Abstract
As peripheral blood has surpassed bone marrow as a predominant source of stem cells for transplantation, use of the cytokine granulocyte colony-stimulating factor (G-CSF) to mobilize peripheral blood stem cells (PBSCs) is increasing. Issues regarding potential genotoxic effects of even short-term, low-dose G-CSF treatment for the healthy donors have been raised. To address the question of chromosomal instability, we used FISH to evaluate the peripheral blood lymphocytes of 22 PBSC donors and 22 matched controls at 5 time points over a 12-month period. The specimens obtained were a pre-G-CSF, followed by collections at the time of PBSC harvest (days 5-7) and at 2, 6, and 12 months after donation. Eight additional PBSC donors provided a single sample at 12 months. Nine loci (mapped to chromosomes 7, 8, 9, 17, 21, and 22) were evaluated for aneuploidy, including 3 mapped to chromosome 7 because of the specific relevance of monosomy 7. Replication timing was evaluated for chromosome 15 and 17 loci. No evidence was found of G-CSF–induced chromosomal instability. This work supports the epidemiologic data that have demonstrated no increased risk for hematologic malignancies in G-CSF–primed PBSC donors.
Introduction
Recombinant human granulocyte colony-stimulating factor (G-CSF) is now widely used for autologous and allogeneic hematopoietic stem cell transplantations. Recent data from the National Marrow Donor Program show that, since 1999, the number of transplantations performed with G-CSF–mobilized peripheral blood stem cells (PBSCs) has increased substantially.1 G-CSF priming affords increased numbers of CD34+ cells with PBSC transplantations associated with more rapid engraftment and reduced infectious complications,2 although no long-term survival advantage for recipients of unrelated donor-mobilized PBSCs has been demonstrated.3
Recently, the issue of potential deleterious long-term effects to healthy donors has become a major focus of discussion and research,4-8 with particular concern for the potential long-term genotoxic effects of short-term G-CSF priming.
In healthy donors who have received low-dose G-CSF short-term, there has been an occasional case report of the development of acute myeloid leukemia (AML) 1-5 years later.9-11 However, providing a more objective view of risk for malignancy among healthy donors have been the studies using patient registries.12-17 In the largest series to date, Pulsipher et al17 found no cases among 2408 donors registered in the National Marrow Donor Program between 1999 and 2004 with a median follow-up after G-CSF administration of 49 months.
Several studies have addressed the effects of G-CSF on DNA, chromosomes, and/or related cellular processes. There are reports of transient increases in tetraploidy,18 in de novo-DNA synthesis, and in relaxation of double-stranded DNA19 after G-CSF treatment. Similarly, transient differences in gene expression profiles have been noted in cells shortly after G-CSF administration, which resolved by 2-6 months after treatment.20,21 Both a transient increase in dysregulation of DNA synthesis (replication asynchrony) in addition to a concerning persistent effect of increased chromosomal loss or gain (aneuploidy) in lymphocytes of healthy donors who received G-CSF were reported by Nagler et al.22
The most frequent clonal chromosome abnormality identified in the abnormal clones of patients with myelodysplastic syndrome (MDS) or AML who have been treated with G-CSF is monosomy 7.10,11 Loss of 7 is also a well-documented recurring abnormality in de novo and therapy-associated MDS and AML, without prior G-CSF therapy.23 However, its relationship with G-CSF has been demonstrated by Sloand et al,24 who showed that G-CSF treatment, in vitro, of cells from patients with aplastic anemia, caused selective expansion of preexisting monosomy 7 clones. (No monosomy 7 cells were detected from aplastic anemia patients without a preexisting clone.) Case reports further describe persons with aplastic anemia who developed transient monosomy 7 in response to G-CSF, which remitted when the G-CSF was withdrawn.25 The present study was designed to prospectively investigate the effects of G-CSF on chromosomal aneuploidy and replication kinetics, focusing on chromosomal regions associated with G-CSF, MDS, or AML.
