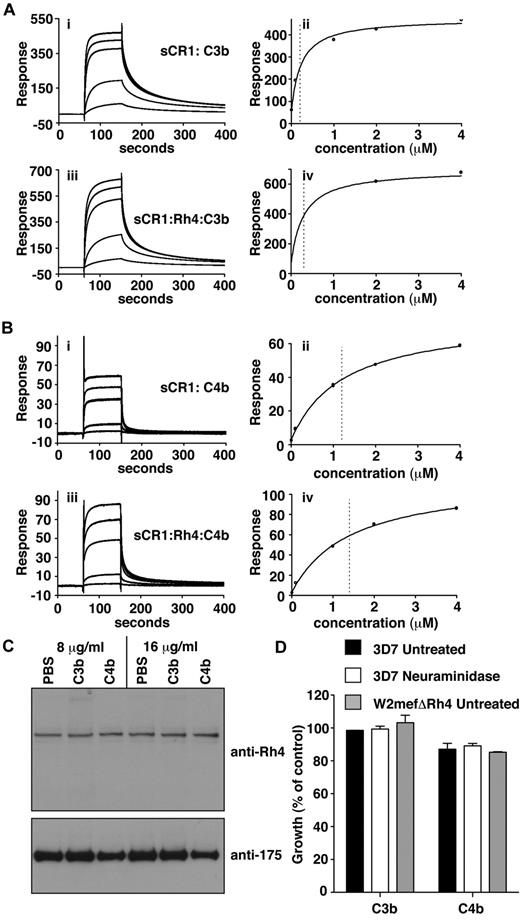
C3b/C4b interaction with CR1 is not perturbed by PfRh4 binding. (A) PfRh4 binding does not affect C3b interaction with sCR1. Duplicate injections of a concentration series of sCR1 (i) or a mixture of sCR1 plus 3-fold molar excess of rPfRh4 (iii) onto a CM5 chip coupled with C3b. KD measurements of sCR1:C3b complex alone (ii) or in the presence of 3-fold molar excess of rPfRh4 (iv) with fitted value indicated by dotted line (B) PfRh4 binding does not affect C4b interaction with sCR1. Duplicate injections of a concentration series of sCR1 (i) or a mixture of sCR1 plus 3-fold molar excess of rPfRh4 (iii) onto a CM5 chip coupled with C4b. KD measurements of sCR1:C4b complex alone (ii) or in the presence of 3-fold molar excess of rPfRh4 (iv) with fitted value indicated by dotted line. In all panels, blank-subtracted sensorgrams are shown. (C) Native PfRh4 binding to erythroid CR1 was not inhibited by C3b or C4b. Competitive binding assays were performed by incubating either C3b or C4b with invasion supernatants at the stated final concentrations (8 μg/mL or 16 μg/mL). Immunodetection of parasite proteins with anti-PfRh4 and anti–EBA-175 antibodies after erythrocyte binding is shown. (D) PfRh4 invasion pathway was not inhibited in the presence of C3b or C4b. Parasite strains W2mefΔRh4 (gray bars) and 3D7 (black bars for untreated, white bars for nm-treated) were tested in growth assays in the presence of 0.5 mg/mL C3b or C4b. Growth (percentage of control) on the y-axis refers to the percentage parasitemia in the presence of CR1 constructs relative to the percent parasitemia with the addition of PBS (arbitrarily set to be 100%).
