Abstract
Primary effusion lymphoma (PEL) is a diffuse-large B-cell lymphoma with poor prognosis. One hundred percent of PELs carry the genome of Kaposi sarcoma–associated herpesvirus and a majority are coinfected with Epstein-Barr virus (EBV). We profiled genomic aberrations in PEL cells using the Affymetrix 6.0 SNP array. This identified for the first time individual genes that are altered in PEL cells. Eleven of 13 samples (85%) were deleted for the fragile site tumor suppressors WWOX and FHIT. Alterations were also observed in the DERL1, ETV1, RASA4, TPK1, TRIM56, and VPS41 genes, which are yet to be characterized for their roles in cancer. Coinfection with EBV was associated with significantly fewer gross genomic aberrations, and PEL could be segregated into EBV-positive and EBV-negative clusters on the basis of host chromosome alterations. This suggests a model in which both host genetic aberrations and the 2 viruses contribute to the PEL phenotype.
Introduction
Primary effusion lymphoma (PEL) is a postgerminal center (GC), diffuse-large B-cell lymphoma (DLBCL) with poor prognosis. It is characterized by an accumulation of tumor cells in the serous cavities of the body and therefore was initially referred to as body cavity–based lymphoma. Since then, isolated instances of solid, lymph node-associated variants have also been described.1 Morphologically, PELs are pleiomorphic and exhibit heterogeneity in cell size and nuclear shape. PEL is an AIDS-defining malignancy, and it usually manifests itself in conjunction with Kaposi sarcoma (KS). However, PEL has also been diagnosed in HIV-negative patients experiencing severe immune-suppression after organ transplantation. PEL is unique histologically as well as in its expression of immunophenotypic markers, mRNA, and microRNA profiles.2-4 As expected for cancer cells, PEL cell lines show gross chromosomal alterations.5,6 Genomewide high-resolution analyses of copy number variants (CNVs) and loss of heterozygosity (LOH), which would aid our understanding of PEL, have not been reported.
All PELs are infected with KS-associated herpesvirus (KSHV).2 KSHV is also the causative agent for KS7 and the plasmablastic variant of multicentric Castleman disease.8 KSHV is required for PEL survival; a subset of viral proteins as well as all viral microRNAs are consistently expressed in all PEL cells.9 Most PELs are also coinfected with EBV, and this results in altered host mRNA transcription compared with EBV-negative PEL cells.10 It has been reported that on overexpression of a dominant-negative form of the EBV EBNA2 protein, some EBV-positive PELs cease to proliferate.11 Yet the contribution of EBV to PEL development remains unclear because both EBV-positive and EBV-negative PEL cell lines grow equally well in culture and form tumors with equal efficiency in immune-deficient mice.12,13
Cancer is thought to arise in a multistep fashion, although not necessarily in a linear manner, in which each step provides a selective advantage in terms of cell proliferation and cell survival in the tumor microenvironment. This leads to cancer type–specific genome signatures such as the classic “Philadelphia” t(9;22)(q34;q11) translocationm resulting in oncogenic BCR/ABL gene fusion in chronic myelogenous leukemia.14 These signatures in turn provide tumor cell–specific targets for therapy (eg, use of imatinib mesylate/Gleevec in chronic myelogenous leukemia). In non–virus-associated cancers, each step in the pathway involves activating or inhibitory mutations in cellular oncogenes or tumor suppressors, respectively. In virus-associated cancers, the virus contributes to one or multiple steps along this pathway, thus reducing the need for specific mutations in host oncogenes or tumor suppressor genes.
Chromosomal imbalances and genomic instability comprise a major contributing factor in malignant diseases. Unlike other lymphomas, no signature translocation or single gene mutation has been associated with PEL to date. The p53 tumor suppressor protein appears functional in PEL cell lines,15 the Myc locus un-rearranged, although the protein is unusually stable,16 and no amplifications or deletions are reported for Bcl-2,2 Bcl-6,17 Ras,2 the catalytic subunit of PI3K,18 phosphatase with tensin homolog or p16/INK4.18 We therefore used the Affymetrix 6.0 single nucleotide polymorphism (SNP)–based microarray to conduct comparative genomic hybridization (CGH) to look at the global genomic profile of PEL cells. This identified PEL-specific gene alterations in the fragile site tumor suppressor genes, fragile histidine triad (FHIT) and WW-domain containing oxidoreductase (WWOX), which were deleted in 11 of 13 (85%) and 12 of 13 (92%) samples, respectively (P ≤ .0005). In addition, we observed alterations in other key signaling pathways albeit at a lower frequency. Because a subset of PEL are coinfected with EBV in addition to KSHV, we asked whether EBV influenced overall genomic instability or was associated with genomic alterations in specific genes. EBV-negative PEL cell lines exhibited significantly increased genomic amplifications compared with EBV-positive, suggesting that the presence of EBV contributes to genomic stability.