Methods
This study involved persons who received G-CSF because they were donating PBSCs for a related family member and healthy controls who did not receive G-CSF or donate PBSCs. The study was approved by the Institutional Review Board of the University of Minnesota's Human Research Protection Program (Human Subjects Code #0606M88190).
PBSC donors who received G-CSF
Bone marrow transplantation physicians at the University of Minnesota Medical Center recruited and consented 20 allogeneic-related PBSC donors for the study; 1 potential subject opted to undergo bone marrow harvest instead of PBSC collection. Two research donors of mobilized PBSCs were also recruited, yielding a total of 22 PBSC subjects.
Three subjects (donors) withdrew from the study before completion: 1 before month 2 for unknown reasons, 1 before month 6 because of pregnancy, and 1 before month 12 because of a recent diagnosis of liver cancer. Nineteen subjects completed the entire 1-year study.
One-year anniversary: PBSC donors.
To supplement the subject pool and address the long-term effects, 8 additional recent (within 1 year) G-CSF–treated donors were identified and consented to provide a single blood sample for aneuploidy analysis at or close to the date of their 1-year anniversary of G-CSF treatment.
Healthy control subjects
Twenty-two control subjects who were not pregnant and with no history of cancer or prior exposure to G-CSF were recruited and demographically matched to donors by sex and age (within 5 years). Blood collections from the donor and his/her matched control were scheduled closely in time. One healthy PBSC donor, whose remote history of prostate cancer was not recognized until after the second sample collection, was permitted to continue on study.
G-CSF administration
Subjects other than controls received 10 μg/kg body weight of G-CSF subcutaneously on each of 5 consecutive days, and PBSCs were collected using the Fenwal CS-3000 Plus Blood Cell Separator by processing approximately 15 000 mL of blood according to institutional standard protocols for PBSC donation and cell collection.26-28 The resulting cells are tested for CD34, with a target yield of 5 million CD34+ cells/kg.
Specimen collection
The baseline (day 0) sample was collected before the first G-CSF injection. The second sample was obtained on G-CSF day 5 to 7 (day of first PBSC collection) and subsequent samples at months 2, 6, and 12.
Regarding compensation, PBSC donors and controls were given a stipend at each collection time point to offset the time and effort required for blood draws and brief health interviews.
Blood samples and testing
Peripheral blood specimens (collected in sodium heparin) for G-CSF donors were drawn on site or at the subject's local clinic and shipped overnight to the research laboratory for the month 2, 6, and 12 samples. Control specimens were all drawn on site. Blood samples were also collected at each time point for complete blood counts and differential. Basic demographic data were collected, and each subject completed a brief health and medication questionnaire at each visit.
Cytogenetic methods
All samples were coded on collection, and the cytogenetics laboratory was not informed of control versus G-CSF donor status or of the sampling time point. The data were decoded and statistically analyzed only after all cytogenetics scoring was completed.
Two methods of processing were performed: peripheral blood lymphocytes were either stimulated with phytohemagglutinin (PHA) and cultured for 72 hours at 37°C before harvesting or were processed directly without prior PHA stimulation or culture. The cultured method was used to be able to more closely replicate the methods used by Nagler et al.22 The direct method was used to study the contribution, if any, of other cell types present with a half-life of < 72 hours that would not be expected to respond to PHA. The direct method also controlled for aneuploidy because of in vitro culture artifact. Cells from the 72-hour cultures were harvested with colcemid arrest, treatment with 0.75M KCL hypotonic solution, and fixation in 3:1 methanol:glacial acetic acid. These harvested cells (which included both metaphase and interphase cells) were spread onto glass slides according to standard cytogenetic protocols. Cells from the direct method were harvested similarly; but as there was no stimulation with PHA or culture, only interphase cells were present.
FISH was used to evaluate 2 cytogenetic variables: aneuploidy and replication timing.