C3b/C4b interaction with CR1 is not perturbed by PfRh4 binding. (A) PfRh4 binding does not affect C3b interaction with sCR1. Duplicate injections of a concentration series of sCR1 (i) or a mixture of sCR1 plus 3-fold molar excess of rPfRh4 (iii) onto a CM5 chip coupled with C3b. KD measurements of sCR1:C3b complex alone (ii) or in the presence of 3-fold molar excess of rPfRh4 (iv) with fitted value indicated by dotted line (B) PfRh4 binding does not affect C4b interaction with sCR1. Duplicate injections of a concentration series of sCR1 (i) or a mixture of sCR1 plus 3-fold molar excess of rPfRh4 (iii) onto a CM5 chip coupled with C4b. KD measurements of sCR1:C4b complex alone (ii) or in the presence of 3-fold molar excess of rPfRh4 (iv) with fitted value indicated by dotted line. In all panels, blank-subtracted sensorgrams are shown. (C) Native PfRh4 binding to erythroid CR1 was not inhibited by C3b or C4b. Competitive binding assays were performed by incubating either C3b or C4b with invasion supernatants at the stated final concentrations (8 μg/mL or 16 μg/mL). Immunodetection of parasite proteins with anti-PfRh4 and anti–EBA-175 antibodies after erythrocyte binding is shown. (D) PfRh4 invasion pathway was not inhibited in the presence of C3b or C4b. Parasite strains W2mefΔRh4 (gray bars) and 3D7 (black bars for untreated, white bars for nm-treated) were tested in growth assays in the presence of 0.5 mg/mL C3b or C4b. Growth (percentage of control) on the y-axis refers to the percentage parasitemia in the presence of CR1 constructs relative to the percent parasitemia with the addition of PBS (arbitrarily set to be 100%).
Interestingly, the addition of C3b or C4b had no detectable effect on native PfRh4 binding or on the CR1-PfRh4 invasion pathway (Figure 5C-D). Hence the binding of PfRh4 to CR1 does not notably affect the ability of CR1 to bind to either C3b or C4b, or presumably to carry out its immune adherence function; and neither does the binding of C3b or C4b obstruct binding of PfRh4 to CR1, despite both host and parasite ligands occupying a similar region of the receptor.
A ternary complex composed of C4b, PfRh4, and CCP1-3
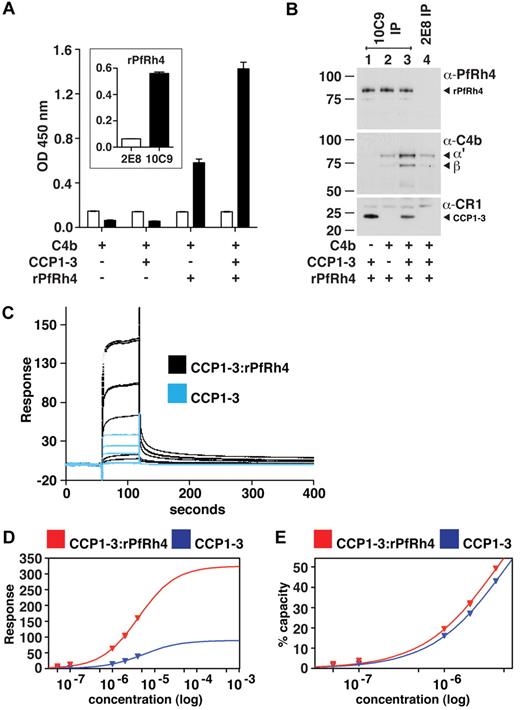
Because PfRh4 binding to sCR1 did not affect the interaction of the latter with C3b and C4b, we hypothesized that CCP1-3 binds both C4b and PfRh4 simultaneously. Using ELISA plates coated with C4b, we did indeed detect binding of rPfRh4 in the presence of CCP1-3 (Figure 6A); however, we also observed a weaker interaction between rPfRh4 and C4b in the absence of CCP1-3. To further investigate, we performed coimmunoprecipitation experiments in which various combinations of CCP1-3, rPfRh4 and C4b were incubated with the anti-PfRh4 monoclonal antibody 10C9 as indicated (Figure 6B). A ternary complex of rPfRh4:CCP1-3–C4b was coimmunoprecipitated by 10C9 (Figure 6B lane 3). The low levels of C4b that also appeared to undergo coimmunoprecipitation with rPfRh4 by 10C9 (Figure 6B lane 2) may be an artifact, given that anti-PfRh4 antibody 2E8, which does not recognize the region encompassed by rPfRh4, was also able to immunoprecipitate low levels of C4b (Figure 6B lane 4).