Methods
Cell culture
The PEL cell lines used in the study are shown in Table 1. All PEL and non-PEL lymphoma and leukemia cells (BJAB, KSHV-BJAB, DG75, BL5, BL8, Thp1, and Thp1-KSHV) were cultured in RPMI 1640 supplemented with 100 μg/mL streptomycin sulfate, 100 U/mL penicillin G (Life Technologies), 2mM l-glutamine, 0.05mM 2-mercaptoethanol, 0.075% sodium bicarbonate, 1 U/mL IL-6 (PeproTech Inc), and 10% FBS and were maintained at 37°C in 5% CO2. All nonlymphoma cells (E1-TIVE, L1-TIVE, HEK293, and HEK293-KSHV) were maintained in DMEM supplemented with 100 μg/mL streptomycin sulfate, 100 U/mL penicillin G, and 10% FBS.
DNA extraction and CGH
Genomic DNA was extracted with the Wizard Genomic DNA Purification Kit (Promega) as per the manufacturer's protocol. Signature profiles were obtained with the 6.0 GeneChip Human Mapping Array that uses > 906 600 known SNP and 946 000 CNV markers (Affymetrix). As control we used the older 500K Affymetrix array. The SNP and CNV data have been deposited in the NIH GEO Datasets archives: GSE25839 and GSE28684.
Data analysis
All analyses were performed with the Partek Genomics Suite v6.0 (Partek Inc). Raw CGH data (.CEL files) were imported and adjusted for background with the use of the Robust Multi-array Average algorithm. CNV was determined by generating copy number values with the use of the Genomic Segmentation algorithm with preset program parameters and was compared with the 270 HapMap baseline (Version 122809) available at http://www.affymetrix.com/support/technical/sample_data/genomewide_snp6_data.affx. Gene lists were generated by determining regions of significance (by estimating t statistics for each probe adjusted for multiple comparisons by MAT algorithm) in multiple samples and annotated with the NCBI Reference Sequence database.19 Further statistical analyses used the R v2.11.1 statistical software environment (R Project for Statistical Computing).
Real-time quantitative PCR analysis
We used real-time quantitative PCR (qPCR) to verify some candidate genes. We selected primers following criteria outlined by D'Haene et al20 from RTprimerDb (www.rtprimerdb.org). Primer sequences used were DERL1 (forward: 5′-TACTCCAGCTACACAAAG-3′; reverse: 5′-AATGAGATACGAGGGTTG-3′), FHIT (forward: 5′-CCAGTGGAGCGCTTCCAT-3′; reverse: 5′-TCCACCACTGTCCCGACTCT-3′), GRID2 (forward: 5′-GCATTTCAGTGTTTTGAAAATTG-3′; reverse: 5′-CCAGTCTGGGCAAACTCATT-3′), WWOX (forward: 5′-GCAATGAAGGCAACAAAGT-3′; reverse: 5′-TTAAAAGACCTGGGGGAAT-3′), and LANA78 (forward: 5′-GGAAGAGCCCATAATCTTGC-3′; reverse: 5′-GCCTCATACGAACTCGAGGT-3′). Cycling conditions were 95°C for 10 minutes followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute followed by melting curve analysis on a Roche LC480 Lightcycler. All reactions were conducted in 5 technical replicates. Purified normal human genomic DNA (Roche) was used as positive control, and water served for the nontemplate negative control. A standard curve was generated with serial (1:4) dilutions of human genomic DNA, starting at 8 μg/mL concentration, against the individual primers. We used robust regression and normalized by primer followed by human diploid DNA (Roche) to obtain relative copy number changes for each sample relative to normal diploid human DNA.