With aneuploidy, the DNA probes (Abbott Molecular) and buffer were mixed according to the manufacturer's instructions and applied to the slides onto which the target cells had been prepared. The slides were incubated at 37°C overnight in a Thermobrite-heated chamber and subsequently washed in 0.4 or 2 × saline sodium citrate, rinsed in sodium phosphate buffer, and counterstained with 4,6-diamidino-2-phenylindole. Slides were evaluated using an Olympus BX61 microscope outfitted with fluorescence filter sets for 4,6-diamidino-2-phenylindole, fluorescein isothiocyanate, and Texas Red, a 175W Ozone-free Zenon light guide illuminator, an interferometer-based CCD cooled camera, and Applied Spectral Imaging FISHview Version 5.5 software.
Nine chromosomal loci (from 6 different chromosomes) were selected for evaluation of aneuploidy (Table 1). Because of the association between in vitro G-CSF treatment and expansion of monosomy 7 clones,24 3 chromosome 7 loci, CEN7 (p10q10), elastin (ELN; 7q11.23), and D7S486 (7q32), were included. Loci for RUNX1T1 (8q22), TP53(17p13), and RUNX1(21q22) were included based on their frequent involvement in de novo and therapy-associated MDS and AML; CEN 17 (17p10q10) because of its use in previous related published studies22 ; and 2 loci, ABL1 (9q34) and BCR(22q11.2), were included to increase the number of different chromosomes evaluated and because of the extensive control and validation data available in our laboratory for these probes.
Scoring of aneuploidy
All subjects were evaluated for aneuploidy. For each probe for each subject, 400 cells were scored, with each of 2 technologists reading 100 cells each from the cultured and direct harvests. A cell was interpreted as aneuploid if the number of fluorescent signals observed deviated from 2. Because of the very low frequencies of cells with gains of signals, for statistical analyses, cells with 3 or 4 signals were grouped together.
With regard to replication timing, this variable was evaluated only for the 22 subjects in the longitudinal study along with 20 controls. Because replication timing can be assessed only in dividing cells, it was performed only on the cultured cells. Three loci were evaluated: SNRPN (15q11.2) a known imprinted locus, and a subtelomeric locus (15q26) and TP53 (17p13), neither of which is documented as imprinted.
Scoring of replication timing
Replication timing is aimed at determining whether or not 2 homologous alleles for a locus replicate synchronously or asynchronously. The number of fluorescent signals present is used to assess replication. A nonreplicated locus should show a single signal; a replicated locus should show 2 signals. If both homologs are in pre-S phase, the expected pattern is one signal for each allele versus 2 signals per allele if both are post-S phase. If the homologous alleles replicate asynchronously, the expected pattern is one signal for one allele and 2 signals for the other. Because replication timing is not used in the clinical laboratory and has not been clinically validated, control ranges and standardized laboratory data were not available. Thus, multiple quality assessment assays were performed on 5 laboratory control individuals. Control cell preparations were used to standardize the scoring criteria and ensure inter-rater reliability. In addition, scoring of replication timing was performed within a limited time frame, thus avoiding any drift of scoring criteria over time.
Statistical analyses
Summary statistics, including mean, SD, median, and range, were calculated for the aneuploidy and replication data. Wilcoxon rank-sum test or signed rank test was used for all 2-sample comparisons, including comparing the aneuploidy rate and asynchrony rate between the G-CSF and control subjects, comparing the direct versus the cultured methods for the aneuploidy data, and the pair-wise comparisons between probes for the replication data. Generalized estimating equations (GEE) method for count data, with or without the study group by time interactions, was used to investigate the difference between the 2 study groups over time. Statistical analyses were performed using SAS Version 9.2 (SAS Institute). P values < .05 were considered statistically significant. A post-hoc power analysis revealed that with power set at 0.8, type I error set at 0.05, and a sample size of 20 versus 20, we would be able to detect differences of 0.6% or greater in aneuploid cells and differences of 4.2% or greater in replication asynchrony between donors and controls. The SD of the 2 variables was set at 0.65% and 4.63%, respectively, based on the observed data.
Results
Hematology data
Baseline and later follow-up (months 2 and beyond) complete blood counts for all subjects were found to be within normal limits. No further comparisons were made between G-CSF donor and control specimens with respect to complete blood count data.