A ternary complex composed of CCP1-3, C4b, and PfRh4. (A) ELISA of ternary complex. Microtiter plates were coated with C4b (1 μg/well). After incubation with rPfRh4, CCP1-3, or both as indicated, bound rPfRh4 was detected using anti-PfRh4 monoclonal antibody 10C9 (black bars) that was raised against rPfRh4. Monoclonal antibody 2E8 (white bars) was raised to the C-terminal end of PfRh4 and recognizes native PfRh4 but not the region encompassed by rPfRh4. The inset shows the reactivity of these monoclonal antibodies in an ELISA using microtiter plates coated with rPfRh4. Data are the mean ± SD for 3 independent experiments. (B) Immunoprecipitation of ternary complex. Purified C4b, CCP1-3, or rPfRh4 were incubated together as indicated (at 0.02 mg/mL). Western blots were performed after immunoprecipitation with anti-PfRh4 10C9 (lanes 1-3) or 2E8 (lane 4) monoclonal antibody, respectively. Immunoprecipitated material was probed with a rabbit polyclonal anti-PfRh4 antibody, a goat polyclonal anti–human C4 antibody and a monoclonal anti-CR1 antibody 1B4, respectively. PfRh4 and C4b samples were run under reducing conditions whereas CCP1-3 was under nonreducing conditions. Arrowheads highlight specific protein bands. Western blot results are representative of 2 or 3 independent experiments. (C) A ternary complex composed of CCP1-3, C4b, and rPfRh4 as inferred from SPR. Measurement by SPR of CCP1-3 binding to C4b immobilized on a CM5 sensorchip in absence (blue lines) and presence (black lines) of a 2-fold molar excess of rPfRh4. CCP1-3 concentrations were 4μM, 2μM, 1μM, 0.1μM, and 0.05μM; single measurements were made at 4μM and 1μM while all others were duplicate measurements. (D) Plots of responses versus concentration of CCP1-3 (either alone, blue, or mixed with rPfRh4, red) from the sensorgrams in panel C are shown, with extrapolations to Rmax values. The much higher responses obtained when rPfRh4 is coinjected with CCP1-3 are consistent with formation of ternary complexes rather than binary complexes. (E) The percentage of maximal binding (estimated from extrapolated Rmax values in panel D) versus concentration of CCP1-3 (either alone, blue, or mixed with rPfRh4, red). The near identical-slope of both binding curves indicates that the affinity of CCP1-3 for C4b, while it cannot be quantified because of subsaturation concentrations of CCP1-3, is not radically altered by rPfRh4 and is consistent with a ternary complex in which C4b and rPfRh4 occupy distinct sites on CCP1-3.
A ternary complex composed of CCP1-3, C4b, and PfRh4. (A) ELISA of ternary complex. Microtiter plates were coated with C4b (1 μg/well). After incubation with rPfRh4, CCP1-3, or both as indicated, bound rPfRh4 was detected using anti-PfRh4 monoclonal antibody 10C9 (black bars) that was raised against rPfRh4. Monoclonal antibody 2E8 (white bars) was raised to the C-terminal end of PfRh4 and recognizes native PfRh4 but not the region encompassed by rPfRh4. The inset shows the reactivity of these monoclonal antibodies in an ELISA using microtiter plates coated with rPfRh4. Data are the mean ± SD for 3 independent experiments. (B) Immunoprecipitation of ternary complex. Purified C4b, CCP1-3, or rPfRh4 were incubated together as indicated (at 0.02 mg/mL). Western blots were performed after immunoprecipitation with anti-PfRh4 10C9 (lanes 1-3) or 2E8 (lane 4) monoclonal antibody, respectively. Immunoprecipitated material was probed with a rabbit polyclonal anti-PfRh4 antibody, a goat polyclonal anti–human C4 antibody and a monoclonal anti-CR1 antibody 1B4, respectively. PfRh4 and C4b samples were run under reducing conditions whereas CCP1-3 was under nonreducing conditions. Arrowheads highlight specific protein bands. Western blot results are representative of 2 or 3 independent experiments. (C) A ternary complex composed of CCP1-3, C4b, and rPfRh4 as inferred from SPR. Measurement by SPR of CCP1-3 binding to C4b immobilized on a CM5 sensorchip in absence (blue lines) and presence (black lines) of a 2-fold molar excess of rPfRh4. CCP1-3 concentrations were 4μM, 2μM, 1μM, 0.1μM, and 0.05μM; single measurements were made at 4μM and 1μM while all others were duplicate measurements. (D) Plots of responses versus concentration of CCP1-3 (either alone, blue, or mixed with rPfRh4, red) from the sensorgrams in panel C are shown, with extrapolations to Rmax values. The much higher responses obtained when rPfRh4 is coinjected with CCP1-3 are consistent with formation of ternary complexes rather than binary complexes. (E) The percentage of maximal binding (estimated from extrapolated Rmax values in panel D) versus concentration of CCP1-3 (either alone, blue, or mixed with rPfRh4, red). The near identical-slope of both binding curves indicates that the affinity of CCP1-3 for C4b, while it cannot be quantified because of subsaturation concentrations of CCP1-3, is not radically altered by rPfRh4 and is consistent with a ternary complex in which C4b and rPfRh4 occupy distinct sites on CCP1-3.