Results
Genetic signature of PEL: FHIT and WWOX
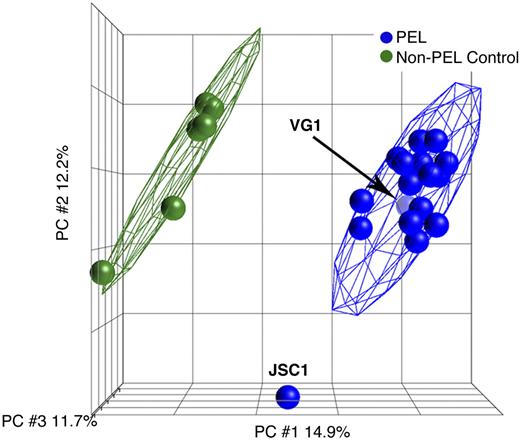
With the use of the high-resolution Affymetrix 6.0 and the 500K SNP array we assessed the genomic signatures of virtually all available PEL cell lines. We used cell lines, because they form a renewable resource and have been characterized phenotypically for growth in culture and tumorigenicity in mice, in vitro and in vivo drug susceptibility, mRNA transcription, miRNA expression, p53 status, and other markers (Table 1 and references therein).3,4,12,13,15 Although the samples displayed a high degree of variability in their genomic signatures, with the use of principle component analysis (PCA) we found that most PEL cell lines (with the exception of JSC1) form a tight cluster. PCA reduced the variability of the data to 3 main and independent components. The first principle component (PC1) accounted for 14.9% of variability, PC2 and PC3 accounted for 12.2% and 11.7%, respectively. As expected PEL cells could easily be distinguished from non-PEL control cell lines (including KSHV-positive endothelial cells that also form xenograft tumors in mice) on the basis of genomic alterations (Figure 1).
PCA of PEL. PCA shows the presence of 2 distinct clusters formed by endothelial cells in green and all the 17 (excluding JSC1) PEL cells in blue, irrespective of whether they were from culture or xenograft. The x-, y-, and z-axes represent PC1, PC2, and PC3, respectively, accounting for 14.9%, 12.2%, and 11.7% of variability in the data.
PCA of PEL. PCA shows the presence of 2 distinct clusters formed by endothelial cells in green and all the 17 (excluding JSC1) PEL cells in blue, irrespective of whether they were from culture or xenograft. The x-, y-, and z-axes represent PC1, PC2, and PC3, respectively, accounting for 14.9%, 12.2%, and 11.7% of variability in the data.
With the use of PCA of only the PEL data we found that all PEL cell lines clustered together irrespective of whether they were grown in culture or as xenografts in mice (data not shown). This showed that PEL cell lines form tumors in mice without acquiring additional mutations in vivo. This phenotype would be expected for a monoclonal tumor, as PEL is believed to be. For our detailed analysis, we excluded the samples from xenografts, to avoid the bias of repeat sampling of some PEL lines, and exclusively focused on the samples from cells in culture, including the outliers. Using only PEL-derived data for PCA afforded us the resolution to identify differences among individual PEL isolates. Ten of 13 PEL cell lines clustered tightly together (data not shown), which reaffirmed the initial, phenotype-based classification of PEL. We also observed 3 unique samples: BCLM and VG1, in addition to the previously noted JSC1.
On analyzing the CNV markers in detail, we identified discrete regions of amplifications and deletions on each chromosome, many of which were shared in > 50% of the samples. Supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) shows the heat map representation of CNV for PEL cell lines. The top panel shows the EBV-positive and the bottom panel EBV-negative PEL cell lines. Chromosomes 7, 8, 12, and the q-arm of chromosome 1 harbored most of the amplifications.
We verified the CNV data with the use of G-band karyotyping of a subset of the samples: BC1, BC3, BCBL1, and BCP1. These samples were chosen to include PELs with or without EBV coinfection and mutation in the p53 gene (as listed in Table 1). On comparing our CNV heat map (supplemental Figure 1) with our karyotyping data (supplemental Figure 2), we see a tight correlation in detecting gross chromosomal aberrations between the 2 methods.
All of the cell lines showed trisomy of chromosome 7 with a subset showing trisomy of chromosome 12 (BC1 and BCP1) and amplification of 1q (BC1 and BCP1), in agreement with previous cytogenetic studies on these cell lines.6,33 In addition, we noticed amplifications and deletions throughout the genome (supplemental Figure 2) and varying among individual cell lines. When we compared this data and previous reports,5,6,33-35 we note the variability of karyotype aberrations. We further note the power of array-based CGH over traditional karyotyping to detect smaller changes that might be more tightly associated with the PEL phenotype than gross variation in karyotype.