Aneuploidy results
All of the values collected for aneuploidy for all of the probes fell within the normal control range established in the clinical cytogenetics laboratory at the University of Minnesota in accord with the guidelines put forth by the American College of Medical Genetics (www.acmg.net). For all 9 probes, absolute aneuploidy (the number of cells of 200 that had loss or gain of signals) ranged from a mean frequency of 1.1 to 9.7 (representing a rate of 0.55% to 4.86% aneuploid cells) for different cell preparation methods and donor groups, at different evaluation visits. The maximum absolute aneuploidy frequency for these groups ranged from 3.0 to 15.0 (1.50%-7.50% aneuploid cells). The mean percentages and SD of aneuploid cells for the direct preparation method for G-CSF–treated and control subjects, across all time points and all probes, are presented in Table 2. With the direct cell preparation method, the lowest mean aneuploidy frequency of 1.00% was observed (combining both G-CSF and control subjects and all evaluation visits) for RUNX1T1 (8q22); the highest mean aneuploidy frequency, 4.00%, was observed for the centromere 17 probe. Similar results were found with the cultured cell preparation method, except that the probes of ELN and BCR were also among the lowest aneuploidy groups. Most of the variability in aneuploidy frequencies appeared to be attributable to the nature of the probe, with the higher rates of aneuploidy seen for the larger centromere probes (such as the cen17), and the lower rates observed for the smaller cosmid probes (such as RUNX1T1). This is probably because of separation of the large signals in interphase, thus appearing as a gain of signal. No value of monosomy or trisomy/tetrasomy was observed that would be interpreted in a clinical setting as suggestive of a pathologic condition.
We conclude that short-term G-CSF stimulation does not induce abnormal levels of chromosome aneuploidy for chromosomes 7, 8, 9, 17, 21, or 22. These results would be expected to generalize to all chromosomes.
Comparisons of G-CSF donors to controls
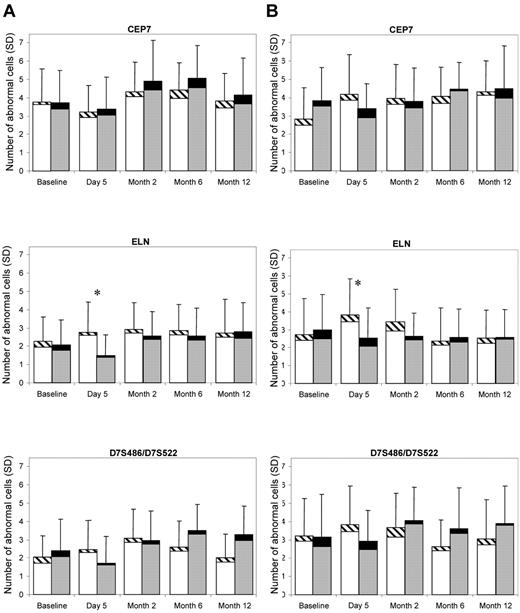
For each probe, for direct and cultured cells, and for each time point, aneuploidy frequencies were compared between G-CSF donors and controls. An example comparison for the 3 chromosome 7 probes from both the direct and the cultured method is illustrated in Figure 1 showing the absolute number of cells (of 200) that had a signal number deviating from the normal value of 2. Overall, there are very low levels of aneuploidy for both donors and controls and little difference between controls and donors for most time points. The larger difference between controls and donors at the day 5 time point is in the opposite direction of that expected for a biologically significant finding; the donors have a decreased rate of aneuploidy relative to controls. However, this could be partially influenced by the baseline aneuploidy levels for the donors and controls. Thus, in addition to the simple 2-sample comparisons performed at each time point, statistical analysis was also performed using a GEE model and testing the interaction of the group and the time variables.
Mean abnormal aneuploidy cell frequency (of a total 200 cells) and SD for the probes of CEP7, ELN, and D7S486/D7S522 in the control samples (left bar at each time point) and the G-CSF samples (right bar at each time point). Within each bar, the bottom part is for loss and the top part is for gain of signals; hence, the full length of the bar represents the frequency of any abnormal (gain/loss) cells. (A) Direct method. (B) Cultured method. *P < .05.