To examine whether a ternary complex could be detected by SPR, a 2-to-1 mixture of rPfRh4 and CCP1-3 flowed over a C4b-coated sensor-chip surface, and a large concentration-dependent response was detected. This response was more than 3.5-fold greater than that obtained for rPfRh4 alone (not shown), and was significantly greater than the response obtained for CCP1-3 alone (Figure 6C). These observations strongly suggest that a complex of rPfRh4:CCP1-3, as opposed to either rPfRh4 or CCP1-3 singly, was the dominant species that bound to the immobilized C4b after coinjection of these 2 proteins. Moreover, the affinity of CCP1-3 for C4b does not appear, albeit based on subsaturating concentrations, to be strongly affected by the presence of rPfRh4 (Figure 6D-E). The simplest explanation for these SPR-based observations is that a ternary complex was formed by CCP1-3 with C4b and rPfRh4.
Decay accelerating activity was disrupted by PfRh4 binding
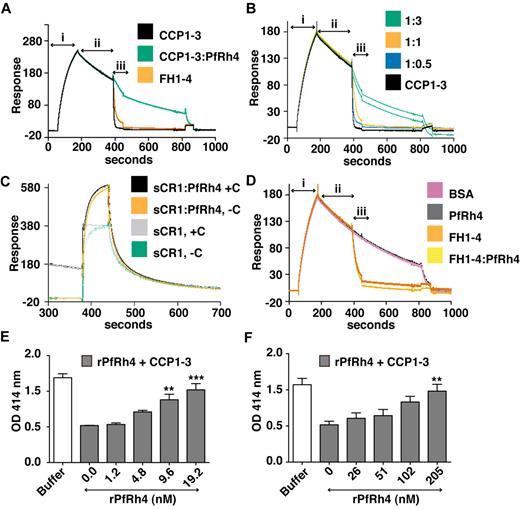
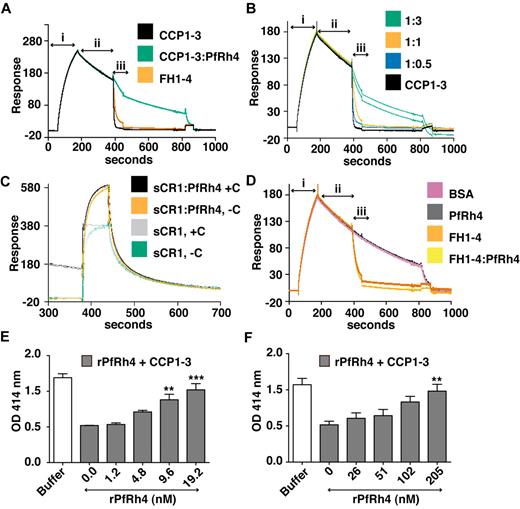
As shown above, PfRh4 does not bind to site 2 of CR1; as expected we did not observe inhibition of cofactor activity using CCP15-17 or sCR1 (supplemental Figure 2). However, CCP1-3 is a key region for CR1's DAA activity, so it was important to examine the changes in activity on PfRh4 binding. To this end, we used a real-time assay based on formation of the alternative pathway convertase on a sensor-chip surface within an SPR instrument (Figure 7A-D).29,36,37 The convertase was formed on the surface by flowing a mixture of factor B and factor D over the immobilized C3b (C3bBb formation, Figure 7A), and the subsequent spontaneous decay of the convertase was monitored over several minutes (spontaneous decay, Figure 7A). Injection of either 1μM CCP1-3 or 1μM factor H CCP1-4 (FH1-4) as a positive control yielded a clear acceleration of the decay rate (analyte injection). Therefore, although CCP1-3 weakly interacts with isolated C3b, it destabilizes the C3bBb complex. On the other hand, a mixture of 1μM CCP1-3 and 5μM rPfRh4 had a minimal effect on the intrinsic rate of C3bBb decay (Figure 7A). A series of CCP1-3:rPfRh4 mixtures (at ratios of 1:0, 1:0.5, 1:1, and 1:3) was prepared and demonstrated a dose-dependent inhibitory effect of rPfRh4 on decay acceleration by CCP1-3 (Figure 7B). A similar inhibitory effect of rPfRh4 on decay acceleration by sCR1 was also observed (Figure 7C, supplemental Figure 3). Decay acceleration by factor H CCP1-4 was not affected by the addition of rPfRh4 (Figure 7D).