The 946 000 CNV-specific probes that are contained in the Affymetrix 6.0 array allowed us, for the first time, to define the genomic signature exhibited by PEL cells at a single gene resolution. Table 2 shows the individual genes with CNV in 12 of 13 PEL cell lines. This represents our most stringent cutoff. Two of these, WWOX and FHIT, represent common fragile site (CFS) genes. The glutamate receptor ionotrophic, delta 2 (GRID2) gene that was deleted exclusively from the EBV-negative PEL cell lines (listed in Table 3) is also classified as a CFS gene. CFSs are regions of the genome particularly susceptible to breaks in metaphase chromosomes. These regions are evolutionarily conserved and tend to encode for genes involved in tumor suppression, replicative stress, and DNA damage repair.36 FHIT, WWOX, and GRID2 map to the CFS FRA3D, FRA16D, and FRA4G on chromosomes 3 (p-arm), 16 (q-arm), and 4 (q-arm), respectively.37-39 Figure 2 shows the detailed distribution of markers around these 3 sites in our PEL samples as well as non-PEL controls. Figure 2A and C represent the PEL samples that were hybridized to the Affymetrix 6.0 and the second, independent, Affymetrix 500K array, respectively. We note that in both cases, irrespective of the density of the markers used, we observe a localized loss of CFS genes in an otherwise normal region of the chromosome. For comparison, the q-arm of chromosome 4 in the same panels (Figure 2A,C) is an example of large-scale chromosomal amplification. These large-scale rearrangements were present in > 1 PEL cell line, but by no means in all cell lines. The control samples in which these markers were assessed in non-PEL tumor samples are shown in Figure 2B and D. Figure 2B contains 6 tumor samples of non–B-cell origin (both KSHV positive and negative) hybridized to the 6.0 SNP array, and Figure 2D shows 5 non-PEL lymphoma samples hybridized to the 500K SNP array. Neither sets of controls show loss of FHIT, WWOX, or GRID2 genes. This shows that the partial or complete loss of FHIT (11 of 13, 85% of PEL cell lines), WWOX (12 of 13, 92% of PEL cell lines), and GRID2 (5 of 7, 71% of EBV-negative PEL cell lines) in an otherwise normal chromosomal setting is particular to PEL. We also investigated other CFS regions such as retinoid-related orphan receptor α (RORA) on chromosome 15 and Parkinson disease 2 (PARK2) on chromosome 6. There was no amplification or deletion detected in these regions (data not shown). This further supported our conclusion that deletion of FHIT, WWOX, and GRID2 cannot be attributed to the fragility of these regions but represent selected mutations characteristic of PEL cell lines.
CFS tumor suppressor genes FHIT, WWOX, and GRID dot plot in PEL. Dot plot representation of markers distributed along the chromosome. (A,C) The loss of FHIT, WWOX, and GRID2 from chromosomes 3, 16, and 4, both in the 6.0 and 500K SNP array are shown. The markers (from each of the 13 PELs) are represented by black dots on a log2 scale (amplification denoted by dots above and deletion by dots below the normal 0 line) with the cytoband at the base of plot. The dots identified by ∧ represent alterations that only occurred in 1 of 13 PELs and were thus considered exceptions. (B,D) Data represent 6 nonlymphoma tumor samples and 5 non-PEL lymphoma controls, respectively.
CFS tumor suppressor genes FHIT, WWOX, and GRID dot plot in PEL. Dot plot representation of markers distributed along the chromosome. (A,C) The loss of FHIT, WWOX, and GRID2 from chromosomes 3, 16, and 4, both in the 6.0 and 500K SNP array are shown. The markers (from each of the 13 PELs) are represented by black dots on a log2 scale (amplification denoted by dots above and deletion by dots below the normal 0 line) with the cytoband at the base of plot. The dots identified by ∧ represent alterations that only occurred in 1 of 13 PELs and were thus considered exceptions. (B,D) Data represent 6 nonlymphoma tumor samples and 5 non-PEL lymphoma controls, respectively.