Mean abnormal aneuploidy cell frequency (of a total 200 cells) and SD for the probes of CEP7, ELN, and D7S486/D7S522 in the control samples (left bar at each time point) and the G-CSF samples (right bar at each time point). Within each bar, the bottom part is for loss and the top part is for gain of signals; hence, the full length of the bar represents the frequency of any abnormal (gain/loss) cells. (A) Direct method. (B) Cultured method. *P < .05.
The P values obtained for comparisons of the G-CSF and control subjects for direct cell preparation aneuploidy data are shown in Table 2. The comparisons of the G-CSF versus control subjects showed no significantly increased aneuploidy for any of the probes examined, with the exception of the ELN probe at day 5. The mean values for this data point (1.4% for controls and 0.8% of G-CSF subjects) were within normal limits for the established laboratory control ranges, and no other time point for this probe or the other chromosome 7 probes was found to be significant. The results for the comparison of G-CSF donors and controls were similarly negative for the cultured method (not shown), except that the G-CSF donors had a marginally significantly higher aneuploidy rate on TP53 than the controls at month 12 (P = .04). We found no consistent increase, over the 12-month time period, in aneuploidy rates among G-CSF donors compared with non–G-CSF–exposed controls. This was also confirmed by the GEE model, both with or without group by time interactions. Similar results were obtained examining monosomy alone or hyperdiploidy.
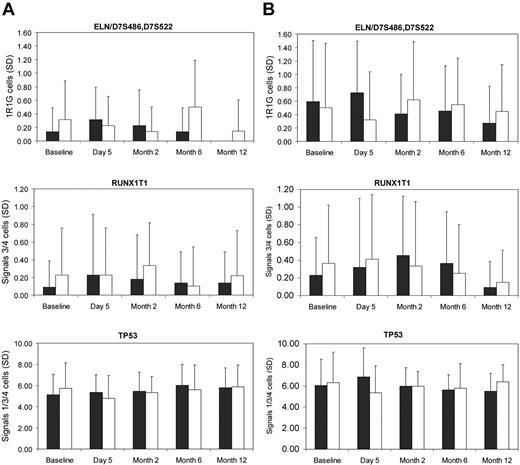
Of note, although no consistent time trend in differences in aneuploidy frequencies between donors and controls were found, data analysis did reveal significant differences between sampling time points for various probes for donors and/or controls (Figure 2). Thus, any differences between time points probably reflect either transient or in vitro technical artifact. All of the variation observed fell within the normal range.
Mean abnormal aneuploidy cell frequency (of a total 200 cells) and SD for the chromosome 7 probes ELN and D7S486/D7S522 scored together for loss to identify cells with monosomy 7), for chromosome 8q22 probe RUNX1T1 scored for gain to identify cells with trisomy 8 or tetrasomy 8, and for the chromosome 17p13 probe TP53 to identify cells with either loss (1 signal) or gain (3 or 4 signals) of TP53. The control samples are represented by the left bar at each time point and the G-CSF donor samples by the right bar at each time point. (A) Direct method. (B) Cultured method.
Mean abnormal aneuploidy cell frequency (of a total 200 cells) and SD for the chromosome 7 probes ELN and D7S486/D7S522 scored together for loss to identify cells with monosomy 7), for chromosome 8q22 probe RUNX1T1 scored for gain to identify cells with trisomy 8 or tetrasomy 8, and for the chromosome 17p13 probe TP53 to identify cells with either loss (1 signal) or gain (3 or 4 signals) of TP53. The control samples are represented by the left bar at each time point and the G-CSF donor samples by the right bar at each time point. (A) Direct method. (B) Cultured method.
Statistical analysis of aneuploidy data also revealed that there were significant differences between cultured and direct preparations for certain probes. Specifically, the direct method yielded higher aneuploidy rates for the centromere probes CEP7, CEP17, and for ABL1, and BCR, whereas the cultured method yielded higher aneuploidy rates for ELN, D7S486, RUNX1T1, and TP53. These differences were observed across subject groups and thus did not influence comparisons between subject groups.