PfRh4 disrupts CR1's decay accelerating activity. (A) Decay acceleration of C3bBb was inhibited on PfRh4 binding. SPR was used to monitor formation of the C3bBb convertase complex as factor D and factor B were flowed together over C3b that was amine-coupled to a CM5 sensor chip (i, C3bBb formation). The subsequent decline in response reflects decay of the complex as Bb is released from the chip surface (ii, spontaneous decay). The rate of decay was accelerated by initiating a flow of CCP1-3 or factor H CCP1-4 (FH1-4; iii, analyte injection), but not when CCP1-3 was in the presence of 3-fold molar excess of rPfRh4. In panels A-D, any convertases remaining were decayed by injecting FH1-4 at ∼ 800 seconds to aid regeneration (B) Decay-acceleration of CCP1-3 was affected in a dose-dependent manner by rPfRh4. The rate of decay was monitored in the presence of increasing concentrations of rPfRh4. A small fraction of inactive rPfRh4 present on the surface triggers nonspecific background binding when the first duplicate injection was assayed. (CCP1-3 in 3-fold molar excess of PfRh4) as manifested in the imperfect reproducibility of the sensorgrams. After saturation of this nonspecific binding capacity, injections were of acceptable reproducibility (all other constructs). (C) Decay-acceleration activity of sCR1 was affected by presence of rPfRh4. Shown are binding responses for sCR1, and for sCR1 in the presence of a 5-fold molar excess of PfRh4, of the convertase C3bBb (+C) or C3b alone (−C). Three C3b-binding sites in sCR1 mediate the overall high binding levels to both the convertase and C3b alone. Only sCR1 binding to the convertase (+C) shows a distinctive association curve that is consistent with an initially enhanced binding of sCR1 to the convertase, followed by 2 simultaneous, overlapping processes: accelerated decay of C3bBb into Bb and surface-bound C3b, and binding of sCR1 to C3b. (D) Decay acceleration by factor H CCP1-4 was not affected by rPfRh4. Assays shown in panels A and C, and in panels B and D were performed on identical biosensor surfaces. (E) Hemolysis assay for the classic pathway. The classic pathway C3 convertase was assembled on the surface of antibody-sensitized sheep erythrocytes. PfRh4 plus CCP1-3 were pre-incubated before mixing with sheep erythrocytes. Cellular lysis was induced by the addition of guinea pig serum and monitored by the O.D. of the supernatant at 414nM. CCP1-3 was used at a concentration of 6.8nM. (F) Hemolysis assay for the alternative pathway. The alternative pathway convertase was prepared using EAC14 cells by the addition of C2 and C3. PfRh4 plus CCP1-3 were preincubated and these mixtures were then added to EAC43 cells and the alternative pathway components factor B, factor D, and properdin and were added and lysis measured as in panel E. A convertase was not formed in the absence of factor B. CCP1-3 was used at a concentration of 34nM. For all panels, **P < .01; ***P < .001).