Table 2 also contains genes that were significantly amplified in 12 of 13 (92%) PEL cell lines based on the Affymetrix arrays. These are DER-like family member 1 (DERL1), ETS translocation variant 1 (ETV1), RAS p21 activator protein 4 (RASA4), thiamin pyrophospho-kinase 1 (TPK1), tripartite motif containing 56 (TRIM56), and vacuolar protein sorting-41, homolog (VPS41). Other genes were deleted or amplified in a smaller fraction of samples. Although these alterations may have biological relevance alone or in combination, the significance of these candidate alterations needs to be established in a larger number of cases.
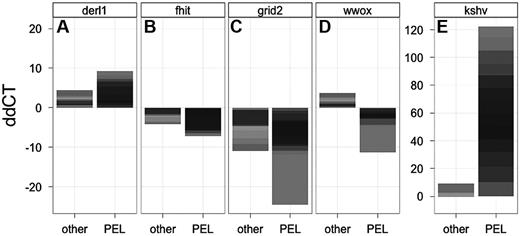
With the use of qPCR we verified a subset of our CNV data: DERL1, as an example of gene amplification, and the 3 fragile site tumor suppressor genes. Figure 3 shows a histogram of the copy number of samples relative to the normal diploid copy number per genome (ddCT) for PEL (n = 13) and non-PEL (n = 11) tumor cell lines. Where normal cells have a ddCT of 0, amplifications and deletions are represented by positive and negative ddCTs, respectively. Note that in this representation the results of individual cell lines are stacked to give a measure of the degree as well as frequency of the genomic aberration.40 Figure 3A represents the DERL1 locus, which was shown to be amplified in PEL by CGH, and we subsequently confirmed this by PCR, as evidenced by a positive ddCT. The same gene was also amplified in a subset of non-PEL tumor cell lines (those with no amplification have a ddCT of 0 and thus do not contribute to the signal). Figure 3B-D shows the results for FHIT, GRID2, and WWOX, respectively. Compared with normal human genomic DNA, PEL cell lines show losses of WWOX, FHIT, and GRID2. These qPCR data confirmed independently our array analysis. FHIT and GRID2 were also deleted in some non-PEL tumor cell lines, as expected for a fragile site. WWOX, however, was exclusively deleted in PEL and not in the other tumor samples (P ≤ .005 by Wilcoxon nonparametric test). If anything, WWOX seemed increased in some non-PEL tumor cells. Again, this would be expected for a random set of tumors, all of which do show some degree of gross genome alterations. It is important to note the scale of the genomic changes, which does not exceed 2-fold for individual amplifications, consistent with large-scale CNV. By contrast, the KSHV probe (Figure 3E) exemplifies the signal differences because of large copy number increases (as denoted by the difference in scale), because KSHV is present in ∼ 50 copies in PEL, but was present in only 5 of our 11 non PEL-tumor lines and here at 1 or < 1 copy per cell.
qPCR verification of CFS tumor suppressor gene loss in PEL. (A-E) The qPCR results for DERL1, FHIT, GRID2, WWOX, and KSHV, respectively, are shown. Shown is the stacked relative level (ddCT) for each gene on the vertical axis and the 2 classes of 13 non-PEL (other) and 13 PEL cell lines on the horizontal axis. The contribution of individual cell lines is indicated by the gray level. Because amplifications and deletions result in only a 2-fold change in signal in the case of cellular genes (and ∼ 50-fold for KSHV because there are ∼ 50 copies of the KSHV genome in each PEL cell) the stacked representation integrates both the degree of change as well as the number of cell lines that contribute to the signal in each group (a similar metric was previously validated).39
qPCR verification of CFS tumor suppressor gene loss in PEL. (A-E) The qPCR results for DERL1, FHIT, GRID2, WWOX, and KSHV, respectively, are shown. Shown is the stacked relative level (ddCT) for each gene on the vertical axis and the 2 classes of 13 non-PEL (other) and 13 PEL cell lines on the horizontal axis. The contribution of individual cell lines is indicated by the gray level. Because amplifications and deletions result in only a 2-fold change in signal in the case of cellular genes (and ∼ 50-fold for KSHV because there are ∼ 50 copies of the KSHV genome in each PEL cell) the stacked representation integrates both the degree of change as well as the number of cell lines that contribute to the signal in each group (a similar metric was previously validated).39
CNV, not LOH, distinguishes EBV-positive from EBV-negative PEL
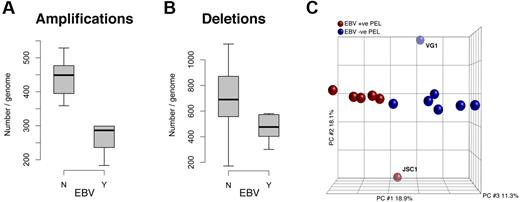
A tally of the CNV markers showed a significantly (P ≤ .05 by t test) greater number of amplifications in the EBV-negative group than in the EBV-positive cell lines (Figure 4A). The data for deletions mirror the trend; EBV-negative cells harbor more deletions than the EBV-positive group (Figure 4B), although the level of significance was lower (P ≤ .1 by t test). Using PCA, we demonstrated the presence of 2 distinct subclusters within PEL cell lines (Figure 4C). These 2 distinct clusters correlate with EBV infection status, JSC1 and VG1 being the outliers (analogous to our initial PCA).