Replication results
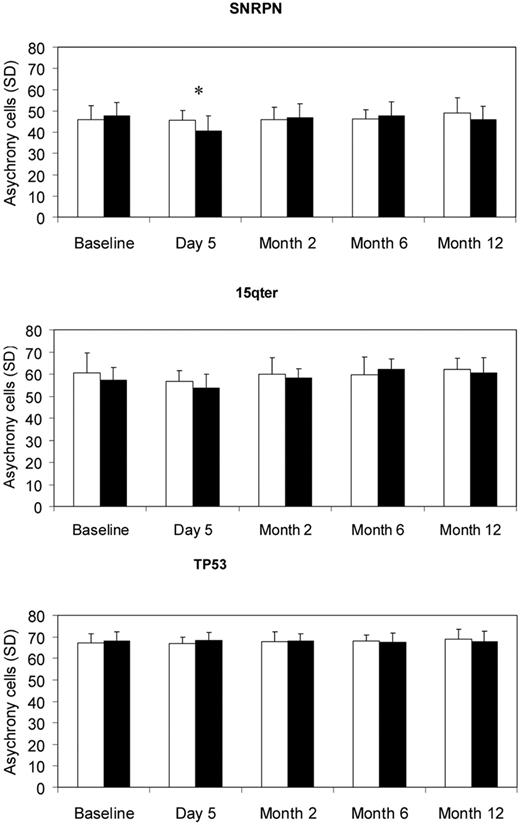
The replication timing results are illustrated in Figure 3. The comparison of G-CSF donor group to the matched controls for each of the 3 probes used to study replication timing showed no significant increase in asynchronous cells for the G-CSF group. However, a significant difference in a reverse direction was observed for SNRPN at day 5 (P = .01). No consistent trend in the difference between the 2 groups was observed over the 12-month time period by the GEE model. Thus, G-CSF treatment did not result in a significant change in replication timing for any of the 3 loci examined.
Mean asynchrony cell frequency (of a total 200 cells) and SD for the probes of SNRPN, 15qter, and TP53 in controls (white bars) and G-CSF subjects (black bars). *P < .05.
Mean asynchrony cell frequency (of a total 200 cells) and SD for the probes of SNRPN, 15qter, and TP53 in controls (white bars) and G-CSF subjects (black bars). *P < .05.
Although there were no differences between the G-CSF and control groups, the 3 probes did significantly differ from each other (P < .001). In contrast to expectation, SNRPN had significantly fewer asynchronous cells than both 15qter and TP53 (both P < .001). For SNRPN, the average frequency of asynchronous cells ranged from 40.6 to 49 (20.32%-24.5%) across all sample time points; for 15qter, the frequency of asynchronous cells ranged from 53.8 to 62.2 (26.9%-31.1%), and for TP53, frequency of asynchronous cells ranged from 66.8 to 69.0 (33.4%-34.5%).
Discussion
This study included a comprehensive, prospective, detailed investigation of genotoxic effects of short-term G-CSF treatment on PBSC donors. Twenty-two G-CSF donors were studied longitudinally over 1 year. The study revealed neither a significant increase in aneuploidy nor any significant alteration in replication kinetics after G-CSF treatment.