PfRh4 disrupts CR1's decay accelerating activity. (A) Decay acceleration of C3bBb was inhibited on PfRh4 binding. SPR was used to monitor formation of the C3bBb convertase complex as factor D and factor B were flowed together over C3b that was amine-coupled to a CM5 sensor chip (i, C3bBb formation). The subsequent decline in response reflects decay of the complex as Bb is released from the chip surface (ii, spontaneous decay). The rate of decay was accelerated by initiating a flow of CCP1-3 or factor H CCP1-4 (FH1-4; iii, analyte injection), but not when CCP1-3 was in the presence of 3-fold molar excess of rPfRh4. In panels A-D, any convertases remaining were decayed by injecting FH1-4 at ∼ 800 seconds to aid regeneration (B) Decay-acceleration of CCP1-3 was affected in a dose-dependent manner by rPfRh4. The rate of decay was monitored in the presence of increasing concentrations of rPfRh4. A small fraction of inactive rPfRh4 present on the surface triggers nonspecific background binding when the first duplicate injection was assayed. (CCP1-3 in 3-fold molar excess of PfRh4) as manifested in the imperfect reproducibility of the sensorgrams. After saturation of this nonspecific binding capacity, injections were of acceptable reproducibility (all other constructs). (C) Decay-acceleration activity of sCR1 was affected by presence of rPfRh4. Shown are binding responses for sCR1, and for sCR1 in the presence of a 5-fold molar excess of PfRh4, of the convertase C3bBb (+C) or C3b alone (−C). Three C3b-binding sites in sCR1 mediate the overall high binding levels to both the convertase and C3b alone. Only sCR1 binding to the convertase (+C) shows a distinctive association curve that is consistent with an initially enhanced binding of sCR1 to the convertase, followed by 2 simultaneous, overlapping processes: accelerated decay of C3bBb into Bb and surface-bound C3b, and binding of sCR1 to C3b. (D) Decay acceleration by factor H CCP1-4 was not affected by rPfRh4. Assays shown in panels A and C, and in panels B and D were performed on identical biosensor surfaces. (E) Hemolysis assay for the classic pathway. The classic pathway C3 convertase was assembled on the surface of antibody-sensitized sheep erythrocytes. PfRh4 plus CCP1-3 were pre-incubated before mixing with sheep erythrocytes. Cellular lysis was induced by the addition of guinea pig serum and monitored by the O.D. of the supernatant at 414nM. CCP1-3 was used at a concentration of 6.8nM. (F) Hemolysis assay for the alternative pathway. The alternative pathway convertase was prepared using EAC14 cells by the addition of C2 and C3. PfRh4 plus CCP1-3 were preincubated and these mixtures were then added to EAC43 cells and the alternative pathway components factor B, factor D, and properdin and were added and lysis measured as in panel E. A convertase was not formed in the absence of factor B. CCP1-3 was used at a concentration of 34nM. For all panels, **P < .01; ***P < .001).
Using hemolysis assays, we also investigated whether rPfRh4 could block the DAA of CCP1-3 for both classic and alternative pathways on an erythrocyte surface (Figure 7E-F). Purified complement components were used to form the convertase on the surface of sheep erythrocytes, and lytic sites were developed with guinea pig serum diluted in EDTA. For these studies, a concentration of CCP1-3 that resulted in approximately 60% inhibition of hemolysis was chosen. For the classic pathway convertase (C4b2a), rPfRh4 inhibited DAA in a concentration-dependent manner (Figure 7E). At a 3-fold molar excess of rPfRh4 over CCP1-3, the percentage of cells that were lysed increased from 44%-86% (P < .001). Similar results were obtained with the alternative pathway convertase (Figure 7F). In this case, a 6-fold molar excess of rPfRh4 over CCP1-3 resulted in an increase in lysis from 47%-92%. These results are consistent with the inhibition of DAA by PfRh4 binding as shown by SPR assays.
Discussion
CR1 plays essential roles in mammalian immunity. On erythrocytes, CR1 is the immune-adherence receptor where it serves to transport C4b/C3b bearing immune complexes to the spleen and liver. On phagocytotic cells, it is particularly efficient at mediating adherence of a C4b-coated or C3b-coated antigen. There is increasing awareness of CR1 as a means of cell attachment and cell entry for a diverse range of pathogens.4-6 Erythroid CR1 may be vulnerable to pathogen exploitation because, although some people have a genetically determined low copy number of CR1 on erythrocytes, complete deficiency is unknown in humans.40 In previous work, 2 groups identified CR1 as a host receptor used for P falciparum invasion of human erythrocytes.7,8 In the current study, the region within CR1 that is recognized by the P falciparum adhesin PfRh4 was mapped to the N-terminal 3 CCP modules of CR1. Our results show that CCP1-3 acts as an effective inhibitor of sialic acid–independent invasion of erythrocytes in a range of P falciparum strains. Interestingly PfRh4 binding to CR1 affected neither its C3b nor its C4b binding. It also did not inhibit cofactor activity, but it did modulate the decay-accelerating activity of CR1.