Quantification of CNV in PEL, separated by EBV status. (A) There are significantly more amplifications in EBV-negative PELs than in EBV-positive PELs. (B) Although there is increased deletion of markers in the EBV-negative population, there difference is less conclusive. (C) PCA shows that PEL cells in culture form 2 distinct groups correlative with their EBV infection status: blue indicates EBV(−) and brown EBV(+) samples. The x-, y-, and z-axes represent PC1, PC2, and PC3, respectively, accounting for 18.9%, 18.1%, and 11.3% of variability in the data.
Quantification of CNV in PEL, separated by EBV status. (A) There are significantly more amplifications in EBV-negative PELs than in EBV-positive PELs. (B) Although there is increased deletion of markers in the EBV-negative population, there difference is less conclusive. (C) PCA shows that PEL cells in culture form 2 distinct groups correlative with their EBV infection status: blue indicates EBV(−) and brown EBV(+) samples. The x-, y-, and z-axes represent PC1, PC2, and PC3, respectively, accounting for 18.9%, 18.1%, and 11.3% of variability in the data.
We further mapped the CNV markers to the RefSeq database19 to identify specific genes. Table 3 shows 23 genes that were most significantly altered in EBV-negative PEL cell lines. We used false-discovery rate that was based on adjustment for multiple comparisons and defined q-values ≤ 0.01 as cutoff. We annotated this list with the use of GeneCards database v3.0.41 On the basis of their known functions, the genes were classified into (1) transcriptional (CDYL, CHD1, MEF2C, RFX2, RREB1, ZEB1), (2) developmental (EPHA-3, -5, GRID2, HBS1L), (3) metabolic (ACSBG2, FUT3), (4) cell signaling (IKBKB, ITPR1), and (5) cell adhesion (CADM2, CDH9, EDIL3, LRFN5, PCDH9) molecules. The oncogene RAF1, tumor suppressor BRCA2, cytoskeletal regulator ARAP2, and MHC receptor HLA-DRB5 were assigned their own categories.
In addition to CNV, the Affymetrix 6.0 SNP markers allowed us to interrogate LOH in PEL for the first time. LOH is indicative of allelic imbalance and can be used to assess overall genomic integrity.42 Unstable genomes harbor a greater extent of LOH compared with those that are genetically stable. The presence of allelic imbalance (LOH) did not correlate with EBV infection status in the PEL cell lines. Both EBV-positive and EBV-negative groups harbor the same degree of copy neutral LOH (indicative of uniparental disomy) and heterozygous gains or losses (data not shown). In summary, CNV (amplifications/deletions) but not LOH separate EBV-positive from EBV-negative PEL cell lines.
Discussion
In this study we defined the genomic signature of PEL cell lines at the individual gene level with the use of Affymetrix SNP6.0 array–based CGH. Most of the genetic changes corresponded to broad amplifications of regions of chromosome 7, 8, 12 and q-arm of 1. This first high-resolution data extend earlier studies that have reported similar observations in a single PEL or in a smaller collection of PEL cell lines with the use of first-generation BAC-mid–based CGH arrays.5,6,33-35 This study for the first time associates 3 fragile site tumor suppressor genes, FHIT, WWOX, and GRID2, with PEL cell lines. PEL cell lines maintained the same genomic signatures whether grown in culture or as xenograft in a SCID mouse, which is consistent with the monoclonal origin of PEL. The presence of EBV, in addition to KSHV, was associated with decreased gross genomic rearrangements in PEL cell lines.