Of particular importance is the finding that low-dose, short-term G-CSF treatment did not induce any increased rates of monosomy 7, as assessed by 3 different probe sets in both uncultured and cultured cells. These data are in good agreement with those of Sloand et al,24 whose in vitro studies indicated that 15-day in vitro treatment with G-CSF at doses of 50 mg/mL did not induce monosomy 7 or preferentially expand a monosomy 7 clone. Similarly, no evidence of increased rates of trisomy 8 were found. Trisomy 8 is another well-documented recurring abnormality associated with MDS and was also reported in the studies29,30 of neutropenia and aplastic anemia patients who received G-CSF in the absence of MDS or AML. Importantly, the lack of a significant effect for chromosome 17 aneuploidy in the present study is in contrast to the findings of Nagler et al,22 who reported significant aneuploidy for chromosome 17 up to 200 to 268 days after G-CSF priming in healthy controls. The reason for this discrepancy is not known as the culture conditions, time of harvest, and probe sets to evaluate chromosome 17 were similar in the 2 studies. More recently, Marmier-Savet et al,31 reported significantly increased aneuploidy for chromosome 8 and 17 just after G-CSF administration, limited to CD34− peripheral blood cells, with lesser, but still elevated levels remaining at 6 months but returning to near baseline at 1 year. These increases were not seen in CD34+ cells or in metaphases from PHA-stimulated cultured lymphocytes. Our data in the present study indicate that different probes with different characteristics (eg, probes that include large amounts of repetitive sequences and that are known to be polymorphic vs probes that are smaller in size and are composed of euchromatin) can perform differently in a FISH assay.
With respect to replication asynchrony, our data showed no significant effect of G-CSF treatment. Because SNRPN is an imprinted locus whereas TP53 and 15qter are not, we would have expected SNRPN to have the highest rate of asynchrony, but SNRPN had the lowest observed rate of asynchrony. The percentages of asynchronous cells seen with SNRPN we observed are slightly below the lower end of the range of 28% to 56% reported by Nagler et al.22 For TP53, our average rate of asynchronous cells is slightly lower than the 37.2% reported by Nagler et al22 for their G-CSF subjects; but in contrast to that study, we did not observe a lower rate in our controls. In contrast to aneuploidy, a cytogenetic variable widely used within clinical practice with both diagnostic and prognostic importance, the measure of replication timing has not been clinically validated. Scoring for this measure can be affected by technical artifacts of hybridization and visualization. There are no published control ranges for replication, thus confounding comparison with those reported by Nagler et al.22 We carefully standardized scoring criteria for replication timing to ensure high inter-rater reliability and observed no significant differences between the controls and G-CSF–treated donors for replication timing of SNRPN, TP53, or 15qter and no trend in replication kinetics over time for either the controls or G-CSF subjects.
We could only examine peripheral blood to assess chromosome instability, rather than bone marrow, the tissue that is directly involved in MDS and AML. Therefore, we also studied direct, uncultured preparations as these should include the mobilized CD34+ cell population. In addition, given reports describing aneuploidy in peripheral blood lymphocytes, the use of peripheral blood would be justified.
Because we demonstrated no increased aneuploidy or replication asynchrony in any G-CSF donors either immediately or over time, our data support the conclusion that G-CSF induces no generalized chromosome instability through the PBSC mobilization procedure; and in this regard, the therapy remains safe for normal donors.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This project was supported by the National Marrow Donor Program and the Department of the Navy, Office of Naval Research (grants N00014-06-1-0704 and N00014-08-1-0058 to the National Marrow Donor Program).
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Office of Naval Research or the National Marrow Donor Program.
Authorship
Contribution: B.H. designed the research, provided oversight for the laboratory work, participated in statistical analysis, and drafted the manuscript; L.O., M. Cain, and E.T. performed the cytogenetics studies; S.P. assisted with study design and oversaw the identification and assessment of research subjects, collection of blood samples, and provision of samples to the laboratory; B.L. designed statistical methods, oversaw statistical analysis, and participated in writing the manuscript; X.L. designed statistical methods, carried out statistical analysis, provided data analysis, and drafted the manuscript; M. Clay participated in research design and oversaw the identification and selection of control subjects and acquisition and provision of blood samples; D.C. and J.M. participated in study design and review of study progress; D.W. participated in research design, oversaw the selection of blood stem cell donors, the care of those donors, administration of the G-CSF, and drafted the manuscript; and J.M. designed research, provided oversight for donor selection and data analysis, and drafted the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey McCullough, Department of Laboratory Medicine and Pathology, MMC 609, University of Minnesota, 420 Delaware St SE, Minneapolis, MN 55455; e-mail: mccul001@umn.edu.