Field isolates from India, the Gambia, Brazil, Tanzania, and Kenya can invade nm-treated erythrocytes, showing that sialic acid-independent invasion pathways are used in the wild.41-45 Using CCP1-3 as an inhibitor, we showed that the PfRh4-CR1 pathway was of broad importance for sialic acid–independent invasion across a wide range of P falciparum strains obtained originally from diverse geographic regions. CCP1-3 also inhibited the binding of PfRh4 to erythrocytes, thus providing a molecular basis for its inhibition of invasion. Furthermore, P falciparum clinical isolates obtained from Kenyan children with acute malaria infection are able to invade nm-treated erythrocytes, and the addition of sCR1 completely inhibits this invasion pathway, indicating that the PfRh4-CR1 invasion pathway is used in the field.46 Additional analyses of field isolates worldwide are needed to fully delineate the overall use of the PfRh4–CR1 invasion pathway in P falciparum infection.
In this study we observed that some P falciparum strains (E8B, FCR3, T994) express PfRh4 but show less invasion into nm-treated erythrocytes. In these strains, sialic acid–independent invasion may not solely depend on PfRh4, but may reflect the efficiency of other sialic acid–independent invasion pathways such as the PfRh5 pathway. It is also possible that either polymorphisms in PfRh4 inactivate the protein, or other parasite ligands are required for optimal function. The former possibility would be similar to the situation in P falciparum strains in which polymorphisms within the EBA-181 and EBA-140 proteins have rendered these ligands nonfunctional in merozoite invasion, despite their apparent expression.39 Of note is the observation that E8B, FCR3, and T994 parasites do not express PfRh2a or 2b, providing an intriguing possibility that these 2 PfRh family members may function cooperatively in invasion.32
It is probably advantageous for PfRh4 to bind to the N-terminal extremity of CR1, as this region lies at the opposite end of the molecule from the membrane-spanning and cytoplasmic domains. Given the probably extended nature of this large (approximately 1800 residue) ectodomain, the N-terminal modules will probably project well beyond the thin glycocalyx of the erythrocyte and represent highly accessible features of the erythrocyte cell surface for parasite recognition, as well as for its binding to C3b and C4b molecules on opsonized surfaces. P falciparum also uses CR1 as a host receptor for rosetting. This occurs via an interaction between the parasite-derived variant erythrocyte–membrane virulence protein, PfEMP-1, and CCP15-17 and CCP8-10. Our studies have shown that the site involved in PfRh4 binding is distinct from the CR1 regions important in PfEMP-1–mediated rosette formation.47,48
Our studies demonstrate that PfRh4 interacts with CR1 via a primary binding site within CCP1-3 and that other regions of CR1 are unlikely to be involved. This conclusion was based on both the experimental binding data collected on representative CR1 fragments, and the high levels of internal similarity reflected in division of CR1 into 4 homologous repeats, that is, CCPs 1-7, 8-14, 15-21, and 22-28 (supplemental Figure 4).17 Thus, binding to CCP4-7 is improbable because there was no detectable binding by PfRh4 to the CCP10-11 fragment (module 11 is nearly identical to module 4), or to CCP15-25 (CCP18-21 share 84% identity with CCP4-7). Binding almost certainly cannot involve CCP8 or CCP9, given their near-identity to CCP15 and CCP16 in the experimentally tested CCP15-17 fragment. Similarly, binding to CCP12-14 seems unlikely, as they are 77% identical to CCP19-21, which, in the CCP15-25 context, were found not to bind PfRh4; likewise, binding to CCP26-28 is highly unlikely, given their 96% identity with CCP19-21. We did not test for binding CCP29 and 30, which share little similarity with other CR1 modules, because of the relatively inaccessible location of this region near the transmembrane domain. Our conclusion that CCP1-3 is the sole binding site for PfRh4 is entirely consistent with the homologous data reviewed above and the observed inhibition of CR1 binding to PfRh4 by CCP1-3.