This first high-resolution CGH analysis of PEL cell lines allowed us to identify individual genes that were deleted or amplified. The 2 most prominent deletions were the fragile site genes WWOX and FHIT. Similar SNP-based array studies have been performed for Burkitt lymphoma (BL) with the use of lower resolution arrays.43,44 The 2 studies reported distinct sets of aberrations. Where Scholtysik et al44 commented on the imbalances affecting the MYC locus, Toujani et al43 found loss of FHIT to be one of the most common losses in BL primary tumors as well as cell lines. WWOX and FHIT have been classified as tumor suppressor genes in multiple cancers, including DLBCL.37,38,45,46 Transgenic mice that lack FHIT or WWOX are more prone to developing tumors, especially those of the lymphoid origin.47,48 Reintroduction of the respective tumor suppressor genes, ectopically or by gene therapy, restores nontumor properties in the mice.49,50 It can be speculated that in PEL, as in other DLBCLs, a loss of function of these genes promotes tumorigenesis. One might even hypothesize that the reintroduction of FHIT or WWOX may be developed into a gene transfer–based therapy for PEL.
We detected 6 nonfragile site genes that were amplified in most of the PEL cell lines. DERL1 and ETV1 have previously been associated with viral and nonviral cancers.51,52 DERL1 protects cells from endoplasmic reticulum (ER) stress–induced apoptosis and is overexpressed in breast cancer.52 Multiple reports have shown associations between solid tumors and ETV1, and alteration in ETV1 is a prognostic marker in tumor progression.51 Yet the specific biochemical function of ETV1 remains to be elucidated. The remaining high-significance amplifications include Ras-p21 activator protein 4 (RASA4), tyrosine protein kinase 1 (TPK1), tripartite-motif containing 56 (TRIM56), and vacuolar protein sorting 41 homolog (VSP41). These are involved in cell signaling, metabolism, and protein maturation. This study, for the first time, suggests that these genes may be involved in cancer.
PEL is believed to be a monoclonal expansion of a post-GC B cell.53,54 Immunophenotypic analysis suggests that PELs comprise a subset of plasma cells. Normally, naive B cells entering the GC undergo B-cell receptor (BCR)–mediated differentiation/activation to form memory and plasma cells. Those that cannot be stimulated because of lack of BCR or crippling mutations are eliminated via Fas-mediated apoptosis. However, in PEL as well as EBV-positive posttransplant-associated lymphomas, a subset of these cells escapes apoptosis and ultimately develops into lymphoma. By some account this escaping population of cells lacking functional BCR is particularly susceptible to EBV infection; EBV infection not only protects them from GC-mediated apoptosis but also contributes to proliferation.55
Mack and Sugden11 reported that EBV is essential for sustained proliferation of some EBV-positive PELs in culture, but EBV-negative PEL cell lines exhibit similar sustained proliferation in culture and tumor-forming potential in mice.13 This study for the first time found a PEL genotype that is associated with EBV infection: EBV-negative PEL cell lines harbor significantly more gross genomic alterations than EBV-positive PEL cell lines. EBV appears to facilitate host chromosomal genomic stability in PEL cells. This phenotype is consistent with EBV-transformed lymphoblastoid cell lines that retain a normal karyotype indefinitely.56 Vaghefi et al57 suggested that for KSHV-negative AIDS lymphomas an inverse correlation exists between EBV infection and the number of chromosomal aberrations. At this point it is unclear whether this is because of a direct stabilizing effect of EBV latent genes or whether the expression of EBV latent genes relieves selective pressure for additional genomic alterations in host oncogene or tumor suppressor gene loci by modulating growth-promoting pathways posttranslationally (which EBV-negative PEL would need to be selected by genomic alterations). Alternatively, the possibility exists that the genomic aberration is a result of EBV-negative PEL cells having traversed the GC51 and EBV-positive PEL not.