By binding to a site within CR1 important for its immune regulatory roles, P falciparum parasites may have an added advantage in that the region is conserved because of its functional importance and not likely to be removed or mutated by the host. However, a consequence of PfRh4 binding to CR1 is the inhibition of DAA. This inhibition of CR1's DAA by PfRh4 may not affect complement regulation for the following reasons: first, cofactor activity was unaffected by PfRh4 binding. Second, the invasion of human erythrocytes by malaria parasites is a relatively rapid process, averaging approximately 30 seconds from initial contact to completion of erythrocyte entry.49 The interaction between CR1 and PfRh4 will therefore be a transient one. Third, the inhibition of CR1's DAA could be compensated for by the similar functional activity of decay-accelerating factor (DAF), another RCA protein found on erythrocytes. We have previously shown that PfRh4 erythrocyte binding is not affected by the addition of anti-DAF antibodies, suggesting that these proteins do not interact and therefore DAF's functional roles should not be affected.8 Furthermore, in this paper we showed that PfRh4 had no effect on the DAA activity of a fluid phase complement regulator, factor H.
Our studies show that PfRh4 forms a ternary complex with CCP1-3 and complement fragment C4b, and therefore PfRh4 binding does not interfere with the immune adherence function of full-length CR1. Our assays did not show evidence of ternary complex formation between CCP1-3, PfRh4, and C3b. The ability of PfRh4 and C4b to bind simultaneously to CR1 may be advantageous to P falciparum for 2 reasons: First, there are substantially fewer molecules of CR1 (100 to 1000 copies per cell) on erythrocytes compared with glycophorins (104 to 106 copies). Thus although erythroid CR1 can bind PfRh4 in the absence of C3b or C4b on the parasite surface, the potential for an avidity effect arising from the presence of C3b or C4b tethered on the parasite surface acting as co-ligands for CR1 could greatly enhance affinity. Second, immune adherence causes clustering of CR1 molecules on the erythrocyte surface, resulting in measurable increase in membrane deformability, a process that may facilitate parasite invasion.50
Our results show that PfRh4 binds to CCP1-3 but not to CCP15-17, which strongly suggests the presence of key binding determinants within CCP1-2. Structure–function analyses of CCP1-2 have identified amino acid residues that are important for classic and alternative pathway convertase decay acceleration and for C3b/C4b binding.23 As PfRh4 interaction reduces decay acceleration activity, but not adherence to C4b or C3b, we were interested in mutants that recapitulated similar properties. Of particular note, Phe82, which completely abrogated decay acceleration for both pathways, has little effect on ligand binding.23 It is thought that Phe82 lies in the interface between CCP1 and CCP2 and is important for the orientation of these modules. Mutation of Thr103, Thr110, and Val111 created a functionally similar mutant protein, although with less effect on decay acceleration. Further structural and mutational analysis of complex formation between PfRh4 and CCP1-3 will be required to identify the precise intermolecular interactions involved.
The current results reveal the functional implications for CR1 of engagement with P falciparum adhesin PfRh4 and illuminate the complex interplay between complement activation and infection. Further studies will be required to determine whether additional Plasmodium adhesins hijack other RCA proteins for invasion into human erythrocytes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by the Victorian State Government OIS and National Health and Medical Research Council IRIISS grants (361646). A.F.C. is a Howard Hughes International Scholar and an Australia Fellow from the National Health and Medical Research Council, Australia. P.B.T.-Q. was supported by the Darwin Trust of Edinburgh. C.Q.S. and M.G. were supported by a Wellcome Trust Grant (081179). R.E.H. and J.P.A. were supported by National Institutes of Health grant AI041592.
National Institutes of Health
Wellcome Trust
Authorship
Contribution: W.-H.T. performed parasite culturing, GIA, ELISAs, and Western blot analyses; C.Q.S. performed SPR and cofactor activity assays; R.E.H. executed the hemolysis assays; M.G. and P.B.T.-Q. purified CR1 proteins; S.L. performed GIA and parasite culturing; and W.-H.T., C.Q.S., A.F.C., P.N.B., and J.P.A. designed and interpreted experiments. All authors contributed to writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan F. Cowman, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia; e-mail: cowman@wehi.edu.au.
References
Author notes
W-H.T. and C.Q.S. contributed equally to this study.