The contribution of EBV to PEL development has remained a matter of debate; EBV-positive and EBV-negative PEL cell lines exhibit the same tumor characteristics in mouse models and in the clinic. Here, we can speculate on a couple different scenarios of PEL tumor progression. One being, a “sequential” scenario, whereby KSHV-infected naive B cells entering the GC are infected with EBV that drives them through the maturation/differentiation process and contributes to their proliferative advantage. Some of the EBV-infected B cells subsequently acquire the PEL-defining FHIT and WWOX gene mutations. As these tumor cells continue to evolve, some of them may lose the EBV episome but acquire additional compensatory genetic mutations, by a yet undefined mechanism. This would give rise to EBV-negative PEL, in other cases EBV is never lost. Alternatively, KSHV-infected naive B cells traversing through the GC remain EBV negative but acquire FHIT and WWOX mutations. Subsequently, these mutant cells are either infected with EBV or acquire additional genetic mutations to form fulminant PEL. This leads to 2 alternative, or “parallel,” tumor development pathways. Such a scenario mirrors the situation in EBV-positive versus EBV-negative BL. Unfortunately, because PEL causes such rapid mortality, no longitudinal samples exist to clinically verify this model.
In Table 3, we also noted individual genes that were correlated with EBV status. A significant number of those were involved with cell adhesion. For known tumor genes, we note amplification of RAF1 or c-RAF and deletion of the tumor suppressor gene breast cancer gene 2, BRAC2, in EBV-negative PEL. c-RAF is the cellular homolog of the murine leukemia viral oncogene, v-RAF-1, a serine/threonine-specific protein kinase involved in the MAPK-ERK pathway, and BRCA2 is associated with DNA damage repair, specifically, double strand breaks and homologous recombination.
Other interesting genes that were significantly amplified in EBV-negative PEL include IKBKB, IKB kinase-β, a key activator of NFκB, and ITPR1, inositol-1,4,5-triphosphosphate receptor 1. NFκB has a known role in PEL tumorigenesis. It is modulated by the KSHV homolog of the cellular FLICE inhibitory protein (vFLIP)52,58 as well as KSHV K15.59 Of note, the EBV latency membrane protein 1 (LMP-1) can also activate NFκB.60 Thus, it seems consistent with the biology of EBV that IKBKB, a cellular activator of NFκB, was amplified in 5 of 7 EBV-negative cells. ITPR1 is involved in regulating calcium homeostasis in the ER, inducing Ca2+ release into the cytosol. Normal B cells undergoing activation and blastic transformation show elevated levels of cytosolic Ca2+.61 It has been shown by Dellis et al62 that EBV LMP-1 can down-regulate ER-associated enzymes to increase levels of cytosolic Ca2+ and promote cellular transformation. Therefore, it can be hypothesized that amplification of ITPR1 in EBV-negative PEL compensates for the absence of EBV-LMP1 to drive B-cell transformation.
Finally, one of the genes deleted exclusively in EBV-negative PEL cell lines, GRID2, is also a fragile site gene. Deletion of GRID2 was observed in 5 of 7 EBV-negative PEL cell lines, whereas it remained unaltered in EBV-positive PEL cell lines. This further speaks to the instability of the host genome in the absence of EBV in PEL cell lines. GRID2 is noted for its role in neurologic development.36 Ours is the second report suggesting a role for GRID2 in tumorigenesis.63
In summary, this represents the first high-resolution CGH analysis of PEL cell lines. We find evidence of genomic instability, which was greater in EBV-negative PEL cell lines. We verify that the PEL-associated genomic signature maps mainly to amplifications in chromosome 7, 8, and 12 and the q-arm of 1. We report the first association of deletion of the fragile site tumor suppressor genes FHIT and WWOX with PEL cell lines and GRID2 with EBV-negative PEL cell lines.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank K. L. Richards for critical reading of the manuscript.
This work was supported in part by the National Institutes of Health grants DE018304, CA019014, CA163217, CA096500, and DE018281; the Centers for AIDS research (CfAR); the AIDS malignancy consortium (grant CA121947); the University Cancer Research Fund (UCRF); and the Leukemia & Lymphoma Society of America (grant R6169-10). B.D. is a scholar of the Leukemia & Lymphoma Society and a Burrows Welcome trust investigator.
National Institutes of Health
Authorship
Contribution: D.R. designed and conducted experiments, analyzed data, and wrote manuscript; S.-H.S. conducted experiments; B.D. designed experiments and analyzed data; and D.P.D. designed experiments, analyzed data, and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dirk P. Dittmer, Curriculum in Genetics and Molecular Biology and Department of Microbiology and Immunology, UNC Lineberger Comprehensive Cancer Center, UNC Center for AIDS Research, University of North Carolina at Chapel Hill, CB# 7290, Mary Ellen Jones Bldg, Chapel Hill, NC 27599-7290; e-mail: ddittmer@med.unc.edu.